Abstract
Soluble Xenopus egg extracts efficiently replicate added plasmids using a physiological mechanism, and thus represent a powerful system to understand vertebrate DNA replication. Surprisingly, DNA replication in this system is highly sensitive to plasmid concentration, being undetectable below ∼10 pM and highly efficient above ∼75 pM. DNA replication at the high plasmid concentration does not require plasmid–plasmid contacts, since replication is not inhibited when plasmids are immobilized in agarose prior to addition of egg extract. The absence of replication at low plasmid concentration is due to a defect in the assembly of pre-replication complexes (pre-RCs). pre-RC assembly requires contact-independent communication between plasmids. Our results show that in Xenopus egg extracts, aggregation of multiple replication forks is not required for efficient replication of plasmid DNA, and they suggest that DNA functions as a co-factor for its own duplication.
INTRODUCTION
Eukaryotes duplicate their chromosomes by initiating DNA synthesis at hundreds to thousands of origins, each of which produces two sister replisomes. The activity of these origins is coordinated to ensure efficient replication during the limited time allotted in S-phase. Work over the past two decades has identified many replication proteins, which function in an ordered, multi-step pathway for highly processive and faithful chromosome duplication (1,2). In the first step, pre-Replicative Complexes (pre-RCs) assemble on chromatin in the G1 phase of the cell cycle via the sequential binding of origin recognition complex (ORC), Cdc6, Cdt1, the MCM2-7 helicase and possibly other factors. Loaded MCM2-7 has the potential to unwind DNA and initiate replication and therefore corresponds to the establishment of a ‘license’ for initiation. In S-phase, the protein kinases Cdk2-Cyclin E and Cdc7–Dbf4 promote loading of GINS, Cdc45 and other proteins onto licensed origins, resulting in helicase activation, origin unwinding, and replisome assembly (‘initiation’). During S-phase, multiple mechanisms block de novo licensing to prevent re-replication (3–5). For example, Cdt1 is inhibited by binding to Geminin and by ubiquitin-mediated proteolysis.
A widespread feature of eukaryotic DNA replication is the localization of DNA synthesis to discrete subnuclear domains called replication factories or foci, where tens to hundreds of replisomes copy DNA simultaneously (6–10). It has been proposed that cells cluster replisomes into foci to promote synchronous and rapid DNA synthesis (10–14). However, replication foci formation has not been disrupted in any experimental system, and what role foci play in DNA replication remains unknown.
Extracts from Xenopus laevis eggs have been used extensively to explore the mechanism of eukaryotic DNA replication, including the composition and dynamics of foci (11,15). Addition of high-molecular weight DNA templates such as sperm chromatin to these extracts leads to the formation of synthetic nuclei, which undergo a single round of DNA replication (16–21). Roughly 200 foci assemble per nucleus, with each focus containing 300–1000 replication forks (17). As seen in mammalian cells, the firing of neighboring origins appears to be coordinated (22,23), suggesting that foci represent functional units of DNA replication in these synthetic nuclei. We previously developed a variation of Xenopus egg extracts in which DNA replication occurs in soluble protein extracts that do not support nuclear envelope formation (24). In this system, simple DNA templates such as circular plasmids undergo efficient replication. It is unknown whether plasmids aggregate to form foci-like structures in this nucleus-free system. Thus, it is presently unclear whether clustering of replisomes into foci is required for replication in soluble Xenopus egg extracts.
Our goal was to determine whether replisome clustering is required for DNA replication in nucleus-free Xenopus egg extracts. We reasoned that reducing the concentration of plasmid substrate might disrupt replisome clustering and replication. Indeed, while plasmid DNA replicated efficiently at high concentration as reported previously (24), DNA synthesis was undetectable at low plasmid concentration. To test directly whether multiple plasmids must come into close contact to undergo replication, we immobilized a high concentration of plasmid in agarose blocks to prevent inter-plasmid interactions. Under these conditions, replication was still 100% efficient, demonstrating that plasmid–plasmid contacts are unnecessary for DNA replication in soluble Xenopus egg extracts. We further show that a high plasmid concentration is required for replication licensing and initiation. In the case of licensing, our data indicate that this event requires positive, contact-independent communication between plasmids over short-ranges. Together, our results demonstrate that efficient replication of plasmid DNA can occur in Xenopus egg extracts in the absence of replisome clustering, and they suggest that DNA acts as a co-factor for its own duplication.
MATERIALS AND METHODS
DNA templates
p2.9 refers to pBluescript II KS(–). p6.6 [pP(whiteout2)], p10.4 (pUC8ySG), and p21.7 (Homing pigeon) were all generously provided by Welcome Bender. pP(whiteout2) is a Drosophila P element transformation vector (from Jeff Sekelsky at UNC, Chapel Hill; http://sekelsky.bio.unc.edu/Research/Vectors/Vectors.htm). pUC8ySG is a pUC8 derived plasmid containing the Drosophila YELLOW gene (25). Homing pigeon is a construct designed for germline transformation of Drosophila (26). Sperm chromatin was prepared as described (27). The sequence of the 27 residue oligo is 5′-CAC TGC TGC CAT GGG GAT GAG TGA TAA-3′. The sequence of the 15 residue oligo is 5′-CGG GAA CAC TCA TAG-3′. The oligo duplexes were prepared in a buffer containing 75 mM NaCl, 1 mM EDTA, 10 mM Tris pH 8 by addition of equimolar amounts of the reverse compliments, incubation at 95°C for 10 min, and cooling to room temperature at a rate of 1°C/min.
Xenopus egg extracts and replication
Extract preparation [high speed supernatant (HSS) and nucleoplasmic extract (NPE)], DNA replication, and calculation of replication efficiency (percentage of input) were carried out as described (27). Any modifications are noted in the text. Extract dilution in buffer was performed with Egg Lysis Buffer (ELB; 2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES pH 7.7, 250 mM sucrose) that was also supplemented with an ATP regeneration system (20 mM phosphocreatine, 2 mM ATP, 5 µg/ml creatine phosphokinase). To inhibit licensing, Geminin was added to HSS at a final concentration of 0.1 ng/µl.
