Abstract
Higher eukaryotes encode various Y-family DNA polymerases to perform global DNA lesion bypass. To provide complete mutation spectra for abasic lesion bypass, we employed short oligonucleotide sequencing assays to determine the sequences of abasic lesion bypass products synthesized by human Y-family DNA polymerases eta (hPolη), iota (hPolι) and kappa (hPolκ). The fourth human Y-family DNA polymerase, Rev1, failed to generate full-length lesion bypass products after 3 h. The results indicate that hPolι generates mutations with a frequency from 10 to 80% during each nucleotide incorporation event. In contrast, hPolη is the least error prone, generating the fewest mutations in the vicinity of the abasic lesion and inserting dAMP with a frequency of 67% opposite the abasic site. While the error frequency of hPolκ is intermediate to those of hPolη and hPolι, hPolκ has the highest potential to create frameshift mutations opposite the abasic site. Moreover, the time (t50bypass) required to bypass 50% of the abasic lesions encountered by hPolη, hPolι and hPolκ was 4.6, 112 and 1 823 s, respectively. These t50bypass values indicate that, among the enzymes, hPolη has the highest abasic lesion bypass efficiency. Together, our data suggest that hPolη is best suited to perform abasic lesion bypass in vivo.
INTRODUCTION
DNA polymerases are grouped into the A-, B-, C-, D-, X- and Y-families. The Y-family DNA polymerases function primarily in the bypass of replication-stalling DNA lesions, a process which can ultimately decrease the possibility of invoking DNA damage-induced apoptosis. In humans, 4 of the 16 identified DNA polymerases are in the Y-family: DNA polymerases eta (hPolη), iota (hPolι), kappa (hPolκ) and Rev1 (hRev1). In numerous biochemical studies (1–3), these enzymes are capable of both error-free and error-prone lesion bypass, depending on the specific lesion. In vivo, hPolη is responsible for the error-free bypass of cis-syn thymine–thymine (TT) dimers (4,5). Moreover, the mutational inactivation of hPolη leads to xeroderma pigmentosum variant (XPV) that predisposes individuals to an increased incidence of sunlight-induced skin cancer (5). hPolη also has been shown biochemically to bypass lesions including abasic sites (AP) (6), 7,8-dihydro-8-oxoguanine (8-oxoG) (7), (+)-trans-anti-benzo[a]pyrene-N2-dG [(+)BPDE-dG] (6), 1,N6-ethenodeoxyadenosine (8), O6-methylguanine (9), N-2-acetylaminofluorene-dG (AAF-dG) (10) and cisplatin-dGpG intrastrand adducts (10). Interestingly, Polι has been shown to incorporate incorrect dGMP opposite a template base dT more efficiently than canonical dAMP (11,12). In vitro, hPolι has been shown to traverse AP sites (12–14), 8-oxoG (14), AAF-dG (14), cis-syn TT dimers (13) and (6-4) TT photoproducts (14,15). Polκ, which is a member of the DinB subfamily and a close relative of Y-family member Sulfolobus solfataricus DNA Polymerase IV (Dpo4), can efficiently elongate mispaired primer termini (16). In addition, hPolκ has been shown to bypass AP sites, 8-oxoG, AAF-dG and (+)BPDE-dG (17). Rev1, which is the only Y-family DNA polymerase to contain a BRCT domain, is classified as a dCMP transferase (18). hRev1 incorporates a dCMP efficiently opposite lesions including AP sites (19,20), 8-oxoG, (+)BPDE-dG, (−)BPDE-dG and 1,N6-ethenoadenine adducts (21). Together, the aforementioned studies indicate that there is a significant overlap in regards to the in vitro lesion bypass spectra of the four human Y-family DNA polymerases. Yet, with the exception of a cis-syn TT dimer, it is unclear which human Y-family enzyme is responsible for the bypass of which lesion(s) in vivo.
One of the most challenging issues in the field of DNA lesion bypass is to identify the in vivo lesion bypass specificity of the Y-family DNA polymerases, especially for those Y-family enzymes that coexist within the same organism. The knowledge of in vitro lesion bypass efficiency and fidelity of a Y-family DNA polymerase may shed light on which lesions it bypasses in vivo. However, the aforementioned in vitro lesion bypass studies have been performed by different laboratories using different DNA substrates under different reaction conditions (22–24). Thus, it is challenging to deduce which human Y-family enzyme bypasses specific lesions in vivo. In order to exclude these variables, we quantitatively assessed the AP lesion bypass abilities of the four human Y-family DNA polymerases using the same in vitro assay under the same reaction conditions. Notably, AP lesions are the most common DNA lesions found in mammalian cells with ∼10 000 spontaneous AP sites generated in each cell every day (25,26). Herein, we have employed a recently developed short oligonucleotide sequencing assay (SOSA) to quantitatively determine the mutational spectra of these human Y-family enzymes in the vicinity of this non-coding DNA lesion. Our in vitro data suggest that hPolη is likely the Y-family DNA polymerase to bypass AP lesions in vivo.
MATERIALS AND METHODS
Materials
Human AP endonuclease and Taq DNA polymerase were purchased from Trevigen and Invitrogen, respectively. [γ-32P]ATP was purchased from GE Healthcare Life Sciences. The oligodeoxynucleotides in Table 1 were purchased from Integrated DNA Technologies and were purified, labeled and annealed as described previously (27).
Table 1.
DNA primer and templatesa
| Primer | |
| 14-mer | 5′-CGCAGCCGTCCAAC-3′ |
| Templates | |
| 51AP | 3′-GCGTCGGCAGGTTGGTTGAGTXG CAGCTAGGTTACGGCAGGACTCAGGTCC-5′ |
| 51CTL | 3′-GCGTCGGCAGGTTGGTTGAGTAGCAGCTA GGTTACGGCAGGACTCAGGTCC-5′ |
| 63CTL | 3′-TTCGTATGGGTAGCGTCGGCAGGTTGGTT GAGTTGCAGCTAGGTTACGGCAGGACTCA GGTCC-5′ |
aX designates an AP site.
Protein purification
The gene encoding the full-length hPolη was cloned into the NdeI and XhoI sites of pET21B to generate a plasmid pET-21B-hPolη. The C-terminal His6-tagged hPolη was induced and expressed in Escherichia coli BL21(DE3) Rosetta cells at 16°C. The protein was purified through a nickel affinity column, a heparin sepharose column and a HiTrap SP column.
The gene encoding the N-terminal 420 amino acid residues of hPolι (hΔPolι) was inserted into the NcoI and XhoI sites of pGST-Parallel1 (28) to produce the plasmid pGST-iota. The N-terminal GST-tag on hΔPolι was used to increase the protein-purification yield as hΔPolι proved to be quite insoluble from cell lysate (data not shown). The GST-tagged hΔPolι was expressed in E. coli BL21(DE3) Rosetta cells at 16°C. The fusion protein in the cleared lysate was bound to a GSTrap column. After washing, the fusion protein on the column was digested with tobacco etch viral protease (TEV) at 4°C overnight. The free hΔPolι was then washed with binding buffer and collected. The pooled fractions were passed through a DEAE column to remove any DNA from E. coli. Finally, hΔPolι was separated from TEV and other impurities using a heparin sepharose column.
