Abstract
Recent deep-sequencing efforts have identified many novel non-conserved small RNAs that are expressed at low levels in certain mammalian cells. Whether these small RNAs are important for mammalian physiology is debatable, therefore we explored the function of one such RNA, human miR-1271. This small RNA is similar in sequence to miR-96, a highly conserved microRNA that when mutated causes hearing loss in humans and mice. Although the miR-1271 and miR-96 sequences differ slightly, our in vitro assays indicate that they have an identical regulatory activity. We have identified brain-expressed mRNAs from genes including, GPHN, RGS2, HOMER1 and KCC2, which share the same miR-96 and miR-1271 regulatory elements. Interestingly, human miR-1271 is expressed abundantly in brain tissue, where miR-96 is not highly expressed. The rodent miR-1271 precursor contains several sequence differences in the precursor stem, which appear to reduce the efficiency of microRNA processing. Our data indicate that although miR-1271 and miR-96 function identically in vitro, they function to some extent uniquely in vivo. Given the expression patterns and nature of the target genes, miR-1271 may have a significant, although non-conserved, role in regulating aspects of neural development or function in humans.
INTRODUCTION
The majority of mammalian protein-coding genes contain microRNA-target motifs in their mature mRNA, which function to regulate gene expression post-transcriptionally (1). There is emerging evidence that specific microRNAs control functionally integrated cellular pathways (1–3). Given their potentially broad regulatory roles, it is not surprising that individual microRNA genes have emerged as important factors in the development of several diseases, including psychiatric and neurological disorders, as well as contributing to the molecular basis of pharmacologic treatments (4–7). Dissecting the gene networks impacted by these small regulators remains difficult, especially in the central nervous system where there is a complex cellular and structural environment and where estimates suggest that 80% of all mRNAs encoded by the mammalian genome are expressed (8). A potential way to gain insight into the role of microRNA regulation in human neurobiology is by characterizing functional variants in microRNA genes or binding sites and the factors that regulate their impact on associated human behavioral phenotypes. This characterization might provide insight into the epistasis of genes involved in neural function and potentially elucidate novel neural regulatory pathways in humans.
We recently described a common human polymorphism in a microRNA target motif in the serotonin receptor 1B (HTR1B) mRNA that attenuated miR-96 microRNA-directed regulation of gene expression in laboratory constructs (9). The microRNA-repressed form of the variant was associated with an increased history of self-reported externalized aggressive behaviors by college-aged students (9) and higher average self reported measures of hostile emotional states in the male portion of the sample (10). These findings suggest that regulation of gene expression by this miR-96 target motif may have potential clinical relevance; therefore it is important to better understand the factors that may regulate gene expression via the HTR1B microRNA-binding element as these may be involved in regulating aspects of neural function relevant to human behavior.
Given that a functional mutation in the miR-96 gene causes progressive hearing loss in adults without other reported behavioral changes (11,12), and that miR-96 neural expression is prominent only in mammalian sensory structures, we used in silico databases to identify a second-potential microRNA, miR-1271, a non-conserved small RNA identified by deep sequencing techniques, that might regulate gene expression via the HTR1B miR-96 motif as it shares a common ‘seed’ sequence with miR-96. As with many small human RNAs identified by deep sequencing, the expression of miR-1271 in the tested tissues was extremely low, and whether it had a significant function in humans was in question. Therefore in this study we compare and contrast the regulatory activities and expression patterns of miR-96 and miR-1271. We have identified mRNAs targeted by the common seed and note that they are enriched in genes involved in neural signaling including, gephyrin (GPHN), regulator of G protein signaling 2 (RGS2), homer protein homolog 1 (HOMER1) and solute carrier family 1 member 1 (SLC1A1). To test for potential in vivo regulatory differences between these apparent homologs, we examine the previously uncharacterized expression of miR-1271 in neuronal and non-neuronal human and rodent tissues.
MATERIALS AND METHODS
Plasmid constructs and RNA reagents
The luciferase test plasmids are a modified version of the psiCHECK-2 (Promega, Madison, WI, USA) plasmid described by Felice et al. (13). Synthetic sense and anti-sense oligonucleotides representing the test element (shown in Supplementary File 1 without restriction sites) were designed to contain SacI and BstEII restriction sites and were ligated into the renilla luciferase gene 3′UTR SacI and BsteII restriction sites. To ectopically express miR-1271, a ∼250-nt portion of human or rat genome containing the miR-1271 sequence was PCR amplified with primers described in Supplementary File 1. The amplified product was digested with BstEII and ClaI restriction enzyme and sub-cloned down-stream of the pSUPER H1 promoter cassette modified to contain the BstEII and ClaI sites. The ‘HuRat’ miR-1271 precursor insert was generated using a synthetic oligonucleotide to introduce four single base changes in the rat gene sequence. The 90-nt sense sequence is shown in Supplementary File 1. The pSUPER H1 expression cassette was sub-cloned into a pENTR11 (Invitrogen, Carlsbad, CA, USA) shuttle vector containing a membrane bound green fluorescent protein (mGFP) reporter gene expressed by a chicken β-actin promoter.
