Abstract
Endonuclease V is an enzyme that initiates a conserved DNA repair pathway by making an endonucleolytic incision at the 3′-side 1 nt from a deaminated base lesion. DNA cleavage analysis using mutants defective in DNA binding and Mn2+ as a metal cofactor reveals a novel 3′-exonuclease activity in endonuclease V [Feng,H., Dong,L., Klutz,A.M., Aghaebrahim,N. and Cao,W. (2005) Defining amino acid residues involved in DNA-protein interactions and revelation of 3′-exonuclease activity in endonuclease V. Biochemistry, 44, 11486–11495.]. This study defines the enzymatic nature of the endonuclease and exonuclease activity in endonuclease V from Thermotoga maritima. In addition to its well-known inosine-dependent endonuclease, Tma endonuclease V also exhibits inosine-dependent 3′-exonuclease activity. The dependence on an inosine site and the exonuclease nature of the 3′-exonuclease activity was demonstrated using 5′-labeled and internally-labeled inosine-containing DNA and a H214D mutant that is defective in non-specific nuclease activity. Detailed kinetic analysis using 3′-labeled DNA indicates that Tma endonuclease V also possesses non-specific 5′-exonuclease activity. The multiplicity of the endonuclease and exonuclease activity is discussed with respect to deaminated base repair.
INTRODUCTION
Endonuclease V (endo V) is a DNA repair enzyme which hydrolyzes the second phosphodiester bond 3′ from a deaminated base lesion (1–4). Inosine (I) derived from adenosine deamination, xanthosine and oxanosine from guanosine deamination, and uridine from cytidine deamination are all substrates in vitro for bacterial endo V (1–3,5,6). The endonuclease activity towards inosine and its role in repair of deaminated inosine damage in vivo have been studied intensely. Biochemical and kinetic analysis reveals that endo V remains bound to inosine-containing DNA after strand cleavage (1,3,7,8). It was proposed that this unique property may act as a sensor to recruit other proteins for downstream repair process (3). Genetic analysis using endo V (encoded by nfi gene) deletion mutants indicates that endo V is a primary repair enzyme for the repair of inosine lesions produced under nitrosative stress conditions (9–11). After the 3′-nicking downstream of the inosine site by endo V, a repair patch needs to be created to remove the lesion. Taking advantage of an oligonucleotide-mediated transformation system, a genetic study demonstrates that repair of inosine in chromosomes of Escherichia coli creates an ∼5 nt repair patch, which is defined by removal of 2 nt from the 5′-side and 3 nt from the 3′-side (12).
The structure and function relationship of this repair enzyme has been investigated intensely using thermostable endonuclease V from the thermophilic bacterium Thermotoga maritima (Tma) as a model system. A systematic site-directed mutagenesis analysis on all seven conserved motifs defines the role of a series of residues in motifs I, III, IV, V, VI, VII in interactions with inosine in DNA (Figure 1A) (13). Biochemical and structural studies reveal that D43, E89, D110 and H214 are part of the active site that coordinates metal binding and catalytic function (14,15). Several conserved amino acid residues in motifs III and IV play an important role in protein–DNA interactions and recognition of deaminated DNA bases.
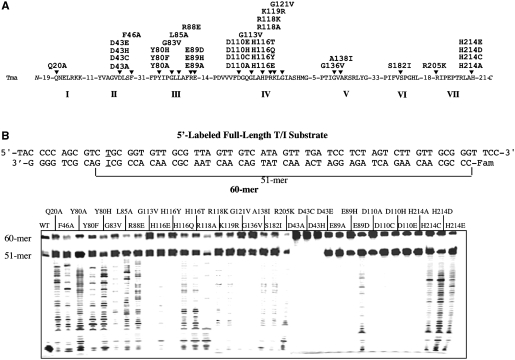
Figure 1.