The gene encoding the truncated fragment (residues 341–829) of hRev1 (hΔRev1) was cloned into plasmid pBAD-REV1S (34–829) (29). The protein was induced with 1% of l-(+)-arabinose and expressed in E. coli BL21(DE3) at 15°C. The N-terminal His6-tagged hΔRev1 was purified using a nickel affinity column, a heparin sepharose column and a HiTrap Q column.
The gene encoding the truncated fragment (residues 9–518) of hPolκ (hΔPolκ) was inserted into the NcoI and XhoI sites of the plasmid pHIS-Parallel1 (28) to create pHIS-hPolkappa-9-518. This plasmid was transformed into E. coli BL21(DE3) Rosetta cells. The N-terminal His6-tagged hΔPolκ was induced and expressed at 19°C, and then was purified using a nickel affinity column, a heparin sepharose column, a HiTrap Q column and a Sephacryl 200 gel filtration column.
The concentrations of the purified recombinant proteins hPolη, hΔPolι, hΔRev1 and hΔPolκ were determined spectrophotometrically at 280 nm using calculated molar extinction coefficients of 70 731, 14 080, 32 430 and 3 1860 M−1cm−1, respectively. As both the N- and C-termini are exposed to solvent and far away from their active sites (30–32), the His6-tag on either end should not affect the activities of the hPolη, hΔPolκ and hΔRev1. All experiments were conducted using the same preparation of each enzyme in order to eliminate variations between experiments.
Reaction buffer
The reaction buffer H contained 50 mM HEPES (pH 7.5 at 37°C), 5 mM MgCl2, 50 mM NaCl, 5 mM DTT, 10% glycerol, 0.1 mM EDTA, and 0.1 mg ml−1 bovine serum albumin (BSA). All reactions were performed at 37°C.
Running start assay
Experiments were performed using a rapid chemical quench flow apparatus (KinTek) by rapidly mixing a solution containing 100 nM 5′-[32P] DNA (14-mer/51AP or 14-mer/51CTL, Table 1) and 100 nM of a human Y-family DNA polymerase preincubated in buffer H with a solution containing all four dNTPs (200 µM each) at 37°C for times ranging from milliseconds to minutes followed by quenching with 0.37 M EDTA. The nucleotide incorporation pattern was resolved by sequencing gel analysis.
Short oligonucleotide sequencing assay
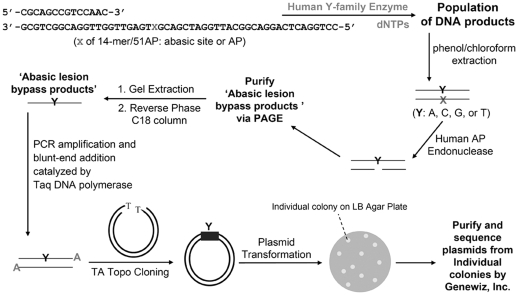
SOSA was performed as previously described (33) with the following modifications. The DNA substrate (14-mer/51AP, Table 1) contained the AP site analog tetrahydrofuran, which was located 22 nucleotides (nt) from the 3′-end of the DNA template. For control SOSA, the DNA substrate (14-mer/63CTL, Table 1) contained a dT in place of the AP site. The resulting full-length purified products were PCR amplified using the following primers: 16mer_AP_upstream, 5′-CACGCAGCCGTCCAAC-3′ and 16mer_AP_downstream, 5′-GCCCTGGACTCAGGAC-3′. The DNA plasmids containing the ligated full-length products were sequenced from bacterial colonies (Genewiz, Inc.). The method is summarized in Scheme 1.
Scheme 1.
Short oligonucleotide sequencing assay.
RESULTS
Our goal was to quantitatively elucidate the relative abilities of the four human Y-family DNA polymerases to replicate through an AP site. Previous studies have indicated that hPolη (6), hPolι (12–14), hPolκ (17) and hRev1 (18,19) are able to bypass an AP site in vitro. However, these in vitro studies have not quantitatively evaluated the AP site bypass efficiency of the four human Y-family enzymes. Moreover, there have been no studies reporting a comprehensive nucleotide incorporation profile for these human enzymes that encompasses nucleotide incorporation events not only opposite the AP site, but also upstream and downstream from this non-coding lesion. Such studies are important in that they will bring us one step closer to possibly determining which of the human Y-family DNA polymerases preferentially performs AP site bypass in vivo. To address these unresolved issues, running start assays were performed to determine the ability of each human Y-family enzyme to elongate a 14-mer/51AP substrate containing an AP site located 22 bases from the 3′-end of the DNA template 51AP (Table 1). The resulting nucleotide incorporation profiles were compared to those obtained with an undamaged DNA substrate 14-mer/51CTL (Table 1) in order to assess the effect of the AP site on DNA polymerase activity.
Running start assays
Recombinant hΔPolι, hΔPolκ and hΔRev1 were used, rather than their respective full-length proteins, in the running start assays because these fragments were able to be expressed and purified from E. coli as soluble and active proteins. Based on the domain structures of these three enzymes (34) and the published X-ray crystal structures of these three enzymes (31,32,35), the purified fragments contain the DNA polymerase core domains. Importantly, hΔRev1 has been shown to possess intact dCMP transferase activity (20) while hΔPolκ and hΔPolι are as active as their full-length counterparts (31,36).
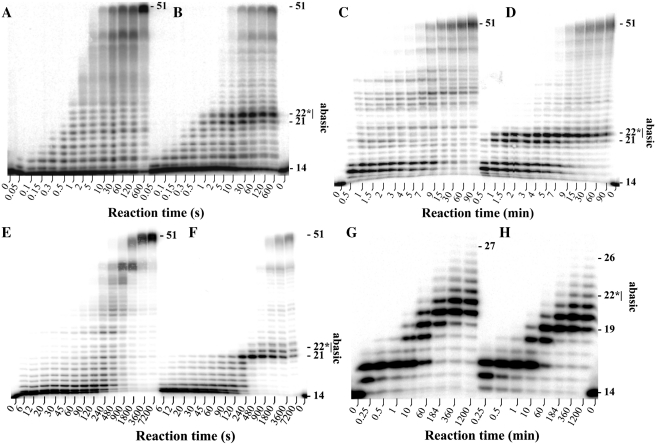
The running start assays for each of the four human Y-family DNA polymerases were performed under the same reaction conditions (see Materials and Methods). These reaction conditions did not deviate significantly from the reaction conditions for individual human Y-family DNA polymerases used in other laboratories (10,11,19,37,38), and therefore should not significantly affect the activity of each enzyme. Overall, all four human DNA polymerases are dissociative in the elongation of both the 5′-[32P]-labeled 14-mer/51AP and the 5′-[32P]-labeled 14-mer/51CTL DNA substrates based on intermediate product accumulation patterns (Figure 1). A general trend was observed in the nucleotide incorporation profile for each DNA polymerase with the exception of hΔRev1 (see below), in that the elongation of the 14-mer/51AP proceeded rapidly until the enzyme encountered the AP site where analysis of the nucleotide incorporation profile indicated two consecutive strong pause sites (Figure 1B, D and F). These consecutive pause sites corresponded to the nucleotide incorporation event directly opposite the AP site and the subsequent extension event, suggesting slow turnover at these sites when compared to the corresponding assays performed with the 14-mer/51CTL substrate (Figure 1A, C and E). Nonetheless, the AP site was bypassed relatively efficiently by hPolη with the full-length product observed at 10 s (Figure 1A and B). The subsequent downstream incorporation was perturbed more significantly than the insertion opposite the AP site, indicating that the AP site modestly affected the elongation of the lesion bypass product catalyzed by hPolη (Figure 1). Comparison of Figures 1D and 1B revealed that hΔPolι appeared to be more perturbed by the AP site than hPolη. There was a difference in the amount of time required for hΔPolι to generate the full-length product between the control (4 min, Figure 1C) and damaged (7 min, Figure 1D) DNA substrates. Analogous assays performed with hΔPolκ showed that this enzyme was also affected by the AP site as revealed by the elongation patterns of the damaged (Figure 1F) and the undamaged (Figure 1E) DNA substrates. Although the nucleotide incorporation patterns of hPolη, hΔPolι and hΔPolκ shared the same two strong pause sites in the vicinity of the AP site; hΔPolκ stalled for longer periods of time at these sites than hPolη and hPolι, generating the full-length product 30 min after reaction initiation (Figure 1F). Further analysis of the nucleotide incorporation profiles indicated that hPolη (Figure 1B) and hΔPolι (Figure 1D) were relatively more efficient at nucleotide incorporation opposite the AP site while hΔPolκ (Figure 1F) was more efficient at catalyzing the subsequent extension step.