Preparation and treatment of DNA in agarose for analysis of radionucleotide incorporation
A 2.25% low-melting point (LMP) agarose (Invitrogen) was prepared with 10 mM Tris pH 8. The melted agarose was maintained at 69°C. Six microlitre of the molten agarose was mixed with 1.5 µl of DNA, also prepared in 10 mM Tris pH 8, by pipetting slowly 20 times. From this mixture, 5 µl drops were deposited on parafilm (Pechiney) and the agarose was allowed to solidify in a humid chamber at 4°C for 30 min. The block was incubated in extracts as described in the text. Upon completion of incubation in NPE, the blocks were treated with 1 mg/ml Proteinase K (Roche), 0.75% SDS, 1 mM EDTA and 10 mM Tris pH 8 overnight at 37°C. For gel electrophoresis, the block was inserted directly into the well. If agarose digestion was required, the block was washed seven times in 1 ml of 1 mM EDTA, 10 mM Tris pH 8 followed by treatment with β-agarase as specified by the manufacturer (NEB).
Preparation and treatment of agarose blocks for fluorescence microscopy
DNA was immobilized in agarose blocks as described above except that M-280 Dynabeads (Invitrogen) were included at 15 fM during block preparation. Blocks were stained with 100 nM YOYO-1 (Invitrogen), and then imaged with an inverted microscope (Olympus IX71) illuminated with an Ar/Kr laser (Coherent I-70C Spectrum). The dye was removed prior to extract addition with five washes in 1 ml of 150 mM NaCl and 0.1% SDS, followed by three washes in 1 ml of ELB. The block was incubated in one volume of HSS for 30 min, followed by two volumes of NPE containing 25 µM biotin-dUTP (Roche) for 90 min. Blocks were then treated with 1 mg/ml Proteinase K (Roche), 0.75% SDS, 1 mM EDTA, 10 mM Tris pH 8 overnight at 37°C and then washed seven times with 1 mM EDTA, 10 mM Tris pH 8. Incorporated biotin-dUTP was stained with Streptavidin coupled to AlexaFluor 647 (SA-647; Invitrogen) and the DNA re-stained with 100 nM YOYO-1 (Invitrogen). Images were acquired as stated above.
Radiolabeling p2.9
p2.9 was incubated in HSS (23.5 nM or 45 ng/µl) for 30 min, followed by addition of two volumes of NPE containing 0.25 µCi/µl of [α-32P]dATP (3000 Ci/mmol) for 2 h. The sample was stopped with 0.75% SDS, 1 mM EDTA, 10 mM Tris pH 8, treated for 4 h at 37°C with 5 µg/ml RNase A (Roche) and digested overnight at 37°C with 1 mg/ml Proteinase K (Roche), followed by phenol/chloroform extraction. The purified DNA was ethanol precipitated and resuspended in 10 mM Tris pH 8.
DNA unwinding assay
The DNA topology assay was carried out as described (28).
RESULTS
DNA replication is sensitive to plasmid concentration
To recapitulate DNA replication in conventional Xenopus egg extracts, sperm chromatin is typically incubated in a low speed supernatant (LSS) of egg cytoplasm. pre-RCs assemble on the chromatin within minutes, followed by nuclear envelope formation, nuclear import of key S-phase promoting factors, and replication initiation (19,21). In this system, DNA replication occurs in the context of nuclear foci, as visualized by punctate PCNA staining and incorporation of modified dNTPs (17,18,20). Subsequently, we developed a system that bypasses the requirement for nuclei in DNA replication (24). In this approach, DNA templates are incubated in a high speed supernatant (HSS) of egg cytoplasm, which supports pre-RC formation, but not nuclear assembly due to the absence of nuclear membrane vesicles. Next, a concentrated nucleoplasmic extract (NPE) is added, which supplies the S-phase factors needed to promote replication initiation. Unlike LSS, the nucleus-free replication system supports 100% efficient and cell-cycle regulated DNA replication of small, circular plasmids.
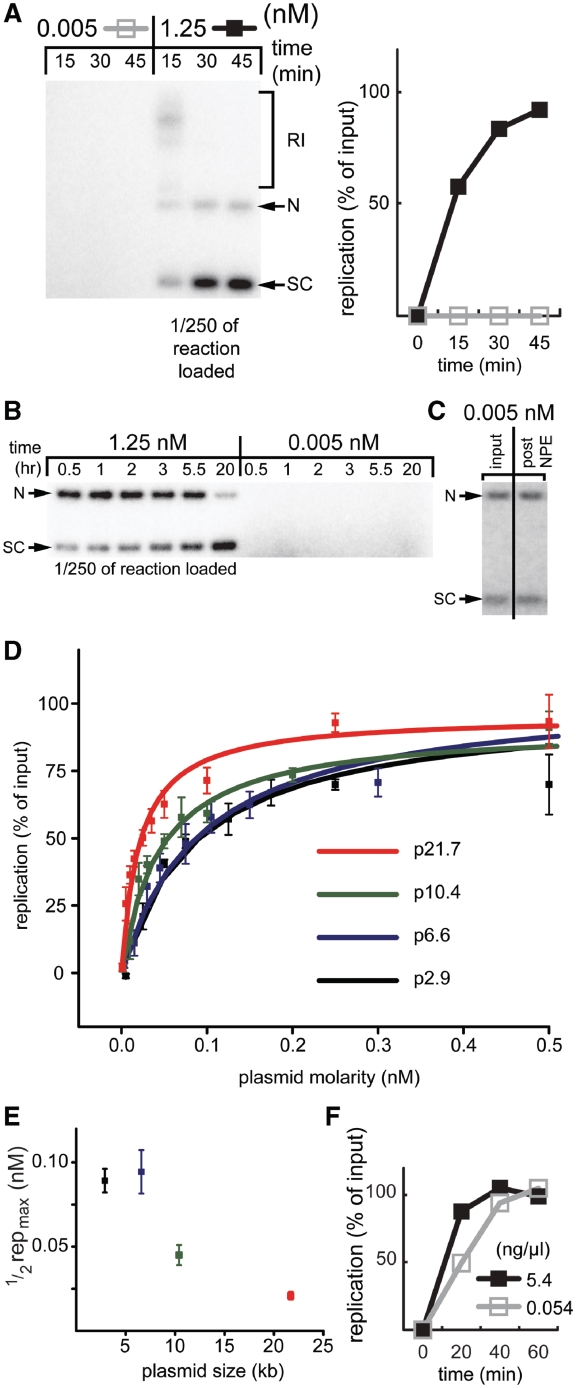
To address whether multiple replication forks must be able to cluster during DNA replication in Xenopus egg extracts, we wanted to determine whether plasmids aggregate during replication in the nucleus-free egg extract system. However, our experiments yielded conflicting results (see ‘Discussion’ section). We therefore designed strategies that are predicted to inhibit plasmid aggregation, and examined the effects on DNA replication. High molecular weight sperm chromatin is replicated from hundreds of thousands of origins (22,23) while plasmids up to 10 kb in size are replicated from just one origin (29). We reasoned that by varying the concentration of a 2.9-kb plasmid, p2.9, we might be able to modulate plasmid aggregation and by extension, DNA replication. Interestingly, while p2.9 replicated with 100% efficiency at a concentration of 1.25 nM (∼2.5 ng/µl), its replication was undetectable at 0.005 nM (∼0.01 ng/µl) (Figure 1A), even after prolonged incubation (Figure 1B). This result was not due to degradation of plasmid in the low concentration reaction (Figure 1C). In a titration experiment, the molarity of p2.9 required for half maximal DNA replication was 0.089 ± 0.007 nM (0.17 ± 0.01 ng/µl, all errors given in standard deviation) (Figure 1D, black line, Supplementary Table S1).