Human miR-1271 precursor RNA was purchased from Ambion (Austin, TX, USA). UCDNA Services, Faculty of Medicine, at the University of Calgary, Alberta, Canada synthesized the precursor RNA for miR-96 [see ref (9)] and miR-600[G] RNA. The sequence of miR-600[G], ACUUACGGACAAGAGCCUUGCUC, was annealed to the passenger strand, miR-600[G]* GGAAGGCUCUUGUCUGUCAGGCA. Argonaute 2 siRNA was from Dharmacon (Lafayette, CO, USA) and Glyceraldehyde 3-phosphate siRNA was from Ambion (Austin, TX, USA).
Cell culture and transfection experiments
HEK293 (ATCC CRL-1573) cells were cultured according to ATCC specifications. For luciferase assay experiments, 2 × 105 or 1 × 105 HEK293 cells were seeded into either 12- or 24-well cell culture plates 24 h prior to transfection. Cells were transfected with 50 ng of luciferase plasmid together with 0–2 nM microRNA precursor using Optimem I media and Lipofectamine 2000 according to manufacture’s instructions (Invitrogen, Carlsbad, CA, USA). Cells were incubated for 24 h then assayed for firefly and renilla luciferase activity using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA). Luminescence was measured with a 20/20n Luminometer (Turner Biosystems, Sunnyvale, CA, USA).
For gene knockdown experiments, HEK293 cells were transfected with AGO2 or GAPDH siRNA at the time of plating at a density of 1 × 105 cells per well (24-well plates). Cells were grown for 12 h then transfected with 50 ng of luciferase plasmid and microRNA. Luciferase activity and mRNA levels were measured after an additional 18 h of incubation.
Western blot analysis of gephyrin
For protein analysis, 48 h after HEK293 cells were transfected with microRNA, cells were washed once with phosphate buffered saline, removed from the plate with SDS loading buffer, and heat denatured for 10 min at 70°C in the presence of 100 mM dithiothreitol. 10 µg of protein was resolved on 10% SDS–PAGE, and transferred to PVDF membrane. The membrane was blocked for 1 h with 3% bovine serum albumin diluted in TBS-T (25 mM Tris, 150 mM NaCl, 1% Tween-20). The membrane was incubated at 4°C overnight or at room temperature for 1 h with goat anti-gephyrin polyclonal antibody (sc-6411; Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA) diluted 1:1000 or α-tubulin mouse monoclonal antibody from Abcam (ab7291; Abcam, Cambridge, MA) at a 1:10 000 dilution. Membranes were washed three times for 5 min with TBS (25 mM Tris, 150 mM NaCl) and then incubated at room temperature for 1 h with HRP conjugated secondary antibody (Santa Cruz Biotechnology, Inc) diluted 1:12 000. Membranes were washed repeatedly with TBS (3×) and TBS-T (3×), then incubated with Amersham ECL reagent (GE Health Care, Piscataway, NJ, USA) and exposed to film.
RT–PCR analysis of gene expression
Forty-eight hours after HEK293 cells were transfected with microRNA, total RNA was extracted with Trizol (Invitrogen) for RT–PCR. Other sources of RNA for RT–PCR included human forebrain cortex (Figure 4A; Advanced Tissue Resource Center, Charlestown, MA, USA), rat forebrain cortex (Figures 4A and 5C; adult Brown Norway) or FirstChoice® Human Total RNA Survey Panel (Figures 4B and 5C; Ambion, Austin, TX).
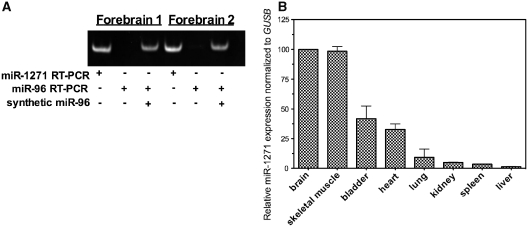
Figure 4.
miR-1271 and miR-96 have unique expression pattern in human nervous system tissue. (A) MiR-1271 is expressed robustly in human frontal cortex while miR-96 is undetectable. Synthetic miR-96 RNA was spiked into the forebrain sample to confirm that miR-96 could be detected with this assay. (B) The expression pattern of miR-1271 in human tissue relative to β-glucuronidase (GUSB). The error bars indicate ± SEM for two experiments with pooled (three individuals) total RNA samples from each tissue.
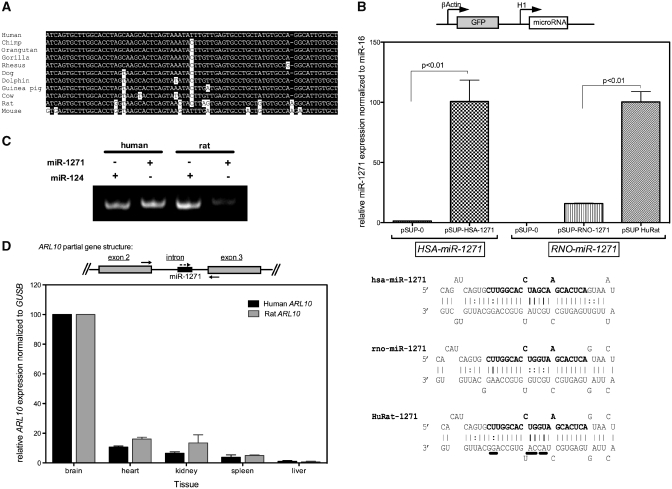
Figure 5.