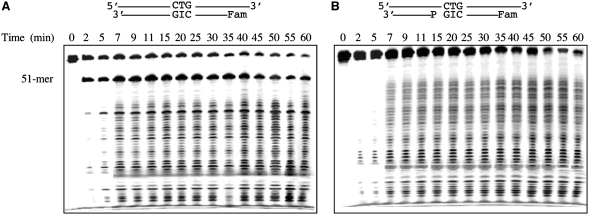
Cleavage activity of Tma endo V on 5′-labeled full-length T/I substrate. (A) Conserved motifs of Tma endo V. Site-directed mutants used in this study were listed above the arrows. (B) Cleavage activity of wt and mutant Tma endo V on 5′-labeled full-length T/I substrate. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate. The 5′-end of the bottom strand was labeled with the Fam fluorophore. The T/I base pair was underlined.
In addition to the endonuclease activity nicking the inosine site, Tma endo V also contains non-specific nuclease activities (3,4,16–18). An important finding from previous systematic site-directed mutagenesis analysis is the revelation of 3′-exonuclease activity from Tma endo V (13). Previous biochemical characterization of the nuclease activity from Tma endo V primarily focused on endonuclease activity on deaminated lesions (2,3,7,13,14,19). In this study, we investigated endonuclease and exonuclease activities from Tma endo V using 5′-labeled, 3′-labeled and 5′-internally labeled inosine-containing DNA. Using 5′-labeled T/I and T/A substrates and 5′-internally labeled T/I substrate, we detected inosine-dependent 3′-exonuclease activity. Using a H214D mutant that primarily retained inosine-dependent endonuclease and 3′-exonuclease activity, we defined the kinetic property of the inosine-dependent 3′-exonuclease activity. Using 3′-labeled T/I and T/A substrates, we detected and defined non-specific 5′-exonuclease activity.
MATERIALS AND METHODS
Reagents, media and strains
All routine chemical reagents were purchased from Sigma Chemicals (St. Louis, MO, USA), Fisher Scientific (Suwanee, GA, USA) or VWR (Suwanee). Deoxyoligonucleotides were ordered from Integrated DNA Technologies Inc. (Coralville, IA, USA). LB medium was prepared according to standard recipes. GeneScan Stop Buffer consisted of 80% formamide (Amresco, Solon, OH, USA), 50 mM EDTA (pH 8.0) and 1% blue dextran (Sigma Chemicals). TB buffer (1×) consisted of 89 mM Tris base and 89 mM boric acid. TE buffer consisted of 10 mM Tris–HCl, pH 8.0 and 1 mM EDTA. Escherichia coli host strain AK53 (mrrB−, MM294) was from our laboratory collection (20). Tma endo V mutant proteins were purified as previously described (7,13).
Oligonucleotide substrates
The sequence of the 6-carboxyfluorescein (Fam)-labeld oligonucleotide is shown in Figure 1B. Oligonucleotides containing inosine were ordered from IDT and purified by polyacrylamide gel electrophoresis. The oligonucleotides were dissolved in TE buffer at a final concentration of 10 µM. The two complementary strands with the unlabeled strand in 1.2-fold molar excess was mixed and incubated at 85°C for 3 min and allowed to form duplex DNA substrates at room temperature for >30 min.
Tma endo V cleavage assays
The cleavage reaction mixtures (10 µl) containing 10 mM HEPES (N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid])–KOH (pH 7.4), 1 mM dithiothreitol, 2% glycerol, 5 mM MnCl2, 10 nM oligonucleotide DNA substrate and 100 nM Tma endo V protein were incubated at 65°C for 30 min. The reactions were terminated by adding an equal volume of GeneScan stop buffer. The reaction mixtures were then heated at 94°C for 3 min and cooled on ice. Samples (3.6 µl) were loaded onto a 16% denaturing polyacrylamide gel containing 7 M urea. Electrophoresis was conducted at 1500 V for 3.2 h using an ABI 377 sequencer (Applied Biosystems). Cleavage products and remaining substrates were visualized using GeneScan analysis software version 3.0. All experiments were repeated independently at least once. Some figures were assembled from multiple gel images.
RESULTS
Cleavage of 5′-labeled inosine- and non-inosine-containing substrates
Endonuclease V is an authentic nuclease involved in repair of deaminated base damage such as inosine. The thermostable ortholog from Thermotoga maritima has been a valuable model system for biochemical investigation given its thermostability and ease of generating site-directed mutants. Previously, we have conducted a systematic site-directed mutagenesis of Tma endo V and generated over 60 site-directed mutants (13). Using the mutants with relatively low binding affinity to inosine-containing DNA, we observed 3′-exonuclease activity of Tma endo V in the presence of Mn2+ metal cofactor. However, the biochemical properties of the exonuclease activity were not defined.