Figure 1.
DNA replication is sensitive to plasmid concentration. (A) p2.9 was incubated in HSS at 0.015 or 3.75 nM final concentration. After 30 min, two volumes of NPE containing [α-32P]dATP were added, reducing the final DNA concentrations to 0.005 and 1.25 nM, respectively. At various times after NPE addition, replication products were separated on a native agarose gel and analyzed by autoradiography. To examine equivalent quantities of DNA from both reactions, only 1/250th of the 1.25 nM reaction was loaded. The signal above background from the entire lane was used to calculate the percentage of replication and the results are graphed on the right. See Walter and Newport (2000) (28) for an explanation of the different replication products. RI, replication intermediates; N, nicked products; SC, supercoiled products. (B) p2.9 was replicated as in (A), except that samples were analyzed for DNA replication at later time points. (C) Low concentration plasmids do not undergo degradation in HSS/NPE. Radiolabeled p2.9 (for radiolabeling, see experimental procedures) was incubated in HSS/NPE as described in (A). The 45-min time point was separated on a gel (right lane) alongside the input (left lane). (D) The effect of plasmid concentration on DNA replication efficiency for four different plasmids. Plasmids were replicated as described in (A). Replication was analyzed 90 min after NPE addition. Data from triplicate experiments were fit with y = replicationmax·[plasmid]/(K + [plasmid]) where K is ½ replicationmax. (E) The plasmid concentration that yields half maximal replication was derived from the curves in (D) and graphed. (F) Sperm chromatin was replicated as in (A) at a final concentration of 0.054 or 5.4 ng/µl and the replication efficiency in each condition was graphed.
We next tested whether larger DNA templates, which support multiple initiations, also require a high DNA concentration to replicate. As for p2.9, the replication efficiency of p6.6 (6.6 kb), p10.4 (10.4 kb), and p21.7 (21.7 kb) was sensitive to the concentration of substrate in the reaction (Figure 1D). However, the plasmid concentration required for half maximal DNA replication decreased as plasmid size increased (Figure 1D and E, Supplementary Table S1). Strikingly, NPE supported complete replication of only 0.054 ng/µl sperm chromatin (17 sperm/µl, Figure 1F). For comparison, the efficiency of plasmid p21.7 replication at a similar concentration (0.05 ng/µl) was only ∼25% (Figure 1D, red line, second data point). Therefore, replication of sperm chromatin is much less sensitive to DNA concentration. The inverse correlation between a DNA substrate’s size and its sensitivity to dilution appears to be consistent with a requirement for replisome aggregation in DNA replication. According to this hypothesis, the need for interactions between substrate molecules declines as the number of replicons per substrate increases and intra-molecular interactions replace inter-molecular contacts.
DNA replication does not require plasmid–plasmid contacts
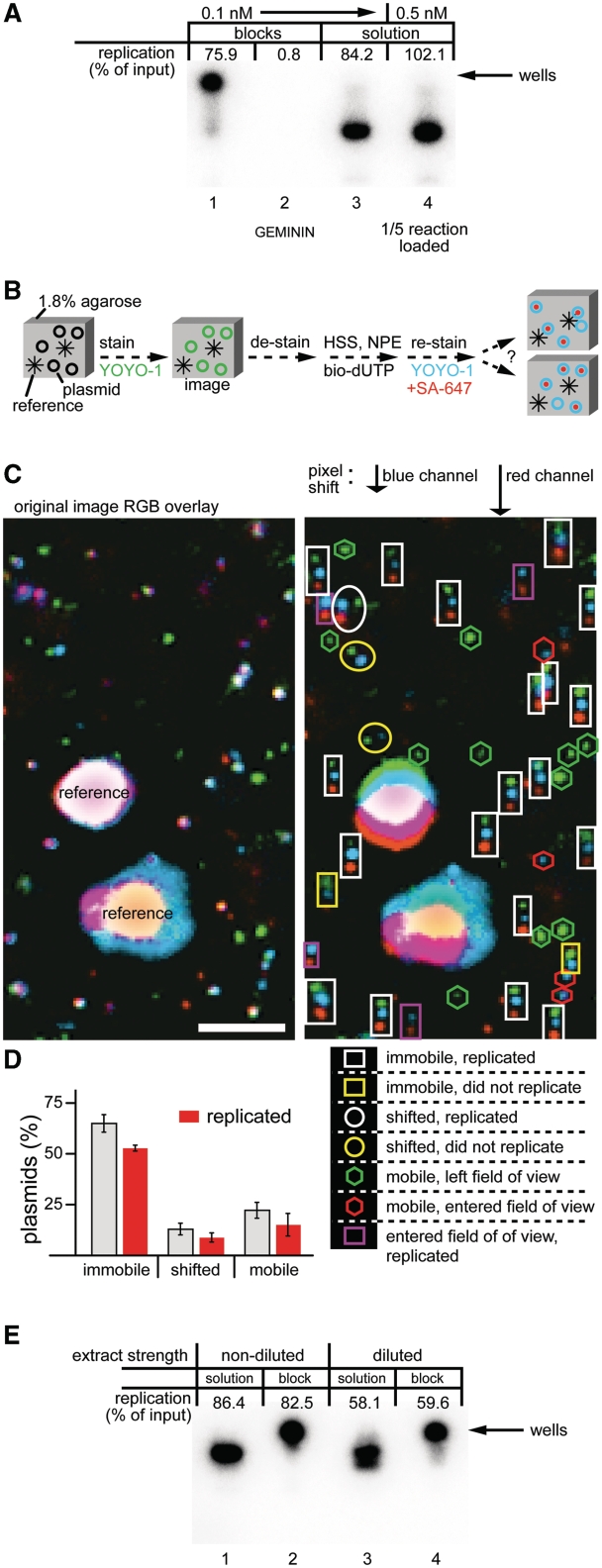
To test directly whether multiple plasmids must interact to achieve DNA replication, we established conditions under which plasmid–plasmid contacts cannot occur. To this end, we prepared 5 µl of a solution containing 1.8% molten agarose and 0.1 nM p10.4 (a concentration that supports efficient replication in solution; Figure 1D; see below, Figure 2A, compare lanes 3 and 4). After polymerization of the agarose, the plasmid is immobilized (30). The agarose block was then incubated in HSS followed by NPE, and we tested whether the trapped plasmid could replicate. Remarkably, the plasmid embedded in agarose replicated as efficiently as plasmid in solution (Figure 2A, compare lanes 1 and 3). Addition of Geminin, which inhibits MCM2-7 loading onto chromatin, abolished replication in the agarose block (Figure 2A, lane 2), as expected for bona fide chromosomal replication. Similar to our results in solution, low concentrations of plasmid did not replicate in the agarose block (see below, Figure 3B, Condition 4), demonstrating that agarose does not bypass the need for high concentrations of plasmid DNA.