The expression of miR-1271 is not conserved in mammals. (A) The mammalian miR-1271 precursor sequence is divergent. Nucleotides with white background differ from the human sequence. (B) By ectopically expressing the human miR-1271 genetic locus, miR-1271 can be induced greater than 50-fold in HEK293 cells. No induction is evident with the parental plasmid, pSUP-0. MiR-1271 induction by the rodent gene is significantly blunted compared to human. By making changes that humanize the precursor sequence (underlined nucleotides, shown below the graph) but leave the mature microRNA sequence unchanged, rodent miR-1271 expression is induced to the level of the human gene. Error bars indicate ± SEM for three experiments. (C) Rodent miR-1271 is expressed at reduced levels in rat brain. A brain expressed microRNA, miR-124, serves as a positive control for microRNA detection in rat brain. (D) Human and rat ARL10 mRNA are expressed with very similar tissue specificity. The partial structure of human and rat ARL10 and miR-1271 gene is shown above the graph showing relative ARL10 expression normalized to GUSB for five tissues. The solid arrows indicate the position of primers used for RT–PCR. The dashed arrow indicates the direction of miR-1271. Error bars indicate ± SEM for two experiments.
Total RNA was reverse transcribed into cDNA using random hexamers and the High Capacity cDNA Archive Kit (Applied Biosystems) in a 20-µl reaction. RNA levels for miR-96/1271 target genes and a normalizing gene, b-glucuronidase, were measured using SYBR Green qPCR and primers designed using Primer3 (14). Primer sequences are listed in Supplemental File 1. One microlitre of each cDNA reaction was used as template in a 10-µl PCR reaction containing 250 nM forward and 250 nM reverse primer, 0.1 × Syber Green (Molecular Probes, Eugene, OR, USA), 1 µg/µl Bovine serum albumin (New England Bio Labs, Ipswich, MA) and 1 × Gene Expression Master Mix [Applied Biosystems (ABI), Foster City, CA] using a ABI 7500 Real Time PCR system. PCR cycles were; 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Levels of β-glucuronidase mRNA were used for normalizing microRNA expression levels between tissues in the FirstChoice® Human Total RNA Survey Panel. The average ΔCt for brain RNA was used as a reference point (i.e. 100%) for normalizing the expression values from the different tissues in the Human Total RNA Survey Panel.
MicroRNA gene expression assays were based on the method described by Varkonyi-Gasic et al. (15). Of the total RNA, 100 ng was reverse transcribed in an 8-µl reaction containing 1 pmole of microRNA gene specific reverse transcription primer (described in Supplementary File 1) and 20 U of Superscript 3 enzyme (Invitrogen Carlsbad, CA, USA). To measure the level of ectopically expressed human or rat miR-1271, the reverse transcription reaction was multiplexed to contain human or rat specific miR-1271 and miR-16 gene specific reverse transcription primers. miR-1271 and miR-16 were then PCR amplified in separate reactions using 1 µl of this common reverse transcription reaction. The average miR-1271/miR-16 ΔCt for the pSUP-HSA-1271 transfected cell RNA was used as a reference (i.e. 100%) for the microRNA induction experiment.
Statistics
Significance (P < 0.05) was determined with a one-sided Student’s t-test unless noted otherwise.
RESULTS
miR-96 and miR-1271 regulate mRNA with similar specificity
The miRBase (16) and TargetScan (1) microRNA databases annotate two human microRNAs, miR-96 and miR-1271, that have considerable sequence homology (Figure 1A). The shared ‘seed’ sequence and the potential interaction of miR-1271 with a previously studied polymorphic element in the human serotonin receptor 1B mRNA suggested that these microRNAs might have similar biological activities. Recent studies indicate that nucleotides outside the seed may exert important control over human microRNA function. For example, the 5′-terminal nucleotide may control microRNA recruitment into an effector protein complex and therefore control microRNA activity (13,17). Also interactions between nucleotides outside the ‘seed’ sequence of the microRNA may provide an additional layer of mRNA target specification (18–20). Given that miR-96 and miR-1271 differ in these sequences, we used a luciferase reporter assay to test for potential similarities and differences in miR-96 and miR-1271 regulatory activity. In this assay, a test element is inserted into the renilla luciferase gene 3′ untranslated region (UTR) and the regulatory activity of that element is measured by normalizing renilla luciferase enzyme activity to that of the firefly luciferase enzyme expressed constitutively from the same plasmid (Figure 1B). A microRNA response is evident from a decrease in the ratio of renilla to firefly activity. As shown in Figure 1B, insertion of the HTR1B microRNA target A-element into the renilla 3′UTR confers a robust repressive effect by both miR-96 and miR-1271. The A>G change associated with the human single nucleotide polymorphism, rs13212041, results in an attenuated response to both miR-96 and miR-1271. A control RNA, miR-600[G] does not regulate either element. Thus, we provide the first evidence that miR-1271 is indeed a functional microRNA and that it regulates the naturally occurring polymorphic HTR1B microRNA-binding element with the same specificity as miR-96.
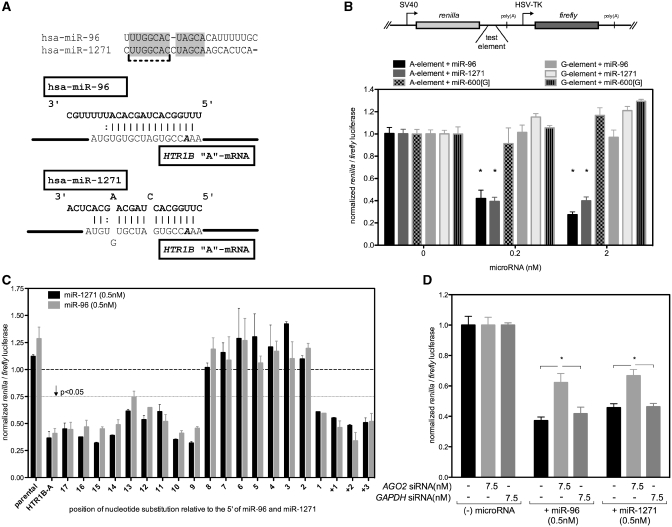
Figure 1.