Figure 1.
Running start assays for human Y-family DNA polymerases: (A) and (B) hPolη; (C) and (D) hΔPolι; (E) and (F) hΔPolκ; (G) and (H) hΔRev1. A preincubated solution of a Y-family DNA polymerase (100 nM) and 5′-[32P]-labeled DNA (100 nM) was mixed with all four dNTPs (200 µM each) for various reaction times before being quenched with 0.37 M EDTA. Reactions using the 14-mer/51CTL substrate are in (A), (C), (E) and (G) while reactions using the 14-mer/51AP substrate are in (B), (D), (F) and (H).
Notably, hΔRev1 was not able to generate the full-length product with either undamaged (Figure 1G) or damaged DNA substrates (Figure 1H), even after a 20-h incubation period at 37°C (data not shown). Additionally, this enzyme was only able to incorporate ∼13 nucleotides on undamaged DNA (Figure 1G), a weak activity that has been reported previously for hRev1 (39). Notably, hΔRev1 quickly catalyzed the first nucleotide incorporation, which was opposite template base dG, and stalled significantly at the next nucleotide incorporation opposite template base dT (Figure 1G and H). This observation was expected as hRev1 has been found to function as a dCMP transferase in vitro (19,20). The AP site did not significantly inhibit the primer elongation, although more accumulation of intermediate 19-mer was observed with the 14-mer/51AP substrate than with the 14-mer/51CTL substrate, indicating that the AP site had some effect on the activity of hΔRev1 (Figure 1H). Since hΔRev1 failed to generate the full-length ‘AP bypass products’ (Figure 1H), we could not quantitatively analyze its bypass specificity using SOSA. The inability to analyze the mutagenic spectrum for hΔRev1 should not diminish the veracity of our results (see Discussion).
Quantitative analysis of the AP bypass efficiencies
We further analyzed the results of the running start assays for hPolη, hΔPolι and hΔPolκ by quantitatively determining their bypass efficiencies (AP bypass%). This was calculated by determining the number of events at a given time t that the enzyme encountered the AP site relative to the number of these encounters that resulted in a lesion bypass event. Thus at reaction time t, an AP ‘encounter’ was defined as an event where the AP lesion was located in the enzyme active site, regardless of whether or not the nucleotide was incorporated. The total AP bypass events (B) was calculated from the concentration of all intermediate products with sizes greater than or equal to the 22-mer in Figure 1. Therefore, at the reaction time t, the total AP ‘encounter’ events (E) equaled the summation of the 21-mer concentration and the total AP bypass events (B). Finally, we defined the AP bypass% at reaction time t as the ratio of the bypass events to the encounter events:
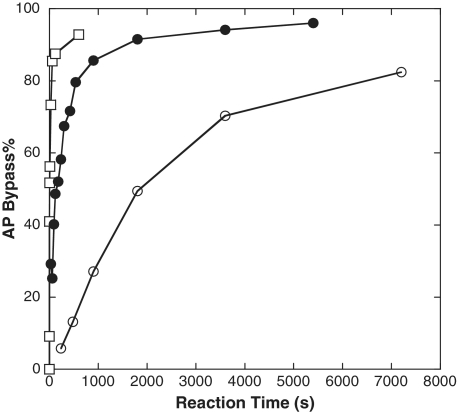
Figure 2 shows the AP bypass% plotted as a function of the reaction time for hPolη, hΔPolι and hΔPolκ. This figure demonstrates that hPolη required the shortest time to bypass the AP site while hΔPolκ required the longest time to traverse the lesion. To quantitatively define the AP bypass efficiency, we defined t50bypass as the time required to bypass 50% of the total AP lesions encountered. The t50bypass values, estimated from Figure 2, were 4.6, 112 and 1 823 s for hPolη, hΔPolι and hΔPolκ, respectively (Table 2). Since hΔRev1 only bypassed the AP lesion after 184, 360 and 1 200 min (Figure 1H) with bypass% values of 14, 20 and 28%, respectively, we did not plot the time-dependent AP bypass% for this enzyme. However, the t50bypass value of hΔRev1 was expected to be >1 200 min and was estimated to be ∼2 150 min based on these three time points. Based on the t50bypass values, hPolη possessed the highest AP bypass efficiency and bypassed an AP site 24-, 396- and >28 043-fold faster than hΔPolι, hΔPolκ and hΔRev1, respectively. The specific activity of the human enzymes to bypass an AP site can be described as the amount of AP site bypassed (nM) per second, and were calculated to be 1.1 × 101, 4.5 × 10−1, 2.7 × 10−2 and 3.9 × 10−4 nM s−1 for hPolη, hΔPolι, hΔPolκ and hΔRev1, respectively.
Figure 2.
Time-dependent AP bypass% during running start assays. The AP bypass% was plotted as a function of reaction time for hPolη (open square), hΔPolι (closed circle) and hΔPolκ (open circle).
Table 2.
The AP bypass efficiencies of the human Y-family DNA polymerases
For comparison, the time (t50) for each enzyme to create 50% of products that extended past a template base dT in the control template 63CTL, which is at the corresponding position of the AP site in the template 51AP, was estimated based on the running start analysis in Figure 1 and is listed in Table 2. The t50bypass/t50 ratios were calculated to evaluate the inhibitory effect of the AP site on DNA synthesis catalyzed by the Y-family DNA polymerases (Table 2). The t50bypass/t50 ratios indicate that the AP site slowed down hΔPolκ the most (87-fold) while it had almost no effect on hΔRev1. The inhibitory effect of an abasic site on hPolη (4.6-fold) and hΔPolι (1.5-fold) were small but notable.