Figure 2.
Efficient DNA replication of immobilized plasmids. (A) The 1.8% agarose blocks (5 µl) containing 0.1 nM p10.4 were incubated with two volumes of HSS, with or without Geminin. After 30 min, the supernatant was exchanged with two volumes of NPE containing [α-32P]dATP. Reactions in solution containing a final concentration of 0.1 or 0.5 nM p10.4 were carried out in parallel. In all cases, DNA replication efficiency was determined 90 min after NPE addition. In lanes 1 and 2, the DNA was not released from the block and thus remained in the well, where the block was loaded. In lane 4, one-fifth of the 0.5 nM reaction was loaded. (B) Cartoon illustrating procedure to determine plasmid position and replication in an agarose block. (C) The 1.8% agarose blocks containing 0.1 nM p10.4 and 2.8 µm beads (to provide reference points) were prepared. Plasmids were stained with YOYO-1, photographed and de-stained. Subsequently, HSS was added, followed by NPE containing biotin-dUTP. After 90 min, the blocks were stained again with YOYO-1 and the biotin-dUTP was detected with AlexaFluor 647 conjugated streptavidin (SA-647). The same position in the block that was imaged before extract addition was located and images were acquired by fluorescence microscopy. The green and blue channels represent the initial and final plasmid positions as determined by YOYO-1 staining, respectively. The red channel represents SA-647 staining of incorporated biotin-dUTP. The blue and red channels were shifted in the y-axis to facilitate analysis. Plasmid classification is shown below the shifted image. (bar = 5 µm) (D) The average percentage of plasmids that was immobile, shifted, or mobile from eight areas of a single block was calculated and graphed. The subset of plasmids in each group that replicated is shown in red. Error bars indicate the standard deviation of the eight areas that were scanned. (E) Replication in diluted extracts. p10.4 (0.1 nM) was replicated in agarose blocks or in solution as described in (A). In the ‘diluted’ condition, HSS and NPE were each diluted 5- and 4-fold with ELB, respectively.
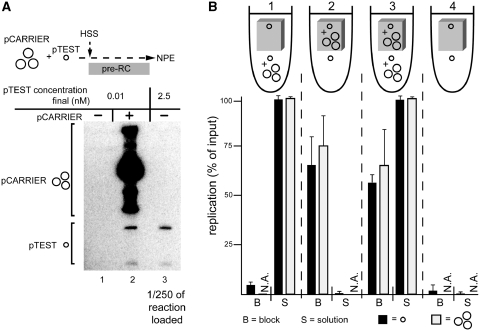
Figure 3.
One plasmid activates replication of another in trans. (A) The effect of pCARRIER on pTEST plasmid replication in solution. In lane 2, pTEST (p2.9, single small circle) and pCARRIER (p10.4, group of large circles) were premixed and incubated in HSS at 0.03 and 1.5 nM final concentrations, respectively. In lanes 1 and 3, only pTEST was incubated in HSS (0.03 and 7.5 nM, respectively). Ninety minutes after addition of two volumes NPE, DNA replication was analyzed by gel electrophoresis and autoradiography. In lane 3, 1/250th of the total reaction was loaded. (B) Effect of pCARRIER on replication of pTEST plasmid in agarose blocks. The 1.8% agarose blocks (5 µl volume) were prepared containing pTEST (p2.9, 0.01 nM) with or without pCARRIER (p10.4, 0.5 nM). HSS was incubated with pCARRIER (p10.4, 1.5 nM final concentration) and pTEST (p2.9, 0.03 nM) or pTEST alone (p2.9, 0.03 nM) for 7.5 min. Ten microlitre of the extract/DNA mixture was then added to the agarose blocks. Relative to the entire HSS/block volume, two volumes of NPE containing [α-32P]dATP were added. At 90 min, the blocks were digested with agarase, and the released replication products were separated on an agarose gel alongside the DNA from the supernatants. Results from three independent experiments were quantified and the averages and standard deviations are graphed below a cartoon of each condition.
We wanted to rule out the possibility that plasmids can interact despite being embedded in agarose. We therefore devised an imaging procedure that determines the positions of plasmids before and after DNA replication (Figure 2B). If individual plasmids are in the same place before extract addition and after replication is complete, they cannot have come into contact, even transiently. Biotin-dUTP was also added to the extract to detect replication upon staining with fluorescent streptavidin. Using this procedure, the location and replication of approximately 2500 plasmids was annotated before and after HSS/NPE addition (one dot = one plasmid, Figure 2C and D). To help interpret the image, the blue channel (YOYO-1, DNA stain after replication) and red channel (streptavidin stain of incorporated biotin-dUTP) were offset from the green channel (YOYO-1, DNA stain before DNA replication) in the vertical direction by 5 and 10 pixels, respectively (Figure 2C, right image). A total of 65% of the plasmids remained completely immobile throughout the experiment (Figure 2C, white and yellow rectangles; Figure 2D). The original location of a further 13% of plasmids could be inferred despite a slight shift in their position (Figure 2C, white and yellow circles; Figure 2D). Twenty-two percent of plasmids could not be re-located after NPE addition (Figure 2C, green and red hexagons, purple rectangles; Figure 2D). These molecules were interpreted to have either left their original position (green hexagons) or moved to the final position either from another position in the same X-Y plane or from another position in the Z-axis (red hexagons, purple rectangles). Therefore, theses plasmids were classified as mobile. Importantly, a greater proportion of immobile plasmids replicated (82%, white rectangles) compared to the shifted (67%, white circles) or mobile (69%, purple rectangles) populations (Figure 2D). Since immobile plasmids, which cannot have aggregated, replicated at least as efficiently as mobile plasmids, our data provide strong evidence that direct plasmid–plasmid interactions are not required for DNA replication.