Gene regulation by miR-1271 is disrupted by the HTR1B A/G 3′UTR polymorphism. (A) Nucleotides that are common to microRNAs, miR-1271 and miR-96 are indicated by grey shading. A dashed line indicates the ‘seed’ sequence, nucleotides 2–8. Shown below is the potential interaction of both microRNAs with an element from the serotonin receptor 1B mRNA 3′UTR. The location of the functional A>G rs13212041 SNP in the 3′UTR opposite nucleotide 3 of the microRNA is indicated by bold italic. (B) The dual luciferase reporter plasmid used to test microRNA response. The expression values in panels (B)–(D) are normalized for each construct to the expression value in the absence of co-transfected microRNA. Similar to miR-96, miR-1271 regulates the expression of the HTR1B rs13212041 A-element, but not the G-element. A control microRNA, miR-600[G], does not regulate the element with the same specificity. **P < 0.005 comparing A-element to G-element in the indicated conditions, and error bars indicate ± SEM for 4-7 experiments. (C) MiR-96 and miR-1271 target HTR1B mRNA with similar specificity. The HTR1B 3′UTR based sequences used to test the nucleotide specificity of miR-96 and miR-1271 are described in detail in Supplementary File 1. The position of each nucleotide change is numbered relative to the microRNA 5′ end, i.e. mutant 1 is opposite nucleotide 1 of each microRNA. + 1 to + 3 indicates HTR1B 3′ UTR nucleotides downstream from potential microRNA annealing site. The responses of each point mutant element to miR-96 and miR-1271 are significantly correlated, r = 0.94, P < 0.0001 (Pearson's correlation). Error bars indicate ± SEM for two experiments. (D) MiR-96 and miR-1271 gene repression is argonaute 2 dependent. Asterisk indicates P < 0.05 and error bars indicate ± SEM for six to eight experiments.
The sequence differences between miR-96 and miR-1271 might allow each microRNA to interact with target mRNA differently and thereby each may differentially regulate a given mRNA. To more finely probe for potential regulatory differences, we interrogated the HTR1B A-element mRNA to identify the nucleotides required for the miR-96 versus miR-1271 microRNAs to regulate gene expression. We used this naturally occurring element as a model test substrate because it responds robustly to microRNA and it has extensive but distinct complementarity to both miR-96 and miR-1271. This allowed us to thoroughly test for potential nucleotide requirements by mapping the element with point mutations. At each position in a 22-nt element, single bases were sequentially changed such that potential Watson–Crick annealing between the response element mRNA and microRNA was disrupted. The regulatory response of each targeted mutant element to miR-96 or miR-1271 was then tested. We observed that miR-96 and miR-1271 regulate the mutated mRNA elements in a strikingly similar fashion (Figure 1C). Each microRNA exerts a repressive effect with all of the test elements that have complete Watson–Crick complementarity between microRNA nucleotides 2–8 and the HTR1B target mRNA sequence. At each mutated position outside nucleotides 2–8 in the 5′ or 3′ direction the response to either microRNA is near wild-type levels. This is interesting, given that the mRNA element has nucleotides at the 5′ end that could potentially anneal to the more variable 3′ regions of both microRNAs in Watson–Crick fashion. Those potential interactions do not appear to be critical for the repressive functions of either microRNA when they act upon the HTR1B model test elements. It is also worth noting that, although there is potential for strong annealing elsewhere in the response element, these regions cannot fully compensate for single changes to nucleotides 2–8. These observations reinforce the functional significance of the human single nucleotide polymorphism, rs13212041, which is opposite the third nucleotide of each microRNA. These findings also highlight the requirement for pairing between the microRNA ‘seed’ and the target mRNA in order for gene silencing to occur for this motif.
Small RNA molecules in animal cells interact directly with conserved RNA-binding proteins called argonautes. The microRNA-directed silencing effects in HEK293 cells appear to rely primarily on the expression of argonaute 2 (AGO2) (21). To test whether the sequence specific regulatory effects we observed were mediated by AGO2, we used siRNA to reduce AGO2 expression, and then tested the repressive effect directed by each microRNA in the luciferase assay. As shown in Figure 1D, pre-treating HEK293 cells with siRNA targeting AGO2 mRNA decreases the silencing effect of each microRNA compared to cells pretreated with siRNA targeting glyceraldehyde-3-phosphate (GAPDH) or no siRNA. The silencing effect of each microRNA was reduced ∼40% relative to mock transfected cells or cells transfected with GAPDH siRNA. We validated gene knock down by using RT–PCR to measure the mRNA of each target gene relative to β-glucuronidase. The average mRNA knockdowns in four experiments were 92% ± 3.5 (SEM) for GAPDH and 74% ± 7.3 (SEM) for AGO2. Our data suggest that the sequence specific regulatory effects of miR-96 and miR-1271 in HEK293 cells are mediated largely by AGO2. The residual repressive activity of each microRNA may be due to incomplete knockdown of AGO2 or perhaps compensation by other argonaute isoforms. It is possible that miR-96 and miR-1271 utilize other argonautes such as AGO1, AGO3 and AGO4 with different regulatory effects and target specificity.