Quantitative analysis of the mutation spectra of AP bypass
We performed SOSA to determine the precise sequences of AP lesion bypass products synthesized by each human Y-family DNA polymerase. Although the t50bypass values for each enzyme varied, our assays allowed adequate time for each enzyme to generate the full-length AP bypass products. Compared with our previously published SOSA (33), we modified the assay (Scheme 1) by increasing the size of the sequencing window from 8 to 22 nucleotides in order to increase the number of incorporation events both upstream (7 nucleotides) and downstream (15 nucleotides) from the AP site in addition to increasing the reliability of the statistical analysis.
hPolη
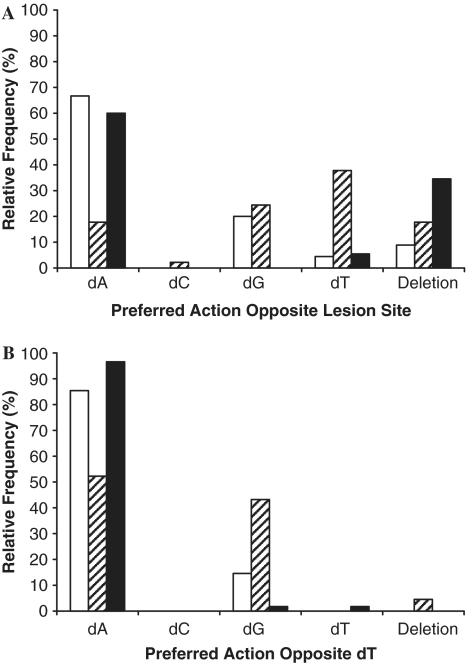
To perform SOSA with hPolη and 14-mer/51AP, we sequenced 45 colonies and the DNA sequences of the 45 AP lesion bypass products are summarized in Supplementary Figure S1A. Our results showed that hPolη incorporated dAMP (30/45 colonies, 66.7%), dGMP (9/45 colonies, 20%) or dTMP (2/45 colonies, 4.4%) opposite the AP lesion (Figure 3A). The preference for dAMP incorporation opposite the AP site was likely due to the well-established ‘A-rule’ (40). The remaining events opposite the AP site (4/45 colonies, 8.9%) were deletion mutations (Figure 3A). These dNMP incorporation frequencies were comparable to the error% measured by M13-based reverse mutation assays (41). In addition, hPolη generated a significant number of substitution, deletion and mixed mutations at positions located both upstream and downstream from the AP site.
Figure 3.
Comparison of preferred actions by human Y-family DNA polymerases opposite the AP site in the damaged template 51AP (A) or the corresponding template base dT in undamaged template 63CTL (B). The results from SOSA were tallied for all events at template Position 0 for hPolη (white bar), hΔPolι (striped bar) and hΔPolκ (black bar).
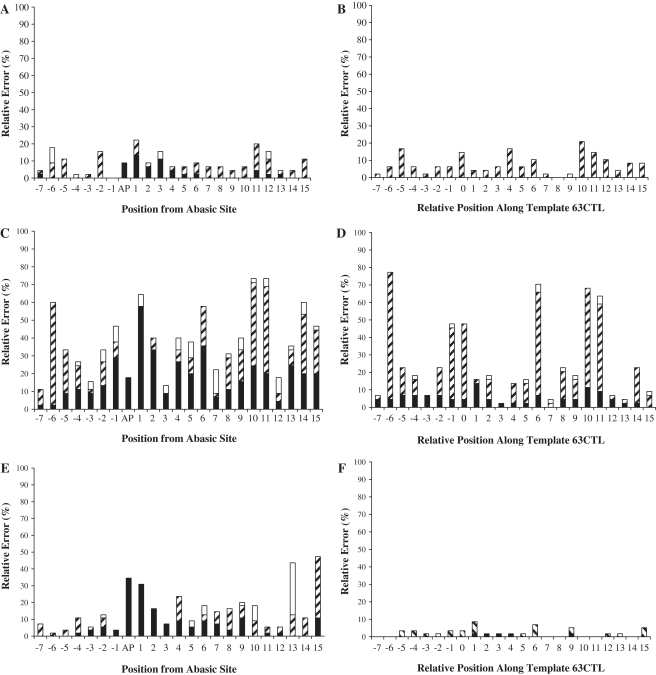
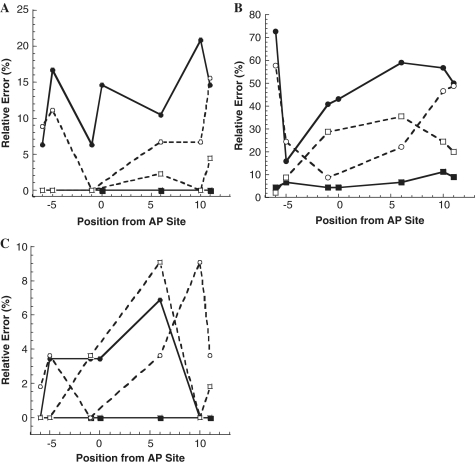
To quantitatively compare mutation frequencies at the AP site and other template positions, we plotted relative error% as a function of template positions (Figure 4A). Each value of relative error% was calculated from the ratio of total number of mutations (insertions, deletions and substitutions) divided by the total number of dNMP incorporations at a specific template position. To compare the contributions of different types of mutations to the value of relative error%, we further calculated the relative base insertion%, substitution% and deletion% at each template position. For calculations at the AP site, since there was no correct dNMP incorporation, relative base substitution% and insertion% were not calculated. In order to compare relative deletion% at the AP site and other template positions, we only presented relative deletion% at the AP site (Figure 4A). Interestingly, Figure 4A indicates that dNMP incorporation was most significantly affected at Position 1 (1 nucleotide downstream from AP site) as evidenced by the highest relative error (∼22.2%) in the sequencing window. Position 1 was also the strongest pause site in Figure 1B. Surprisingly, the most error-free incorporation occurred at the template base position preceding the AP site (Position -1, Figure 4A). To further examine the sequence-dependence of the relative error%, we plotted the relative error% versus the positions of template bases dTs (Positions -6, -5, -1, 0, 6, 10 and 11; Figure 5A). Obviously, the relative base deletion% increased after hPolη encountered the AP lesion. In contrast, the relative base substitution% was higher at positions further away from the AP site and dropped to 0% at Position -1. When opposite template bases dGs (Positions -7, -4, -2, 1, 4, 8, 9, 14 and 15), the opposite trends of relative base deletion% and substitution% were observed with hPolη (Supplementary Figure S2A). While the relative base substitution% trend for template bases dAs (Positions -3, 3, 7 and 12) was similar to the one observed in Figure 5A, the relative base deletion% increased only in the vicinity of the AP site (Supplementary Figure S3A). Notably, there were no upstream template bases dCs on 51AP. Based on these template base-dependent analyses, it is apparent that there is no clear pattern for relative error% during translesion DNA synthesis catalyzed by hPolη.
Figure 4.
Histogram of relative error% as a function of template position. At each position along the DNA template, the relative base insertion% (white bar), substitution% (striped bar) and deletion% (black bar) are shown to reveal total relative error% and the contribution of each type of mutation simultaneously. The AP site in 51AP is indicated as ‘AP’ along the X-axis, and the corresponding template base dT in 63CTL is denoted as ‘0’ along the X-axis. For values opposite the AP site, an error was scored only for those nucleotide incorporation events that resulted in a deletion. AP bypass analyses for hPolη (A), hΔPolι (C) and hΔPolκ (E) are shown. DNA synthesis with the control template 63CTL was also analyzed for hPolη (B), hΔPolι (D) and hΔPolκ (F).
Figure 5.
Relative error% as a function of template position from the AP site for hPolη- (A), hΔPolι- (B) and hΔPolκ- (C) catalyzed nucleotide incorporation events opposite template bases dTs. The plots show the relative error% for deletion (open square) and substitution mutations (open circle) as a function of the template position from the AP site. The relative error% for deletion (closed square) and substitution mutations (closed circle) as a function of the control template position in 63CTL is also shown for comparison. Only incorporations opposite template base dTs were analyzed.