This study attempts to understand the exonuclease activity in greater detail using 36 mutants and the wild-type (wt) Tma endo V (Figure 1A). Among them, D43, E89, D110 and H214 are part of the active site that coordinates metal binding and catalysis (14). We initially tested the nuclease activity using a 5′ fluorescently labeled inosine-containing substrate (Figure 1B). The inosine was placed at the 11th position from the 3′-side so that endo V cleavage would generate a 51-mer, since it cuts at the 3′-side 1 nt from inosine. If a mutant exhibited 3′-exonuclease activity, a long ladder below the 51-mer was anticipated. Under the assay condition in which Mn2+ was used as a metal cofactor, all the mutants except D43A, D43C and D43H showed a strong 51-mer band, resulting from the specific inosine-dependent endonuclease activity (Figure 1B). The lack of activity in the D43 mutants ruled out the possibility that the observed activity was due to nuclease contamination from the expression host. Due to non-specific nuclease activities, the wt enzyme cleaved the specific 51-mer to small low molecular weight products (Figure 1B). Similar to the previous observation (13), some mutants produced a ladder that was indicative of 3′-exonuclease activity (Figure 1B). The active site mutants in general did not show the 3′-exonuclease activity except E89D, H214C, H214D and H214E (Figure 1B). Some mutants such as G113V, G121V and G136V previously known for their relatively low activity also showed minimal ladder formation (13). Consistent with the substantial reduction of binding affinity to inosine-containing DNA, H116E and in particular H116Y had little exonuclease activity (Figure 1B). Overall, the uniform ladders generated from some of the mutants indicated that Tma endo V possesses 3′-exonuclease activity.
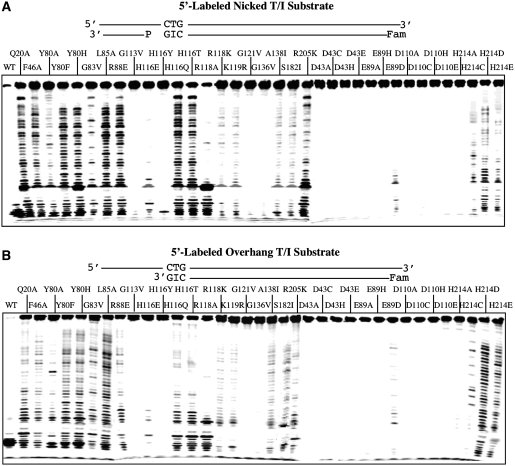
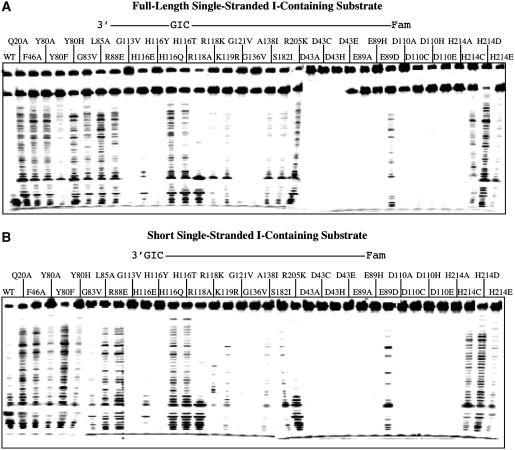
To further confirm the 3′-exonuclease activity, we assayed DNA cleavage using a synthesized nicked inosine-containing DNA with (Figure 2A, nicked T/I substrate) or without the 3′-downstream complementary DNA (Figure 2B, overhang T/I substrate). Regardless of which substrate tested, they all showed a very similar laddering pattern, suggesting that the 3′-exonuclease activity did not depend on the 3′-downstream complementary sequence. We then tested whether the same pattern would hold for single-stranded inosine-containing substrates. Indeed, the wt enzyme and the mutants all showed a similar laddering pattern with the full-length and the short single-stranded inosine-containing substrates (Figure 3).