One possible explanation for why plasmid immobilization does not perturb replication in Xenopus egg extracts is that replication factors in this system are so concentrated that plasmid clustering is dispensable for replication. This model predicts that the replication of immobilized plasmids should be more sensitive than plasmids in solution to extract dilution. To test this prediction, plasmids in solution and in an agarose block were incubated in HSS and NPE that were undiluted or diluted 5- and 4-fold, respectively. This level of extract dilution reduced replication efficiency of plasmids in solution by approximately one-third, indicating that replication factors started to become limiting (Figure 2E, compare lanes 1 and 3). However, under the diluted conditions, replication in a block and in solution were affected equally (Figure 2E, compare lanes 3 and 4), supporting our conclusion that plasmid aggregation is not essential for DNA replication, even when replication factors are limiting.
Short-range stimulation of one plasmid’s replication by another plasmid
The experiments described above suggest that in the absence of any plasmid–plasmid contacts, plasmid concentration controls DNA replication efficiency. To better understand the basis of this effect, we performed mixing experiments using a high concentration of a large plasmid (called ‘pCARRIER’) and a low concentration of a small plasmid (called ‘pTEST’). Thus, a sub-threshold concentration of pTEST (p2.9, 0.01 nM) was incubated either alone or in the presence of pCARRIER (p10.4, 0.5 nM) (Figure 3A). As expected, pTEST alone did not replicate, whereas in the presence of pCARRIER, both pTEST and pCARRIER replicated (Figure 3A, compare Conditions 1 and 2). When the plasmids were immobilized in an agarose block, we obtained the same result as in solution (Figure 3B, compare Condition 2, block, with Condition 1, block). These data illustrate that one plasmid can promote DNA replication of a second plasmid in the absence of physical contact. The same result is obtained when a small plasmid (p2.9) is used as pCARRIER and a large plasmid (p21.7) is used as pTEST (data not shown).
We next addressed whether the stimulation of pTEST plasmid by pCARRIER plasmid occurs over long distances. To this end, we embedded pTEST alone (p2.9, 0.01 nM) in an agarose block and surrounded it with extract that was pre-mixed with a high concentration of pCARRIER (p10.4, 1.5 nM). pTEST in the block did not replicate under these conditions (Figure 3B, Condition 1, block). This was not due to sequestration of essential replication factors by pCARRIER outside the agarose block, since a high concentration of pCARRIER inside the agarose block replicated efficiently in this condition (Figure 3B, Condition 3). Thus, when the plasmids are physically segregated into separate reaction volumes, stimulation of one plasmid by another is not observed. Therefore, high concentrations of DNA are only able to promote DNA replication over short distances.
DNA concentration-dependent replication cannot be explained by mass action or titration of non-specific inhibitors only
One potential explanation for our results is that at low plasmid concentration, an essential replication factor does not bind efficiently to the DNA substrate. However, according to the law of mass action, the fraction of DNA that is bound to the factor is independent of the DNA concentration itself (Supplementary Figure S1). Therefore, mass action cannot explain the sensitivity of DNA replication to DNA concentration.
Another possible explanation for our results is that Xenopus egg extracts contain a non-specific inhibitor of replication that is neutralized by DNA in a concentration-dependent manner. This model predicts that pre-exposing an extract to a high concentration of pCARRIER to neutralize the inhibitor would support replication of pTEST added subsequently at low concentration. This prediction applies to an inhibitor that binds irreversibly to DNA, as well as one that is in dynamic equilibrium with DNA; in both cases the pre-incubation and continued presence of pCARRIER will, throughout the entire reaction volume, lower the effective concentration of free inhibitor that is available to affect pTEST. As shown in Figure 3B (Condition 1), pTEST embedded in an agarose block did not replicate upon incubation with an extract that had been pre-incubated with pCARRIER. Based on this result, we conclude that at least one step in DNA replication is regulated by DNA concentration in a manner that is independent of an inhibitor. This step must be regulated by positive, short-range communication between plasmids (Figure 3).
High plasmid concentration is required for replication licensing
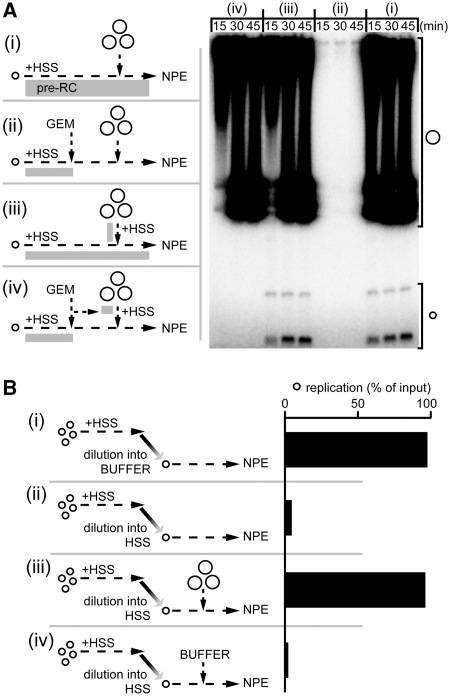
We next asked which of the events underlying DNA replication fails at low plasmid concentration. We first investigated pre-RC formation (‘licensing’), which was achieved by specifically limiting the DNA concentration during the licensing period. pTEST (p2.9, 0.015 nM) was incubated in HSS for 20 min, which is normally sufficient time to allow licensing [results not shown and see (31)]. At t = 20 min, Geminin was added to prevent any further licensing, followed by the addition of pCARRIER (p10.4, 0.65 nM; Figure 4A, scheme ii). If licensing does not occur on pTEST alone, then the subsequent addition of pCARRIER after the licensing period has been terminated is not expected to promote pTEST replication. Indeed, pTEST did not replicate in this sequence, suggesting that licensing is DNA concentration-dependent (Figure 4Aii). The failure to observe DNA replication was not due to the serial addition of pTEST and pCARRIER plasmids, since omitting Geminin led to efficient replication of pTEST (Figure 4Ai). Moreover, the replication defect of pTEST in scheme (ii) was not related to the absence of pCARRIER replication: Adding pre-licensed pCARRIER that could replicate did not rescue pTEST replication when the licensing period for pTEST DNA was terminated before pCARRIER addition (Figure 4Aiv). We conclude that licensing fails at low plasmid concentration. Independent evidence for this conclusion was obtained from single molecule experiments in which we found that the addition of pCARRIER to extract was essential to promote the binding of MCM2-7 to λ DNA bound to the surface of a microfluidic flow cell, as measured by immunofluorescence (A. Kochaniak et al., unpublished results).