For further evaluation of the effect of the AP site on upstream and downstream nucleotide incorporations catalyzed by hPolη, we performed SOSA using a control DNA substrate 14-mer/63CTL which has a template base dT at the corresponding position of the AP site in 14-mer/51AP (Table 1). The control template 63CTL is 12 nucleotides longer than 51AP in order to facilitate PAGE separation of the full-length products from the control template which cannot be cleaved by the human AP endonuclease (Scheme 1). Based on the sequences gathered (Supplementary Figure S1B), hPolη incorporated mostly correct dAMP (85.4%) and occasionally misincorporated dGMP (14.6%) opposite a template base dT at Position 0 (Figure 3B). However, the mutations made around Position 0 changed from mostly multi-base deletions and single-base substitutions of dT and dA in the presence of the AP site to mostly single-base substitutions of dG and dT in the absence of the AP site (Supplementary Figures S1A and B). As shown in Figure 4B, hPolη created mutations along template 63CTL with an average relative error% of ∼6.7% at each template position. This value is similar to the average error% in Figure 4A. However, the only observed mutations that occurred with template 63CTL were base substitutions (Figure 4B and Supplementary Figure S1B). The relative base substitution error% with template bases dTs (6–21%) was sequence dependent (Figure 5A). Similar trends were also observed with template dAs and dGs (Supplementary Figures 2A and 3A). Overall, the base substitution frequency of hPolη was calculated to be 7.5 × 10−2 with 14-mer/63CTL (Table 3). This value was comparable to the error rate of 7.1 × 10−2 for combined dT:dG and dA:dC mutations measured by the M13-based forward mutation assays (42) and to 5.6 × 10−2 measured by steady-state kinetic studies (43). Interestingly, differences in the mutagenic data in Figure 4A and B suggest that the base deletions and insertions observed with template 51AP were most likely caused by the presence of the AP site. Both 2.2% deletion error frequency at Position -7 and 8.9% insertion-error frequency at Position -6 (Figure 4A) may also suggest that hPolη could detect the presence of the AP site before the lesion entered its active site.
Table 3.
Error rates of the four human Y-family DNA polymerases
| Enzyme | DNA | Event | Insertion Errora | Insertion Error Ratiob | Deletion Errorc | Deletion Error Ratiod | Substitution Errore | Substitution Error Ratiof |
|---|---|---|---|---|---|---|---|---|
| hPolη | 14/63CTL | Totalg | 0 | N/A | 0 | N/A | 7.5 × 10−2 | N/A |
| Upstreamh | 0 | N/A | 0 | N/A | 6.5 × 10−2 | N/A | ||
| Downstreami | 0 | N/A | 0 | N/A | 7.9 × 10−2 | N/A | ||
| hPolη | 14/51AP | Totalg | 4.4 × 10−3 | – | 2.6 × 10−2 | – | 6.4 × 10−2 | 0.85 |
| Upstreamh | 0 | – | 3.5 × 10−3 | – | 5.9 × 10−2 | 0.91 | ||
| Downstreami | 6.5 × 10−3 | – | 3.6 × 10−2 | – | 6.7 × 10−2 | 0.85 | ||
| hΔPolι | 14/63CTL | Totalg | 1.2 × 10−2 | N/A | 5.1 × 10−2 | N/A | 1.9 × 10−1 | N/A |
| Upstreamh | 6.5 × 10−3 | N/A | 5.8 × 10−2 | N/A | 2.2 × 10−1 | N/A | ||
| Downstreami | 1.5 × 10−2 | N/A | 4.7 × 10−2 | N/A | 1.8 × 10−1 | N/A | ||
| hΔPolι | 14/51AP | Totalg | 7.0 × 10−2 | 5.8 | 1.8 × 10−1 | 3.5 | 1.7 × 10−1 | 0.89 |
| Upstreamh | 4.1 × 10−2 | 6.3 | 1.1 × 10−1 | 1.9 | 1.8 × 10−1 | 0.82 | ||
| Downstreami | 8.3 × 10−2 | 5.5 | 2.2 × 10−1 | 4.7 | 1.7 × 10−1 | 0.94 | ||
| hΔPolκ | 14/63CTL | Totalg | 0 | N/A | 3.9 × 10−3 | N/A | 1.9 × 10−2 | N/A |
| Upstreamh | 0 | N/A | 0 | N/A | 2.0 × 10−2 | N/A | ||
| Downstreami | 0 | N/A | 5.7 × 10−3 | N/A | 1.8 × 10−2 | N/A | ||
| hΔPolκ | 14/51AP | Totalg | 3.2 × 10−3 | – | 5.4 × 10−2 | 14 | 6.3 × 10−2 | 3.3 |
| Upstreamh | 0 | – | 1.5 × 10−2 | – | 3.1 × 10−2 | 1.6 | ||
| Downstreami | 4.8 × 10−3 | – | 7.1 × 10−2 | 12 | 7.9 × 10−2 | 4.4 |
‘–’ means the ratio cannot be calculated because the denominator is 0. ‘N/A’ means not applicable.
aCalculated as Σ(base insertions)/[(sample size) × (number of bases in a specific event)].
bCalculated as {Σ(base insertions)/[(sample size) × (number of bases in a specific event)]}51AP/{Σ(base insertions)/[(sample size) × (number of bases in a specific event)]}63CTL.
cCalculated as Σ(base deletions)/[(sample size) × (number of bases in a specific event)].
dCalculated as {Σ(base deletions)/[(sample size) × (number of bases in a specific event)]}51AP/{Σ(base deletions)/[(sample size) × (number of bases in a specific event)]}63CTL.
eCalculated as Σ(base substitutions)/[(number of samples) × (number of bases in a specific event)].
fCalculated as {Σ(base substitutions)/[(sample size) × (number of bases in a specific event)]}51AP/{Σ(base insertion)/[(sample size) × (number of bases in a specific event)]}63CTL.
gTotal events include all dNMP incorporation events during DNA synthesis of full-length products except those that occurred at Position 0 in Figure 4.
hUpstream events include all dNMP incorporation events during DNA synthesis of full-length products that occurred before an enzyme encountered Position 0 or ‘AP’ in Figure 4.
iDownstream events include all dNMP incorporation events during DNA synthesis of full-length products that occurred after an enzyme traversed Position 0 or ‘AP’ in Figure 4.