Figure 2.
Cleavage activity of Tma endo V on 5′-labeled nicked and overhang T/I substrates. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate. The sequences of the oligonucleotides are shown in Figure 1B. (A) Cleavage activity of wt and mutant Tma endo V on 5′-labeled nicked T/I substrate. (B) Cleavage activity of wt and mutant Tma endo V on 5′-labeled overhang T/I substrate (51-mer).
Figure 3.
Cleavage activity of Tma endo V on 5′-labeled single-stranded and short I-containing substrates. (A) Exonuclease activity of Tma endo V mutants on 5′-labeled single stranded I-containing substrate. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate. (B) Exonuclease activity of Tma endo V mutants on 5′-labeled short I-containing substrate. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate.
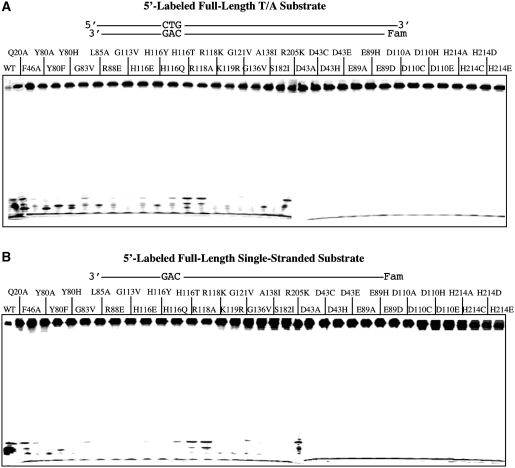
Since all the substrates tested so far contained a single inosine in the sequences, we set out to determine how the wt enzyme and mutants cleave non-inosine-containing DNA. Using a double-stranded substrate in which the T/I base pair was replaced with a T/A base pair, we measured the non-specific nuclease activities. As expected, the wt Tma endo V degraded the substrate to low molecular weight fragments due to non-specific activities [Figure 4A and (2)]. The majority of the binding defective mutants showed varying degrees of non-specific activities as indicated by the low molecular weight fragments near the bottom of the gel (Figure 4A, Q20A-R205K). The active site mutants exhibited little low molecular weight fragments (Figure 4A, D43A-H214E), suggesting that the non-specific activities were minimized. A very similar pattern was observed in the single-stranded non-inosine-containing substrate (Figure 4B). A significant difference of the 3′-exonuclease pattern between the inosine-containing and non-inosine-containing substrates was noted in some active site mutants (Figures 1 and 4). In particular, H214D mutant generated the well-formed ladder only with inosine-containing DNA, not with non-inosine-containing DNA. This result indicates that the 3′-exonuclease activity requires the presence of an inosine site in the DNA.
Figure 4.
Cleavage activity of Tma endo V on 5′-labeled non-inosine substrates. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate. (A) Cleavage activity of wt and mutant Tma endo V on 5′-labeled full-length T/A substrate. (B) Cleavage activity of wt and mutant Tma endo V on 5′-labeled full-length single-stranded substrate.
The minimal non-specific activity exhibited by the H214D mutant allowed us to better characterize the 3′-exonuclease activity without the interference of non-specific activity as seen in the wt enzyme. Taking advantage of this unique nuclease property, we studied the kinetics of the 3′-exonuclease activity using the H214D mutant. At the initial time points, we observed significant cleavage at the inosine site to generate the 51-mer fragment and a small amount of lower molecular weight fragments on a T/I substrate (Figure 5A, 2–5 min). After that, a long ladder was produced by the 3′-exonuclease cleavage (Figure 5A). The intensities of bands below 51-mer position were initially low but then became stronger as the reaction progressed to approximately below 43-mer position (Figure 5A). To verify this kinetic property, we then performed a time course analysis using the nicked inosine-containing substrate. The cleavage of the nicked inosine-containing substrate essentially showed the same laddering pattern (Fig. 5B). These results indicated that the H214D mutant did not dissociate frequently from the DNA in the initial 3′-exonuclease action. After removing ∼8-nt, the frequencies of dissociation increased, which resulted in an intensified ladder. The apparent rate constant of the inosine-dependent 3′-exonuclease activity for the nicked inosine-containing substrate was 0.025 per min.