MiR-96 and miR-1271 regulate human genes involved in neural signaling
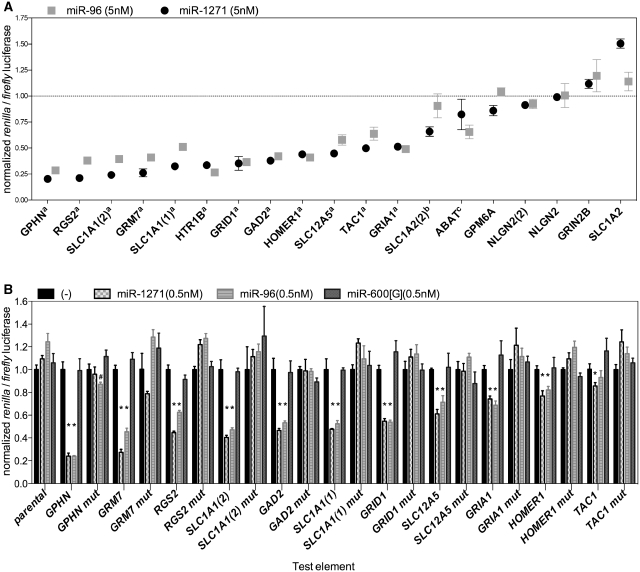
Given the similarities we observed in the regulation of the model test elements, we hypothesized that miR-96 and miR-1271 could potentially target the same repertoire of genes. To test this hypothesis, we focused on a set of genes that are involved in processes related to neuronal excitation e.g. glutamate signaling, synapse structure and membrane electrical potential. Indeed, we noted that many of the potential miR-96 and miR-1271 targets annotated by the TargetScan (1) and miRanda (22) algorithms are related to neural signaling. Potential miR-96/miR-1271 targets include; two glutamate transporters (SLC1A1, SLC1A2), four glutamate receptors (GRIN2B, GRID1, GRIA1 and GRM7), two proteins that regulate the activity of glutamate receptors (RGS2 and HOMER1) (23,24), two enzymes involved in glutamate and γ-amino butyric acid metabolism (ABAT and GAD2), and proteins such as neuroligin 2 (NLGN2) and gephyrin (GPHN) that are involved in the post-synaptic structure of inhibitory synapses (25,26). We also observed this motif in the mRNA of the tachykinin 1 gene (TAC1), which encodes the neuropeptides substance K and substance P, and the mRNA of SLC12A5 (also known as KCC2), the gene that encodes the potassium chloride symporter 2. KCC2 is important in the development and control of the chloride ion gradient and in the regulation of interneuron migration (27,28). The motif is also present in the mRNA for glycoprotein M6a (GPM6A), a stress-regulated protein expressed on the glutamatergic axons of neurons in adult rat forebrain (29). We sub-cloned fragments of the 3′UTR containing the putative miR-96/miR-1271 target elements from each of these genes into the luciferase reporter construct to screen for potential miR-96 and miR-1271 regulatory effects. Upon co-transfection with microRNA, the majority (14/19) of test element plasmids had a reduced ratio of renilla to firefly luciferase expression indicating potential regulation by miR-96 and miR-1271 (Figure 2A). Three genes had two potential target sites, so in sum we observed that 75% (12/16) of the tested genes had an mRNA element that responded to co-transfected miR-96 and miR-1271. The miR-96 and miR-1271 repressive effects on each test element were strikingly similar and highly correlated. We further validated that the repressive effects of miR-1271 and miR-96 required a seed sequence match in the mRNA for the genes that showed a repressive response in the initial screen by mutating the third nucleotide of the target site. As shown in Figure 2B, the repressive effect of each microRNA was attenuated with test elements that had a single nucleotide mismatch to nucleotide three of the microRNAs. The regulatory response was also specific to miR-96 and miR-1271, a control microRNA miR-600[G] does not regulate the test elements.
Figure 2.
MiR-96 and miR-1271 show similar regulation of target elements from multiple human genes involved in neuron function. (A) The response of each test gene element to the indicated co-transfected microRNA is normalized to the activity of the test construct in the absence of added microRNA. aP < 0.05 for both miR-96 and miR-1271, bfor miR-1271 only and cfor miR-96 only. The effects of each microRNA are significantly correlated, r = 0.92, P < 0.00001 (Pearson's correlation). Error bars indicate ± SEM for two experiments. (B) The repressive effects of miR-96 and miR-1271 require a seed sequence match. Results for 11 targets that showed repressive effects of miR-1271/96 are illustrated. The repressive effect of both miR1271 and miR-96 microRNAs is attenuated by a single nucleotide mismatch in the target site aligned with nucleotide 3 of the microRNA (designated as xx mut). Error bars indicate ± SEM for three to six experiments. *P < 0.05 compared to minus microRNA condition. Hash (#) indicates a repressive effect (P < 0.05) for the mutant GPHN element that is significantly different than the wild type. The response of each test gene element to the indicated co-transfected microRNA is normalized to the activity of the test construct in the absence of added microRNA.