hΔPolι
To quantitatively analyze the mutagenic profiles of AP bypass catalyzed by hΔPolι, we sequenced 45 colonies and the SOSA results are shown in Supplementary Figure S1C. Interestingly, hΔPolι lacked strong preference when incorporating a dNMP directly opposite the AP site as indicated by the relative nucleotide incorporation percentages for dAMP (8/45 colonies, 17.8%), dGMP (11/45 colonies, 24.4%), dCMP (1/45 colonies, 2.2%) and dTMP (17/45 colonies, 37.8%) in addition to the deletion events (8/45 colonies, 17.8%; Figure 3A). Thus, hΔPolι did not follow the ‘A-rule’ at the AP site. These SOSA results were similar to other published kinetic studies (14,15) with only one difference: they found that dGMP incorporation opposite an AP site is slightly more efficient than dTMP. In addition, our SOSA assay also identified base deletions opposite the AP site (Figure 3A). At positions both upstream and downstream of the AP lesion, hΔPolι generated a plethora of base substitution, deletion and insertion mutations that varied from 10 to 80% of the total flux opposite each template position in the sequencing window (Figure 4C). There were more mutagenic events opposite template bases dTs (Figure 5B) than opposite template bases dAs (Supplementary Figure S3B). As reported previously (11,44), we also observed the unusual ability of hΔPolι to efficiently incorporate dGMP opposite dT at Position -6 where 58% (26/45 colonies) of the total dNMP incorporation events were dGMP substitution mutations while only 40% involved correct dAMP incorporation (Figure 4C). Perhaps most intriguingly, as hΔPolι approached the AP site, there was a sequence-dependent increase in the number of deletion mutations, and a corresponding sequence-dependent decrease in deletion mutations as the polymerase proceeded downstream from the AP site with a maximum deletion mutation% (26/45 colonies, 58%) observed at Position 1 (Figures 4C and 5B). This trend was inversely correlated to the number of substitution mutations generated by hΔPolι. Additionally, Positions 0 and 1 were also the strongest pause sites in Figure 1D. Notably, the results for hΔPolι indicated an extraordinarily high probability of generating frameshift mutations during AP bypass with a probability of 13% (6/45 colonies) to 58% (26/45 colonies) of the total dNMP incorporation events catalyzed within two template bases of the AP site (Figure 4C).
The effects of the AP site on DNA synthesis catalyzed by hΔPolι were more evident when comparing the results in Supplementary Figure S1C with the results for the control template 63CTL (Supplementary Figure 1D). While mostly multi-base deletions were observed in the immediate area of the AP site, hΔPolι created mostly multi-base substitutions with dGMP being the most frequently incorporated incorrect nucleotide on control template 63CTL. Opposite template bases dTs, the inversely correlated, sequence-dependent variation observed with template 51AP was not observed with template 63CTL (Figure 5B). The relative error% for base deletions was lower than base substitutions along template 63CTL and displayed no sequence bias. Similar trends are also observed upon comparison of incorporation events opposite template bases dAs and dGs (Supplementary Figures 2B and 3B), although the overall error rate of hΔPolι opposite dA was relatively low. While relative error frequencies at specific positions on template 63CTL were as high as 76% (Figure 4D), hΔPolι synthesized one completely error-free product out of 44 sequenced products (Supplementary Figure S1D). Among the 44 dNMP incorporation events opposite template base dT at Position 0, hΔPolι made two base deletions and incorporated dAMP, dGMP, dTMP and dCMP 23, 19, 0 and 0 times, respectively (Figure 3B). Again, the data demonstrated that hΔPolι formed the base pairs of dG:dT and dA:dT with similar efficiency (11,44). Overall, the base insertion, substitution, and deletion frequencies with undamaged template 63CTL were calculated to be 1.2 × 10−2, 1.9 × 10−1 and 5.1 × 10−2, respectively (Table 3). The base substitution error rate is similar to 1.0 × 10−1 measured by steady-state kinetic studies (45). When comparing the relative error% along the templates 63CTL (Figure 4D) and 51AP (Figure 4C), the relative error% for hΔPolι in the presence of an AP site increased at almost every template position.
hΔPolκ
We sequenced 55 AP lesion bypass products formed by hΔPolκ and the SOSA results are shown in Supplementary Figure S1E. Opposite the AP lesion (Figure 3A), 60% (33/55 colonies) of the nucleotide incorporation events were dAMP incorporations, 8.5% (3/55 colonies) were dTMP incorporations, 34.5% (19/55 colonies) were no incorporations and 0% (0/55 colonies) were dCMP and dGMP incorporations, suggesting that hΔPolκ, like hPolη, followed the ‘A-rule’ to select an incoming dNMP opposite the AP site. This conclusion is consistent with previously published results derived from single dNMP incorporation assays (17). The strong pause sites observed in Figure 1F were the same positions that created the most deletion mutations shown in Figure 4E. Notably, there were a significantly higher number of substitution and frameshift mutations at template positions downstream from the AP lesion (Figure 4E), which was not observed with hPolη (Figure 4A) and hΔPolι (Figure 4C). When following the dTs along template 51AP, the inversely correlated relative base substitution% and deletion% observed with hΔPolι was also observed with hΔPolκ (Figure 5C). Opposite template bases dGs and dAs, hΔPolι followed similar patterns of relative base substitution% and deletion% (Supplementary Figures S2C and 3C).
With undamaged 14-mer/63CTL (Table 1), our SOSA data (Supplementary Figure S1F) indicated that hΔPolκ made significantly less errors with control 14-mer/51CTL (Figure 4F) than with damaged 14-mer/51AP (Figure 4E). Although hΔPolκ created multi-base deletions in the immediate vicinity of the AP site, mostly single-base substitutions were observed when the AP site was replaced with base dT (Supplementary Figure S1F). For the 61 sequenced full-length products in Supplementary Figure S1F, the base insertion, substitution and deletion error rates were calculated to be 0, 1.9 × 10−2 and 3.9 × 10−3, respectively (Table 3), and the base substitution error rate was similar to 1.4 × 10−2 measured by steady-state kinetic studies (46). Among the 61 dNMP incorporation events opposite dT at Position 0, hΔPolκ only misincorporated one dGMP and one dTMP and did not make any other mutations (Figure 3B). Opposite template bases dTs in 63CTL, there were no base deletions and few base substitutions (Figure 5C). Similar trends were also observed when hΔPolκ incorporated dNMPs opposite template bases dGs and dAs (Supplementary Figures S2C and 3C). When comparing Figure 4E and F, it was clear that the relative error% at each template position increased significantly in the presence of an AP site.
All Y-family human enzymes
Within the distinct characteristic fidelity of each of the three human Y-family enzymes, there were mutational hotspots observed for both templates 51AP and 63CTL. All three DNA polymerases created a substantial amount of mutations at Positions 0 and 1 in the presence of the AP site with deletion mutations having the highest frequency (Figure 4). Furthermore, these positions created the strongest pause sites during running start analysis (Figure 1). There were also common distal positions downstream from the AP site (Positions 10 and 11) where these enzymes created mostly base substitutions. At Positions 10 and 11, the most frequent mutation was a base substitution to either dT or dG for all enzymes (Supplementary Figure S1). Similarly, both hPolη and hΔPolι displayed high frequencies of mutations at Positions -6 and -5 on both templates 51AP and 63CTL. At these positions where slight enzyme pauses occurred during running start analysis (Figure 1), these two enzymes created mostly a base substitution to dG (Supplementary Figure S1). Thus, the slight pause sites were directly correlated to mutational hotspots observed by SOSA. Notably, while downstream nucleotide incorporation events were not significantly affected, the observation of increased deletion and insertion mutations upstream from the AP site may be suggestive of a mechanism by which hPolη was affected by the AP site several nucleotides prior to encountering the lesion in its active site (Figure 4A). This upstream AP effect was also observed with both hΔPolι and hΔPolκ (Figure 4). In addition, the AP lesion affected downstream nucleotide incorporation catalyzed by hΔPolι and hΔPolκ (Figure 4).