Figure 4.
Two steps in replication are sensitive to plasmid concentration. (A) Licensing fails at low plasmid concentration. As indicated by the cartoons (i–iv), pTEST plasmid (p2.9, single small circle) was incubated with HSS and then subjected to four different reaction schemes. In scheme (i), after 30 min, pCARRIER (p10.4, group of large circles) was added followed within another 30 min by NPE. (ii), same as (i), except that Geminin was added 10 min before pCARRIER addition. (iii), same as in (i) except that pCARRIER was incubated for 30 min separately in HSS before being added to pTEST/HSS. (iv), same as (iii) except that Geminin was added for 10 min to pTEST and pCARRIER before they were mixed. Before addition of two volumes of NPE, the concentration in HSS of pTEST was 0.015 nM and of pCARRIER was 0.65 nM. At the indicated times after NPE addition, the replication products were separated by gel electrophoresis as shown on the right. (B) (i and ii) HSS inhibits events downstream of licensing at low DNA concentration. As illustrated on the left, pTEST (p2.9; group of small circles) was incubated in HSS at a concentration of 3 nM for 30 min followed by a 200-fold dilution into buffer (i) or HSS (ii) prior to addition of two volumes of NPE containing [α-32P]dATP. Replication was measured by gel electrophoresis and autoradiography 60 min after NPE addition, and results are graphed on the right. (iii and iv) pCARRIER neutralizes the inhibitory effect of HSS on post-licensing events. pTEST (p2.9, group of small circles) was incubated at a concentration of 1.5 nM in HSS for 30 min, followed by a 100-fold dilution in fresh HSS. p10.4 CARRIER DNA at a final concentration of 0.65 nM (iii) or buffer (iv) was added after the dilution in HSS, and prior to addition of NPE. The replication products were analyzed as in (Bi, ii).
A cytosolic inhibitor suppresses initiation at low plasmid concentration
If licensing is the only step that is sensitive to DNA concentration, then DNA replication should proceed at any plasmid concentration after licensing has occurred. To test this possibility, a high concentration of p2.9 (3 nM) was licensed in HSS followed by a 200-fold dilution in buffer to a final concentration of 0.015 nM, followed by the addition of NPE (Figure 4Bi). Under these conditions, p2.9 replicated efficiently, demonstrating there is no intrinsic requirement for high DNA concentration once licensing has occurred (Figure 4Bi). Unexpectedly, when the plasmid was diluted with HSS instead of buffer prior to addition of NPE, there was no DNA replication (Figure 4Bii). However, when the NPE was supplemented with a high concentration of pCARRIER, DNA replication was restored (Figure 4Biii), whereas addition of buffer had no effect (Figure 4Biv). The data suggest that HSS inhibits replication downstream of licensing, and that this inhibition is antagonized by carrier DNA. Further analysis demonstrated that the event blocked by HSS is origin unwinding (Supplementary Figure S2). Together, our data indicate that HSS contains an inhibitor of initiation that is neutralized at high DNA concentration.
The lack of licensing at low DNA concentration is not due to an inhibitor
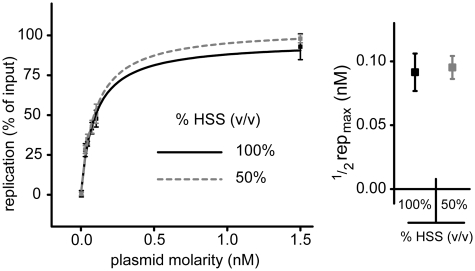
So far, we showed that both licensing and initiation are sensitive to DNA concentration but that initiation does occur at low DNA concentration when no HSS is present (Figure 4Bi). Importantly, since extracts that were pre-incubated with a high concentration of pCARRIER do not promote replication of pTEST in trans (Figure 3B, Condition 1), there is at least one DNA concentration-dependent step that is not blocked by a titratable inhibitor. Together, the data imply that this step is licensing. To further test this idea, we examined licensing in diluted HSS. If a non-specific inhibitor represses licensing in HSS, then 50% dilution of HSS should reduce the concentration of the inhibitor 2-fold and thereby also reduce the concentration of DNA required to achieve half maximal replication 2-fold. However, we observed no change in the DNA concentration required for licensing in diluted HSS (Figure 5). Importantly, reducing the HSS concentration by half is sufficient to functionally dilute the inhibitor of the initiation step, thus validating our approach (Supplementary Figure S3). Figure 5, therefore, further supports the notion that the DNA concentration-dependence of licensing does not involve the neutralization of an inhibitor. In summary, licensing and initiation are both sensitive to DNA concentration, but for different reasons. Initiation is dependent on high DNA concentration, but only when HSS is present, indicating that initiation is blocked by a cytosolic inhibitor that is neutralized by DNA. In contrast, we found no evidence that licensing is controlled by an inhibitor, suggesting that this reaction is intrinsically dependent on high DNA concentration.
Figure 5.
HSS dilution does not rescue replication at low DNA concentration. p6.6 was incubated for 30 min in 100% HSS or HSS diluted 50% v/v with buffer, followed by NPE addition. Replication was analyzed 90 min after NPE addition. Data from triplicate experiments were fit with y = replicationmax · [plasmid] / (K + [plasmid]) where K is ½ replicationmax. The p6.6 concentration that produces half-maximal replication was derived from the curves and graphed on the right.