Figure 5.
Time course analysis of cleavage activity by Tma endo V-H214D on 5′-lableled full-length and nicked T/I substrates. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM H214D and 10 nM substrate. Reactions were stopped on ice at the indicated time points, followed by addition of an equal volume of GeneScan stop buffer. The sequences of the oligonucleotides are shown in Figure 1B. (A) Time course analysis of cleavage activity by H214D on 5′-labeled full-length T/I substrate. (B) Time course analysis of cleavage activity by H214D on 5′-labeled nicked T/I substrate.
Cleavage of internally labeled inosine-containing substrate
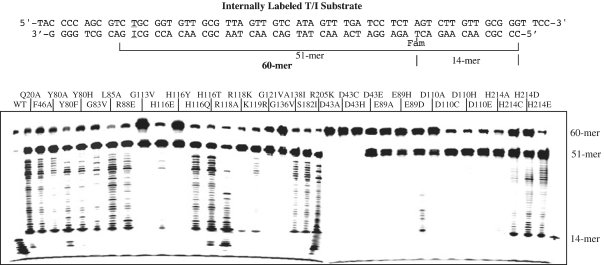
To better understand the 3′-exonuclease activity, we designed an internally labeled substrate in which the fluorescent group was moved from the 5′-end to the 14th position thymine (Figure 6). The overall cleavage pattern was similar to the 5′-end labeled T/I substrates. All the mutants except D43A, D43C and D43H generated a 51-mer specific inosine-dependent cleavage product at the 51-mer position (Figure 6). The same set of binding and active site mutants showed a 3′-exonuclease ladder. A significant deviation from the cleavage of the internally labeled T/I substrate is that for many mutants, in particular H214C, H214D and H214E, the ladder halted at around 14-mer position (Figure 6). The implication of these observations will be discussed later.
Figure 6.
Cleavage activity of Tma endo V on internally labeled T/I substrate. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate. The thymidine at position 14 from 5′-end of the bottom strand was labeled with the Fam fluorophore.
Cleavage of 3′-labeled inosine- and non-inosine-containing substrates
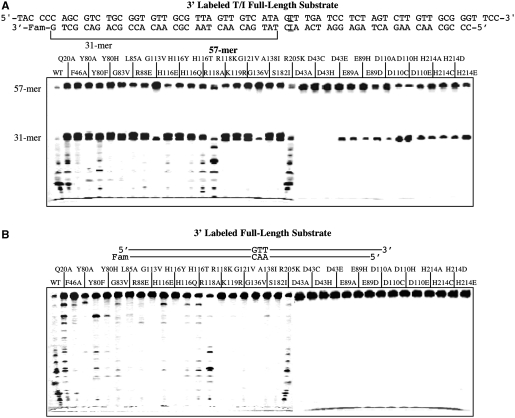
To characterize any potential nuclease activities that initiate from the 5′-side, we switched the fluorescent label to the 3′-side of the oligonucleotide substrate with the inosine placed at the 33th position from the 3′-label (Figure 7A). As expected, the endonuclease cleavage at the inosine site generated the 31-mer product (Figure 7A). Again, the wt enzyme degraded the product to low molecular weight fragments, as seen in the bottom of the gel. Many mutants, in particular Q20A, H116T, R118A and R205K, still showed non-specific activity, but only R118A and R205K showed more extensive degradation of the 31-mer. G113V, H116Y, R118K, K119R, G121V, G136V and S182I generated little low molecular weight fragments, indicating attenuated non-specific activity (Figure 7A). The active site mutants showed varying degrees of specific endonuclease cleavage at the inosine site but little non-specific activity.
Figure 7.
Cleavage activity of Tma endo V on 3′-labeled full-length T/I and T/A substrates. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate. The 3′-end of the bottom strand was labeled with the Fam fluorophore. (A) Cleavage activity of wt and mutant Tma endo V on 3′-labeled full-length T/I substrate. (B) Cleavage activity of wt and mutant Tma endo V on the 3′-labeled full-length T/A substrate.