DISCUSSION
A mammalian genome is estimated to undergo ∼100 000 modifications per day (47). It is obvious that the four human Y-family DNA polymerases have to bypass different types of DNA lesions efficiently in order to rescue and facilitate DNA replication. While S. solfataricus Dpo4 has served as an excellent model for characterizing the Y-family, it has become apparent that DNA polymerases even within the same family possess structural and functional attributes that render them fundamentally different from one another (48). Thus, it is extremely likely that the Y-family DNA polymerases, especially those from the same organism, possess distinct lesion bypass specificities in vivo. We hypothesized that one Y-family DNA polymerase may play a dominant role on the insertion or extension bypass of a specific lesion while other DNA polymerases may play a secondary role. In order to maintain genetic stability, we further hypothesized that the in vivo bypass of a widespread lesion such as the AP site requires a relatively efficient and faithful Y-family DNA polymerase that causes few frameshift mutations. In this article, we quantitatively analyzed all four human Y-family enzymes for their abilities to perform AP bypass. The resulting analyses are discussed herein.
Human Rev1 is not the DNA polymerase to bypass AP lesions in vivo
For the following reasons, Rev1 does not appear to play an important DNA polymerase role in AP lesion bypass in vivo. First, previous studies have shown hRev1 to function as a dCMP transferase that uses a novel mechanism involving the side chain of Arg357 in the N-digit domain that specifically interacts with the base of dCTP through hydrogen bonds to direct dCMP incorporation (32). This property is supported by our recently published kinetic studies on hΔRev1 which preferentially incorporated dCMP opposite all four possible template bases (49). This low fidelity of hRev1 could severely compromise genomic stability. Second, among the four human Y-family DNA polymerases, hΔRev1 possesses the lowest AP bypass efficiency (see Results) and may not be able to timely bypass numerous AP sites generated per cell per day (25,26). Third, due to the ability of Rev1 to specifically interact with DNA polymerases η, κ and ι, but not with other DNA polymerases such as β or µ (50–52) coupled with the observation that Rev1 will switch its interaction partner based on the relative concentrations of the competing Y-family DNA polymerases (50), it has recently been hypothesized that Rev1 may have an important role in mediating the DNA polymerase switching process by serving as a protein scaffold. Finally, additional evidence for the scaffold role of Rev1 were provided by demonstrating that Rev1 is required for DNA-damage induced mutagenesis in yeast, but its dCMP transferase activity is not required for AP site bypass in vivo (53).
hPolη possesses the highest AP bypass efficiency in vitro
Using similar running start analysis performed previously for Dpo4 (54), our data demonstrated that hPolη, hΔPolι and hΔPolκ were able to bypass a site-specifically placed AP site and subsequently generate the full-length bypass product. Based on the t50bypass values for hPolη, hΔPolι and hΔPolκ estimated in Figure 2, hPolη possessed the highest AP bypass efficiency and bypassed an AP site 24- and 396-fold faster than hΔPolι and hΔPolκ, respectively. Relative to the control DNA substrate, the presence of an AP site had only a slight inhibitory effect on the polymerase activity of hPolη and hΔPolι but significantly slowed the hΔPolκ-catalyzed DNA synthesis. Moreover, further analysis of the nucleotide incorporation profile indicated that hPolη and hΔPolι were relatively more efficient at nucleotide incorporation opposite the AP site while hΔPolκ was more efficient at catalyzing the subsequent extension step. Such observations agree with previous reports that Polκ has a unique ability to extend mispaired and aberrant primer termini (16,55). This superior extension ability can be explained by the presence of a unique N-terminal extension of the palm domain, which helps stabilize the Polκ•DNA complex at the primer–terminus junction (31).
hPolη has a lower mutagenic potential than hΔPolι and hΔPolκ during AP bypass
Analysis of the mutation spectra of AP bypass observed via SOSA revealed several general trends: (i) opposite the AP site, all three enzymes made frameshift mutations with the following order of deletion frequency: hPolη < hΔPolι < hΔPolκ (Figure 3A); (ii) at template positions other than Position 0 (Table 3), all three human Y-family enzymes had higher deletion and insertion frequencies with the damaged template 51AP than with the control template 63CTL (Figure 4); (iii) the base deletion and insertion frequencies of each of the three human Y-family enzymes increased significantly after the enzyme encountered the AP site (Table 3); (iv) with both templates 51AP and 63CTL, hΔPolι made more insertion, deletion and substitution mutations than hPolη and hΔPolκ (Table 3); and (v) hPolη was the least likely human Y-family polymerase to generate frameshift mutations in the vicinity of the AP site.
Sensing of a lesion by DNA polymerases
Figure 3A shows that hPolη preferentially incorporated dAMP opposite the AP site, a preference that has been observed previously in some mammalian cells lines (48,50,51). Additionally, while downstream nucleotide incorporation events were not significantly affected, the observation of increased deletion and insertion mutations upstream from the AP site (Figure 4A) may be suggestive of a mechanism by which hPolη is affected by the AP site several bases prior to encountering the lesion within its active site. This observation was also made for AP translesion DNA synthesis catalyzed by both hΔPolι (Figure 4C) and hΔPolκ (Figure 4E). This ability to sense DNA lesions has been reported previously for Pyrococcus furiosus (Pfu) DNA polymerase, which has been shown to sense uracil in a DNA template at least four bases upstream from the lesion (56). In addition, S. solfataricus DNA polymerase B1 has been shown to be affected by uracil and hypoxanthine four bases upstream of these lesions, although the inhibitory effect is not as substantial as that reported for Pfu DNA polymerase (56,57). How a polymerase senses a DNA lesion is unclear. Based on crystal structures of eukaryotic Y-family DNA polymerases (32,58–60), the 5′ single-strand portion of the DNA template from the primer and template junction is threaded through the interface between the finger domain and polymerase-associated domain (PAD), or little finger domain, and interacts with amino acid residues in the interface. The presence of a damaged template base may affect these interactions, facilitating the polymerase to sense the upstream lesion and make more mutations.
Potential deletion mechanism of hPolι
The results for hΔPolι indicated an extraordinarily high probability of generating frameshift mutations during AP bypass (Figure 4C). Interestingly, a recent crystal structure of hPolι with an AP-containing DNA substrate indicates that the protrusion of the AP site would be sterically forbidden in hPolι by the presence of Lys60 and Glu97 of the finger domain and Ser307 of the PAD (61). Thus, it is possible that hΔPolι utilizes a different AP site bypass mechanism, such as a primer misalignment, to generate the large number of frameshift mutations observed within the sequencing window. In addition, the inactivation of hPolη, i.e. XPV cell lines, is known to have the phenotype of hypermutability after exposure to DNA-damaging conditions (62). The hypermutability that was observed for hΔPolι (Figure 4C), which was the second most efficient enzyme to bypass an AP site in vitro, suggests that hPolι may substitute for the defective hPolη during the bypass of AP sites in the XPV cell lines (63).
Impact of an AP lesion on hΔPolκ
hΔPolκ, which is shown to generate -1 deletions even on undamaged DNA (64,65), was intermediate to hPolη and hΔPolι, generating a moderate number of base deletion mutations immediately downstream from the AP site in a manner similar to that reported for its Y-family homolog Dpo4 (55). Furthermore, the embedded AP site affected the accuracy of dNMP incorporation catalyzed by hΔPolκ for at least 15 bases downstream from the AP site (Figure 4E), which was similar to the pre-steady state kinetic data reported for Dpo4 (54). Together, these data indicate that the mutagenic effects of an AP site on DNA synthesis catalyzed by Polκ and Dpo4 are similar.