A high concentration of double-stranded oligonucleotides stimulates licensing
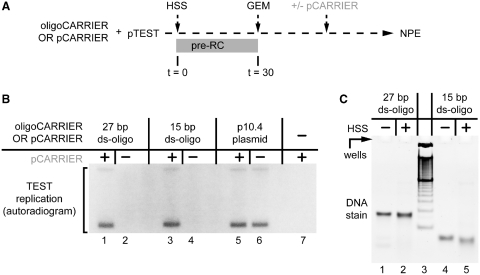
To understand whether the DNA concentration dependence of licensing involves positive interactions between pre-RCs, we asked whether the carrier DNA must be licensed to promote licensing of pTEST. To this end, we examined whether duplex oligonucleotides (‘oligos’) that are too short to recruit MCM2-7 complexes (32) could promote licensing of pTEST. HSS containing pTEST (p2.9, 0.01 nM) was supplemented with different duplex oligos (‘oligoCARRIER’; Figure 6A). The oligoCARRIERs were added at a concentration of 20 ng/µl in HSS because efficient replication (and licensing) is normally supported by 15–45 ng/µl of plasmid regardless of size (data not shown). After terminating the licensing period with Geminin, pCARRIER (p10.4, 1 nM) was added with NPE to overcome the HSS-dependent inhibition of initiation. The DNA replication measured in this sequence thus reflects the effect of each oligoCARRIER on licensing. When 27- or 15-bp duplex oligoCARRIERs were used, they were as efficient as a circular plasmid, p10.4, in promoting licensing of pTEST (Figure 6B, compare lanes 1, 3, and 5). This activity was not due to ligation of the oligos into longer species during incubation in HSS (Figure 6C, lanes 2 and 5). Shorter oligo duplexes were not tested because they are thermodynamically unstable. When oligoCARRIER was left out of the reaction, pTEST failed to replicate (Figure 6B, left gel, lane 7). When pCARRIER was omitted after licensing, pTEST did not replicate in the presence of oligoCARRIERs (Figure 6B, lanes 2 and 4). Therefore, although duplex oligos can stimulate licensing, they are unable to support the DNA-sensitive initiation step, presumably because dsDNA oligos are rapidly degraded in NPE (data not shown). Our results demonstrate that ds-oligos, which are too short to assemble their own pre-RCs, can stimulate licensing in trans.
Figure 6.
Short double-stranded oligonucleotides stimulate licensing. (A) Scheme to determine rescue of pTEST licensing. pCARRIER or oligoCARRIER was premixed with pTEST and incubated in HSS. After 30 min, Geminin was added, and after a further 10 min the reaction was supplemented with pCARRIER or buffer (p10.4, 1 nM, grey lettering to indicate post-GEMININ addition), followed by NPE. (B) Effect of oligoCARRIER on pTEST licensing. HSS containing pTEST (p2.9, 0.01 nM) was supplemented with a 27-bp duplex oligo (lanes 1 and 2, 20 ng/µl, 1.1 µM), 15-bp duplex oligo (lanes 3 and 4, 20 ng/µl, 2.0 µM), or pCARRIER (p10.4) (lanes 5 and 6, 20 ng/µl, 2.9 nM). Thereafter, the experiment proceeded as described in (A). Ninety minutes after NPE, the replication products were separated by gel electrophoresis and quantified by autoradiography. (C) Just prior to Geminin addition, the DNA/HSS mix from the 27- and 15-bp duplex oligo samples described in Figure 6B were treated with RNAse, phenol–chloroform extracted, separated by native PAGE, and imaged with Sybr-Gold staining (right gel, lanes 2 and 5), alongside input DNA minus HSS (lanes 1 and 4) and a 10-bp ladder (lane 3).
DISCUSSION
The organization of chromatin into higher order structures within eukaryotic nuclei has been proposed to facilitate enzymatic processes underlying genome maintenance and gene expression (10,12,14,33). By incubating plasmid DNA in a concentrated NPE, we and others previously showed that the formation of nuclei is not required for efficient, cell-cycle regulated chromosomal DNA replication (24), checkpoint activation (34), or DNA inter-strand crosslink repair (35) in Xenopus egg extracts. These results, however, did not rule out the possibility that plasmid aggregation might be required for these processes. To determine whether plasmids aggregate in Xenopus egg extracts, we imaged extracts containing plasmids that were labeled with fluorescent nucleotides or pre-stained with an intercalating agent. However, the two approaches yielded contradictory results [data not shown, see also (19)]. We therefore tested directly whether plasmid aggregation is needed for DNA replication by encasing plasmids in agarose. Strikingly, these immobilized DNA templates replicated just as efficiently as plasmids in solution (Figure 2A). Even in diluted extracts, when replication factors became limiting, immobilizing the plasmid templates did not perturb replication (Figure 2E). We further showed that immobilizing plasmids in agarose did not disrupt two mechanisms used to prevent re-replication in Xenopus egg extracts, Geminin-mediated inhibition of Cdt1 (Figure 2) and replication-dependent Cdt1 ubiquitylation and proteolysis (data not shown). Moreover, the repair of DNA interstrand-crosslinks (35) was not inhibited in agarose blocks (unpublished results). These results show that replisome clustering is not essential for the basic enzymatic steps underlying plasmid replication or several important genome maintenance pathways.
It is tempting to speculate from these results that nuclear replication foci are not required for DNA replication in Xenopus egg extracts. Indeed, DNA replication in NPE appears to involve a highly physiological mechanism. Thus, plasmids are chromatinized before synthesis begins (36–38), and no proteins are known that are dispensable for plasmid replication in NPE but required for nuclear replication. Furthermore, since NPE is extracted from nuclei using a procedure that entails dilution, the concentration of replication factors in the nucleus-free system is necessarily lower than in nuclei. Therefore, if the function of foci is to sequester essential replication factors, replication in NPE should be more, not less, sensitive to the loss of replisome aggregation than an intact nucleus. Nevertheless, we cannot exclude the possibility that replisome aggregation is essential for chromosomal DNA replication in the context of the nucleus. A definitive answer to what function foci play in chromosomal replication will require methods that specifically disrupt these structures within nuclei.
Surprisingly, we also discovered that DNA replication is exquisitely sensitive to the concentration of plasmid present in the reaction (Figure 1). By adjusting the plasmid concentration at different stages of replication, we identified two steps in this process that are dependent on high DNA concentration, licensing and initiation (Figure 4). Initiation is blocked by a cytosolic inhibitor, which is counteracted by high DNA concentration (Figure 4B). Whether this inhibitor represents a non-specific inactivator of dilute DNA templates or a physiological regulator of replication is presently unclear. In contrast, our data strongly suggest that there is an intrinsic requirement for high DNA concentration during licensing that does not involve neutralization of an inhibitor. In support of this interpretation, pre-incubation of extract with pCARRIER and the continued presence of pCARRIER in solution did not rescue replication of pTEST embedded in agarose (Figure 3B). Also, HSS dilution did not change the DNA concentrations required for replication (Figure 5). Neither of these outcomes is predicted if licensing were controlled by a stoichiometric inhibitor. Instead, the data indicate that above a certain threshold concentration, plasmids communicate with each other at a distance and thereby stimulate licensing (Figure 2).