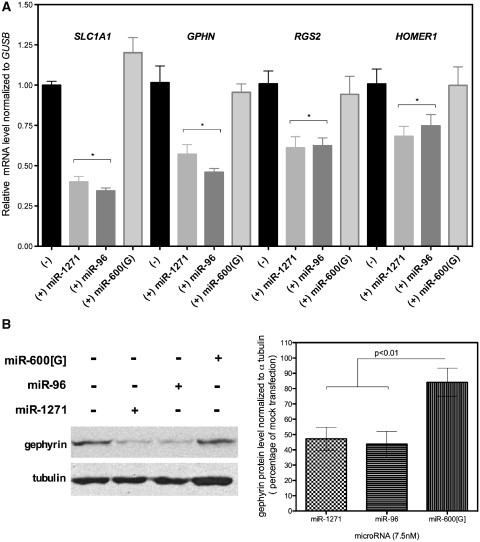
HEK293 cells express several of the genes examined in Figure 2 whose elements were regulated in the luciferase assay. Therefore, we examined whether the level of endogenous mRNA for four of these genes were reduced in HEK293 cells in the presence of exogenous miR-96 or miR-1271. As shown in Figure 3A, the mRNA from SLC1A1, GPHN, RGS2 and HOMER1 genes was significantly reduced in cells transfected with miR-96 or miR-1271 compared to mock transfected cells or cells transfected with a control microRNA. The level of SLC1A1, GPHN, RGS2 and HOMER1 mRNA was reduced ∼60, 50, 40 and 30%, respectively, in either miR-96 or miR-1271 transfected cells. The miR-96 and miR-1271 effects on levels for each of these mRNAs were similar. We additionally tested whether the similarities we observed for regulation of mRNA were also manifested at the protein level by examining the protein expression for gephyrin, one of the more abundantly expressed mRNAs. After 48 h, HEK293 cells transfected with miR-96 or miR-1271 had significantly less gephyrin protein than mock transfected cells or cells transfected with a control microRNA (Figure 3B). There was a significant 2-fold decrease in gephyrin protein level in cells transfected with either miR-96 or miR-1271. There was no significant difference in the level of gephyrin protein in cells transfected with a control microRNA. Therefore, we conclude that miR-96 and miR-1271 potentially regulate the same genes including many that are involved in neuron function.
Figure 3.
miR-1271 and miR-96 regulate neural gene expression in HEK293 cells. (A) The level of target gene mRNA in HEK293 cells is reduced by miR-1271 and miR-96 transfection. The normalized level of the indicated target mRNA relative to β-glucuronidase (GUSB) is significantly less in cells transfected with 7.5 nM of either miR-1271 or miR-96 compared to mock transfected cells. The mRNA levels were unchanged when the cells were transfected with a control microRNA (miR-600[G]). Asterisk indicates P < 0.05 and error bars indicate ± SEM for three to four experiments. (B) Gephyrin protein is reduced ∼55% in cells transfected with either miR-96 or miR-1271. The right panel summarizes the immunoblot densitometry ± SEM for four experiments, where 100% is the level of gephyrin/alpha-tubulin in mock-transfected cells.
Human miR-1271 and miR-96 are expressed in different human tissues
Although, miR-96 and miR-1271 can function similarly in HEK293 cells, their potential regulatory activity in vivo is dependent upon their own pattern of expression, which may differ temporally and spatially. The expression of miR-96 has been extensively characterized in vertebrates. In mammals, although miR-96 is detectable in brain, it is considerably more abundant in sensory organ components of the central nervous system including the retina, olfactory bulb and inner ear, and therefore miR-96 is considered a sensory organ specific microRNA (30–32). Less is known about the expression of human miR-1271. The online repository for microRNA sequence data, miRBase (16), only notes that miR-1271 has been identified experimentally at low copy number by deep sequencing of small RNA from human embryonic stem cells (33) and human teratoma cell lines (34). Given that many of the target mRNAs we tested are widely expressed in subsets of brain neurons we hypothesized that miR-1271 would be brain expressed. Indeed, as shown in Figure 4A, we detect robust expression of miR-1271 in two human frontal lobe cortex samples compared to miR-96, which, as expected based on prior reports, was very low to undetectable. Because we did not have RNA from human sensory organs (i.e. tissues that would be expected to contain abundant levels of miR-96), synthetic miR-96 was spiked into the human brain RNA template prior to RT–PCR at a concentration that would correspond to ∼6000 copies per cell. When the synthetic RNA is spiked into template RNA samples, miR-96 is readily detected. The synthetic miR-96 RNA positive control supports our conclusion that there is a striking difference in miR-1271 and miR-96 expression in human brain tissue.
We next tested whether miR-1271 was as abundant in other human tissues, including non-neuronal tissues. As shown in Figure 4B, miR-1271 expression was highest in brain and skeletal muscle, followed by heart and bladder. In brain and skeletal tissues, the level of miR-1271 relative to β-glucuronidase was equal, while the levels in heart and bladder were 60% and 70% less, respectively. Among the tissues we tested, the level of miR-1271 was lowest in liver (<5% of brain). An interesting feature of this expression pattern is that the highest ranked tissues, brain, skeletal muscle, heart and bladder, all contain a high level of excitable cells such as neurons and muscle (skeletal, cardiac and smooth).