Potential role of the little finger domain on a Y-family enzyme’s lesion bypass abilities
The abovementioned differences in the AP bypass efficiency and mutagenic frequency among human Y-family DNA polymerases are a fundamental function of the ability of the enzymes to incorporate a dNMP directly opposite the AP site. This ability has been shown to be influenced by the PAD or little finger domain, which can modulate the activity of the enzyme by modifying the relative position of the DNA within the polymerase active site (66,67). As a proof of principle, the bypass specificity has been shown to be significantly influenced by the little finger domain in a study that swapped this domain in two Y-family polymerases and observed a switch of the bypass specificities (68). Thus, our data collectively demonstrate the functional differences between DNA polymerases from the same phylogenetic family.
In summary, our study serves as an initial attempt to understand the precise mutagenic profile of each of the four human Y-family polymerases in response to an AP site. The data presented suggest that the most likely Y-family polymerase to bypass an AP site in vivo is hPolη, due to (i) the extremely high frameshift potential of hΔPolι, (ii) the higher potential of hΔPolκ to create frameshift mutations as opposed to directly incorporating dNMP at the AP site, (iii) the weak polymerase activity observed opposite the AP site by hΔRev1 coupled with its reported preference to function as a scaffolding protein and (iv) the low frameshift potential and extremely high AP bypass efficiency of hPolη. Future studies with a variety of different DNA lesions are forthcoming, and will certainly provide a plethora of useful information regarding lesion bypass specificity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation Grant (MCB-0960961 to Z.S.); National Institute of Health Grant (GM079403 to Z.S.); American Heart Association Great River Affiliate Predoctoral Fellowship Grant (GRT00014861 to S.M.S.); Presidential Fellowship from Ohio State University (to K.A.F.). Funding for open access charge: National Science Foundation Grant (MCB-0960961); National Institute of Health Grant (GM079403).
Conflict of interest statement. None declared.
REFERENCES
- 1.Boudsocq F, Ling H, Yang W, Woodgate R. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair (Amst) 2002;1:343–358. doi: 10.1016/s1568-7864(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 2.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann AR. Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat. Res. 2002;509:23–34. doi: 10.1016/s0027-5107(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 5.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 8.Levine RL, Miller H, Grollman A, Ohashi E, Ohmori H, Masutani C, Hanaoka F, Moriya M. Translesion DNA synthesis catalyzed by human pol eta and pol kappa across 1,N6-ethenodeoxyadenosine. J. Biol. Chem. 2001;276:18717–18721. doi: 10.1074/jbc.M102158200. [DOI] [PubMed] [Google Scholar]
- 9.Haracska L, Prakash S, Prakash L. Replication past O(6)-methylguanine by yeast and human DNA polymerase eta. Mol. Cell. Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol. Cell. Biol. 2000;20:7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc. Natl Acad. Sci. USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 16.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc. Natl Acad. Sci. USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 19.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J. Biol. Chem. 2001;276:15051–15058. doi: 10.1074/jbc.M008082200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor JS, Wang Z. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 2002;30:1630–1638. doi: 10.1093/nar/30.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 23.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 24.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase eta. Proc. Natl Acad. Sci. USA. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindahl T, Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleoic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 27.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 28.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 29.Masuda Y, Kamiya K. Role of single-stranded DNA in targeting REV1 to primer termini. J. Biol. Chem. 2006;281:24314–24321. doi: 10.1074/jbc.M602967200. [DOI] [PubMed] [Google Scholar]
- 30.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol. Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 31.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 32.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J. Mol. Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiala KA, Suo Z. Sloppy bypass of an abasic lesion catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 2007;282:8199–8206. doi: 10.1074/jbc.M610719200. [DOI] [PubMed] [Google Scholar]
- 34.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 35.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa T, Uematsu N, Mizukoshi T, Iwai S, Iwasaki H, Masutani C, Hanaoka F, Ueda R, Ohmori H, Todo T. Mutagenic and nonmutagenic bypass of DNA lesions by Drosophila DNA polymerases dpoleta and dpoliota. J. Biol. Chem. 2001;276:15155–15163. doi: 10.1074/jbc.M009822200. [DOI] [PubMed] [Google Scholar]
- 37.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 38.Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol kappa, a DNA polymerase encoded by the human DINB1 gene. J. Biol. Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- 39.Masuda Y, Kamiya K. Biochemical properties of the human REV1 protein. FEBS Lett. 2002;520:88–92. doi: 10.1016/s0014-5793(02)02773-4. [DOI] [PubMed] [Google Scholar]
- 40.Strauss B, Rabkin S, Sagher D, Moore P. The role of DNA polymerase in base substitution mutagenesis on non-instructional templates. Biochimie. 1982;64:829–838. doi: 10.1016/s0300-9084(82)80138-7. [DOI] [PubMed] [Google Scholar]
- 41.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 42.Glick E, Chau JS, Vigna KL, McCulloch SD, Adman ET, Kunkel TA, Loeb LA. Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase eta. J. Biol. Chem. 2003;278:19341–19346. doi: 10.1074/jbc.M300686200. [DOI] [PubMed] [Google Scholar]
- 43.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 44.Washington MT, Johnson RE, Prakash L, Prakash S. Human DNA polymerase iota utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol. Cell. Biol. 2004;24:936–943. doi: 10.1128/MCB.24.2.936-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson RE, Prakash L, Prakash S. Biochemical evidence for the requirement of Hoogsteen base pairing for replication by human DNA polymerase iota. Proc. Natl Acad. Sci. USA. 2005;102:10466–10471. doi: 10.1073/pnas.0503859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000;28:4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 48.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown JA, Fowler JD, Suo Z. Kinetic basis of nucleotide selection employed by a protein template-dependent DNA polymerase. Biochemistry. 49:5504–5510. doi: 10.1021/bi100433x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. Embo. J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 52.Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 53.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 2007;282:8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 55.Wolfle WT, Washington MT, Prakash L, Prakash S. Human DNA polymerase kappa uses template-primer misalignment as a novel means for extending mispaired termini and for generating single-base deletions. Genes Dev. 2003;17:2191–2199. doi: 10.1101/gad.1108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 57.Gruz P, Shimizu M, Pisani FM, De Felice M, Kanke Y, Nohmi T. Processing of DNA lesions by archaeal DNA polymerases from Sulfolobus solfataricus. Nucleic Acids Res. 2003;31:4024–4030. doi: 10.1093/nar/gkg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 59.Vasquez-Del Carpio R, Silverstein TD, Lone S, Swan MK, Choudhury JR, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Structure of human DNA polymerase kappa inserting dATP opposite an 8-OxoG DNA lesion. PLoS ONE. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain R, Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Replication across template T/U by human DNA polymerase-iota. Structure. 2009;17:974–980. doi: 10.1016/j.str.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA Synthesis across an Abasic Lesion by Human DNA Polymerase iota. Structure. 2009;17:530–537. doi: 10.1016/j.str.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuasa M, Masutani C, Eki T, Hanaoka F. Genomic structure, chromosomal localization and identification of mutations in the xeroderma pigmentosum variant (XPV) gene. Oncogene. 2000;19:4721–4728. doi: 10.1038/sj.onc.1203842. [DOI] [PubMed] [Google Scholar]
- 63.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 65.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 66.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 67.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.