What underlies this communication? We postulate the existence of a diffusible activator of licensing, which is converted from a latent to an active state by DNA. The activated factor diffuses to its target, where it performs its licensing function in trans. Accordingly, in the presence of high DNA concentration, a critical level of activator accumulates, enabling licensing. At low DNA concentrations, the latent factor is not converted at a sufficient rate, and it never reaches critical concentration. In this view, large plasmids replicated efficiently at lower molarities (Figure 1D) because they contain more DNA, supporting more activator conversion per plasmid. It is likely that sperm chromatin is largely insensitive to dilution because this extremely large DNA template is sufficient to generate a high local concentration of activator.
When we immobilized a low concentration of pTEST in agarose and surrounded it with extract containing a high concentration of pCARRIER, we did not observe any DNA replication of pTEST (Figure 3B). Replication of pTEST occurred only when the two templates were interspersed, thereby minimizing the distance between the molecules (Figure 3B). These data show that the stimulatory effect of DNA on replication only functions over short distances and suggest that the putative activator has a short half-life and diffuses only a small distance before being switched off. In the future, it will be important to identify the relevant factor(s) and understand how its activity is regulated by DNA.
An interesting question addresses why chromosomal replication might have evolved to depend on high DNA concentration. One possible reason is to help prevent the cytoplasmic replication of extrachromosomal DNA, such as the minicircles, which are formed de novo in preblastula Xenopus embryos and then lost during development (39,40). DNA concentration dependent replication might also help to prevent viruses from using the cellular replication machinery to replicate their DNA in the cytoplasm. Importantly, licensing occurs in telophase, before the nuclear envelope has re-assembled (41). Since nuclear and cytoplasmic factors are intermixed at this stage of the cell-cycle, restricting licensing to chromosomes by DNA-mediated signaling may be particularly important to avoid replication of extrachromosomal DNA species.
Our discovery that DNA replication is concentration-dependent in Xenopus egg extracts has important practical implications for the study of DNA replication. For example, we have recently employed Xenopus egg extracts to study replication of lambda DNA molecules that are immobilized on the surface of a microfluidic flow cell (Yardimci et al., submitted for publication). Since the concentration of lambda DNA in the flow cell is extremely low, replication only occurs when the licensing extract is supplemented with carrier DNA. If the need for high DNA concentration is a general feature of metazoan DNA replication, attempts to achieve cell-free DNA replication with extracts from other organisms will have to be performed using adequate DNA concentrations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grant (GM62267) and Leukemia and Lymphoma Scholar Award (to J.C.W.). Funding for open access charge: National Institutes of Health grant (GM62267).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Anna Kochaniak and Joseph Loparo for their thoughtful discussions and input.
REFERENCES
- 1.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 2.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 3.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 4.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diffley JF. DNA replication: building the perfect switch. Curr. Biol. 2001;11:R367–R370. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- 6.Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 8.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 10.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 11.Newport J, Yan H. Organization of DNA into foci during replication. Curr. Opin. Cell Biol. 1996;8:365–368. doi: 10.1016/s0955-0674(96)80011-1. [DOI] [PubMed] [Google Scholar]
- 12.Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Djeliova V, Russev G, Anachkova B. Dynamics of association of origins of DNA replication with the nuclear matrix during the cell cycle. Nucleic Acids Res. 2001;29:3181–3187. doi: 10.1093/nar/29.15.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi JY, Stein JL, Lian JB, Javed A. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 2003;13:584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Almouzni G, Wolffe AP. Nuclear assembly, structure, and function: the use of Xenopus in vitro systems. Exp. Cell Res. 1993;205:1–15. doi: 10.1006/excr.1993.1051. [DOI] [PubMed] [Google Scholar]
- 16.Blow JJ. Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J. 2001;20:3293–3297. doi: 10.1093/emboj/20.13.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills AD, Blow JJ, White JG, Amos WB, Wilcock D, Laskey RA. Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. J. Cell Sci. 1989;94(Pt 3):471–477. doi: 10.1242/jcs.94.3.471. [DOI] [PubMed] [Google Scholar]
- 18.Hutchison C, Kill I. Changes in the nuclear distribution of DNA polymerase alpha and PCNA/cyclin during the progress of the cell cycle, in a cell-free extract of Xenopus eggs. J. Cell Sci. 1989;93(Pt 4):605–613. doi: 10.1242/jcs.93.4.605. [DOI] [PubMed] [Google Scholar]
- 19.Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 20.Cox LS, Laskey RA. DNA replication occurs at discrete sites in pseudonuclei assembled from purified DNA in vitro. Cell. 1991;66:271–275. doi: 10.1016/0092-8674(91)90617-8. [DOI] [PubMed] [Google Scholar]
- 21.Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- 22.Herrick J, Stanislawski P, Hyrien O, Bensimon A. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J. Mol. Biol. 2000;300:1133–1142. doi: 10.1006/jmbi.2000.3930. [DOI] [PubMed] [Google Scholar]
- 23.Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract are 5-15 kilobases apart and are activated in clusters that fire at different times. J. Cell Biol. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 25.Morris JR, Geyer PK, Wu CT. Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev. 1999;13:253–258. doi: 10.1101/gad.13.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–3992. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- 27.Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol. Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- 28.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 29.Lucas I, Chevrier-Miller M, Sogo JM, Hyrien O. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 2000;296:769–786. doi: 10.1006/jmbi.2000.3500. [DOI] [PubMed] [Google Scholar]
- 30.Dean WW, Dancis BM, Thomas CA., Jr The trapping of circular DNA in agarose gels. Anal. Biochem. 1973;56:417–427. doi: 10.1016/0003-2697(73)90207-8. [DOI] [PubMed] [Google Scholar]
- 31.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J. Cell Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 33.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Stokes MP, Van Hatten R, Lindsay HD, Michael WM. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 2002;158:863–872. doi: 10.1083/jcb.200204127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskey RA, Mills AD, Morris NR. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- 37.Gottesfeld J, Bloomer LS. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982;28:781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- 38.Almouzni G, Mechali M. Assembly of spaced chromatin involvement of ATP and DNA topoisomerase activity. EMBO J. 1988;7:4355–4365. doi: 10.1002/j.1460-2075.1988.tb03334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen S, Menut S, Mechali M. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol. Cell. Biol. 1999;19:6682–6689. doi: 10.1128/mcb.19.10.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen S, Mechali M. Formation of extrachromosomal circles from telomeric DNA in Xenopus laevis. EMBO Rep. 2002;3:1168–1174. doi: 10.1093/embo-reports/kvf240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 2001;20:4263–4277. doi: 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.