A second striking difference between miR-96 and miR-1271 is the difference in degree of conservation between vertebrates. As shown in Figure 5A, the potential miR-1271 precursor sequences are divergent among mammals. More distant species, in particular, have a number of nucleotide substitutions (rat = 9, mouse = 10) that occur throughout the potential gene. A similar length sequence encompassing miR-96 is identical between human and rats, while the mouse sequence varies by only one nucleotide (this difference is not in the mature miR-96 sequence). We viewed the between-species differences in the miR-1271 gene precursor sequence as potentially significant because the precursor sequence forms a specific secondary structure essential for proper microRNA processing and expression. Therefore, we hypothesized that the changes in the human sequence may stabilize microRNA precursor secondary structure and subsequent processing events leading to enhanced microRNA expression. To test this, we compared the expression of microRNA from the human and rat miR-1271 genetic locus. We sub-cloned a ∼250-nt portion of the potential precursors into an H1 promoter controlled expression vector. The expression vector also contained a green fluorescent protein (GFP) gene as part of a different transcriptional unit, which enables cell tracking and assessment of transfection efficiency between plasmids. After 48 h, the level of human miR-1271 was increased more than 50-fold in HEK293 cells transfected with pSUP-HSA-miR-1271 compared to cells transfected with the parental vector, pSUP-0 (Figure 5B). Expression of miR-1271 by the rat locus was significantly less than the human locus. To test whether this blunted expression from the rat precursor sequence was potentially from a defect in the hairpin structure, we created a precursor sequence (HuRat) that had 4 nt changes that potentially humanize the hairpin structure without changing the sequence of the mature rat microRNA (Figure 5B). We observed that the humanized version of the rat precursor produces microRNA levels similar to that of the human gene. The human cell line we were using, HEK293, may have lacked factors necessary for expression of the rat gene; therefore we also examined endogenous miR-1271 expression in rat brain. As shown in Figure 5C, we detect considerably less miR-1271 in rat brain compared to human. To verify our ability to detect microRNA expression in this tissue we amplified the brain specific microRNA, miR-124 (35) as a positive control and observed robust miR-124 expression. An alternative basis for low-miR-1271 expression in rat brain compared with human brain might be that the rodent miR-1271 precursor is expressed in a different tissue pattern in rat versus humans. Human miR-1271 and the potential rat miR-1271 are each encoded within intron 2 of ADP-ribosylation factor-like 10 (ARL10). Intronic microRNAs are expressed from their host gene as a single transcriptional unit; therefore, we tested whether the host gene in each species was expressed similarly (36,37). As shown in Figure 5D, ARL10 mRNA is expressed with the same specificity in human and rat tissues. The expression of ARL10 is highest in brain for each species and lowest in liver. It is possible that the wild-type rodent precursor is processed to a biologically active small RNA sequence that is not detected by our assay. However, our data do indicate that the subtle differences in the microRNA precursor sequence that occur between mammalian species have significant effects on miR-1271 processing and expression. Therefore, the expression of the functional miR-1271 may vary between mammals and confer unique regulatory effects in humans.
DISCUSSION
Our findings provide an intriguing example of two human microRNAs, miR-96 and miR-1271 that have the potential to act redundantly yet are expressed and perhaps function in different human tissues. The expression of the highly conserved miR-96 gene has been extensively characterized in mammals where it is expressed in sensory organs (30–32), while miR-1271 is a small RNA that was only recently identified at low copy number in human non-neuronal tissue. It could be argued that this low level of expression renders miR-1271 with limited biological activity, yet our studies on human miR-1271 expression and function indicate that it likely has significant, although non-conserved, biological importance.
We used in silico databases to determine that the miR-96/miR-1271 target motif is present in the mRNA from several genes that are involved in neural signaling and cell excitation, suggesting that either of these microRNAs might have a role in regulating aspects of human neural development or function. One target protein that we have studied, gephyrin, is an important structural anchor for GABAA and glycine receptors. Loss of gephyrin expression results in reduced post-synaptic glycine receptor numbers and altered GABAA receptor subunit localization (26,38). Other genes such as SLC12A5, HTR1B and the glutamate receptors described in this paper also have significant functions in regulating cell excitation. Although not tested in this article, TargetScan (1) also predicts the miR-96/miR-1271 target motif in the mRNA of other genes important for cell excitation, including four voltage-gated sodium channel subunits (SCN1A, SCN2A, SCN3A and SCN9A) and four voltage-gated calcium channel subunits (CACNA1C, CACNA2D2, CACNB1 and CACNB4). Given that miR-1271 is highly expressed in tissues that contain excitatory cells such as brain and muscle, one function of miR-1271 might be to regulate the development or maintenance of these processes. An in-depth analysis of all potential targets may reveal additional potential pathways or process regulated by miR-1271. Regulation of cell excitation and neural transmission are interesting characteristics that unify many genes in the miR-96/1271 target gene set. However, the conclusions that can be drawn from this observation may be limited by the nature of the microRNA prediction algorithm and the mRNA transcript database used as a source of potential targets. Perhaps in this dataset, these genes are over-represented as potential microRNA targets because their mRNA transcripts are more extensively characterized and annotated. However, it would be interesting to see whether human variation in the mRNA target site of any of these genes, as we have identified in HTR1B, influence behavioral phenotypes relevant to psychiatric disorders. Such a finding may yield additional clues to the function of human miR-1271 in vivo.
Contrary to many well-characterized human microRNA genes such as miR-96, the nucleotides in the miR-1271 gene precursor are not highly conserved in mammals. We have shown that by substituting four nucleotides in the rat gene to generate a potentially ‘humanized’ secondary structure, the expression of a miR-1271 gene product is significantly enhanced. Therefore, the precursor sequence differences that occur between mammals may cause the small RNAs generated from this locus to be processed with different efficiencies or into different biologically active forms that confer unique species-specific regulatory effects. According to miRBase, the only other experimentally identified mammalian miR-1271 has been from dog lymphocyte RNA (39). This expressed small RNA sequence resembles human miR-1271 (Figure 5), however it is truncated by two nucleotides on the 3′ end. We have observed that this sequence maps with 100% identity to at least three regions of the canine genome (Chr4:40143926-40143945, Chr15:54162445-54162464 and Chr38:4899026-4899045). It is unclear the frequency at which this RNA was detected and whether it is a bona fide product of the locus that is homologous to the human miR-1271 gene or perhaps other regions of the canine genome. A more rigorous examination of this genomic locus in all mammalian species may provide insight into the mechanisms involved in the genesis of this microRNA. These differences between mammals suggest that it is possible for closely related species and perhaps even individuals of the same species to differ in the number and identity of microRNA genes. Yet, for such a microRNA gene to become fixed in one species genome, it must impart some benefit to fitness through regulatory interactions with functional target mRNA motifs. MiR-1271 expression in humans may have developed to some extent as a consequence of regulating mRNAs from genes with miR-96 targets that are conserved in vertebrates and also coincidently expressed with miR-1271.
The degree that human miR-1271 functions in human tissues that have high-miR-96 expression, such as eye, olfactory bulb or inner ear remains untested. A more thorough analysis of these tissues is warranted given the recent reports showing that heritable mutations in the miR-96 gene seed sequence are responsible for two independent cases of familial non-syndromic progressive hearing loss (11). Individuals bearing a mutated miR-96 gene might have a less pronounced sensory deficit phenotype due to the potential compensatory activity of miR-1271. Another potential way to distinguish between the activities of each microRNA is to identify the proteins that interact with them. This may provide insight into any distinct functional roles not revealed in our studies.
In sum, we believe that human miR-1271 has regulatory functions that are relevant to the biology of neurologic or psychiatric disorders. We hope that our findings promote a more thorough analysis of the function of novel human small RNA genes discovered by deep sequencing techniques. Although often detected at low copy number, these genes may have important functions that are restricted to certain tissues or developmental time periods.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [P60AA03510 (Alcohol Research Center) and F31 DA023341 (to K.P.J.)]. Funding for open access charge: National Institutes of Health grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Linda Burian for her expert technical assistance, Dr Lisa Conti for rat tissues, Dr Kristian Hedstrom for reagents and Drs Henry Kranzler and Asis Das for their helpful comments during the course of this work.
REFERENCES
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 3.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34:1395–405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum. Mol. Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 6.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 8.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 9.Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol. Psychiatry. 2009;14:381–189. doi: 10.1038/mp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conner TS, Jensen KP, Tennen H, Furneaux HM, Kranzler HR, Covault J. Functional polymorphisms in the serotonin 1B receptor gene (HTR1B) predict self-reported anger and hostility among young men. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;153B:67–78. doi: 10.1002/ajmg.b.30955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 12.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felice KM, Salzman DW, Shubert-Coleman J, Jensen KP, Furneaux HM. The 5′ terminal uracil of let-7a is critical for the recruitment of mRNA to Argonaute2. Biochem. J. 2009;422:329–341. doi: 10.1042/BJ20090534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 15.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 18.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y. The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS ONE. 2008;3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kammermeier PJ, Ikeda SR. Expression of RGS2 alters the coupling of metabotropic glutamate receptor 1a to M-type K+ and N-type Ca2+ channels. Neuron. 1999;22:819–829. doi: 10.1016/s0896-6273(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 24.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 25.Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Essrich C, Lorez M, Benson JA, Fritschy JM, Luscher B. Postsynaptic clustering of major GABAA receptor subtypes requires the gamma 2 subunit and gephyrin. Nat. Neurosci. 1998;1:563–571. doi: 10.1038/2798. [DOI] [PubMed] [Google Scholar]
- 27.Rivera C, Voipio J, Payne JA, Ruusuvuori E, Lahtinen H, Lamsa K, Pirvola U, Saarma M, Kaila K. The K+/Cl- co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature. 1999;397:251–255. doi: 10.1038/16697. [DOI] [PubMed] [Google Scholar]
- 28.Bortone D, Polleux F. KCC2 expression promotes the termination of cortical interneuron migration in a voltage-sensitive calcium-dependent manner. Neuron. 2009;62:53–71. doi: 10.1016/j.neuron.2009.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper B, Fuchs E, Flugge G. Expression of the axonal membrane glycoprotein M6a is regulated by chronic stress. PLoS ONE. 2009;4:e3659. doi: 10.1371/journal.pone.0003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 33.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nygaard S, Jacobsen A, Lindow M, Eriksen J, Balslev E, Flyger H, Tolstrup N, Moller S, Krogh A, Litman T. Identification and analysis of miRNAs in human breast cancer and teratoma samples using deep sequencing. BMC Med. Genomics. 2009;2:35. doi: 10.1186/1755-8794-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirsch J, Wolters I, Triller A, Betz H. Gephyrin antisense oligonucleotides prevent glycine receptor clustering in spinal neurons. Nature. 1993;366:745–748. doi: 10.1038/366745a0. [DOI] [PubMed] [Google Scholar]
- 39.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.