We then tested the nuclease activities on the 3′-labeled non-inosine substrate. The wt enzyme exhibited non-specific activities to degrade the DNA to low molecular weight fragments (Figure 7B). Consistent with the results from the inosine-containing substrate, Q20A, H116T, R118A and R205K showed stronger non-specific activity than other binding defective mutants (Figure 7B, Q20A-R205K). The active site mutants exhibited minimal non-specific activity (Figure 7B, D43A-H214E).
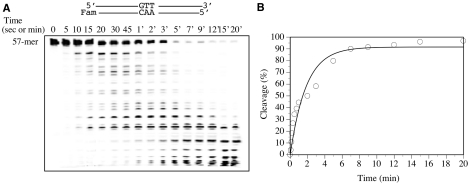
To further understand the kinetic property of the non-specific activity, we performed a time course analysis on the 3′-labeled non-inosine substrate. At the initial 5 s time point, we observed a small amount of high molecular weight fragments generated from degradation of the non-inosine substrate (Figure 8A). With increased incubation time, the intensities of the fragments also increased. After 1 min incubation, the high molecular weight fragments started to disappear, while the bands of low molecular weight started to intensify. These results indicated that Tma endo V possesses exonuclease activity that initiates non-specific degradation from the 5′-end. The apparent rate constant of the non-specific 5′-exonuclease activity was 0.55 per min (Figure 8B). The caveat is that although the apparent rate constant may be dominated by the 5′-exonuclease activity, contribution from some non-specific endonuclease activity cannot be ruled out.
Figure 8.
Time course analysis of cleavage activity by wt Tma endo V on 3′-labeled full-length T/A substrate. Cleavage reactions were performed as described in ‘Materials and Methods’ section with 100 nM enzyme and 10 nM substrate. Reactions were stopped on ice at the indicated time points, followed by addition of an equal volume of GeneScan stop buffer. (A) Time course analysis of cleavage activity by wt Tma endo V on 3′-labeled full-length T/A substrate. (B) Plot of time course analysis of cleavage activity by wt Tma endo V on 3′-labeled full-length T/A substrate. The experimental errors were estimated to be <10%. The cleavage data were fitted into a first-order equation using DeltaGraph version 5 (SPSS Inc. and Red Rock Software Inc.).
DISCUSSION
Endonuclease V is a versatile DNA repair enzyme with multiple nuclease activities on different DNA substrates, including deaminated bases and mismatched base pairs. Previous studies have indicated that endo V possesses multiple non-specific nuclease activities in addition to the inosine-specific endonuclease activity that nicks the inosine-containing strand at the 1 nt downstream of the lesion from the 3′-side (3,4,16–18). However, the nature of the multiple nuclease activities is not defined. This study takes advantage of the site-directed mutant repertoire we accumulated previously and uses different end labeled and internally labeled oligonucleotide substrates to investigate the nuclease activities. The biochemical data presented here allows us to dissect the nature of the multiple nuclease activities.
Inosine-dependent endonuclease activity
Endo V is known as a deaminated base repair enzyme. The inosine-dependent endonuclease activity is well documented and is a relevant enzymatic activity in vivo (1,3–5,8,9,11,21,22). The inosine-dependent endonuclease activity is the primary enzymatic activity that is well maintained within the protein structure (13,14). This is evidenced by the retention of the activity by most of the Tma endo V mutants tested except three active site mutants (D43A, D43C, D43H) (Figures 1 and 3). The inosine-dependent endonuclease activity allows endo V to initiate the removal of inosine in DNA.
Inosine-dependent 3′-exonuclease activity
Previously, we detected 3′-exonuclease activity from Tma endo V using binding defective mutants (13). Here we establish that the 3′-exonuclease activity is inosine-dependent. Support for this notion primarily comes from biochemical analysis of some active site mutants. H214C, H214D and H214E failed to show any ladder on the 5′-labeled non-inosine substrate but formed a distinct ladder on all 5′-labeled inosine-containing substrates (Figures 1–4), suggesting that the 3′-exonuclease activity relies on the primary cut at the inosine site to initiate the secondary enzymatic action. The observation of DNA cleavage activity on the single-stranded inosine-containing substrate but not the non-inosine-containing DNA indicates that the 3′-exonuclease does not rely on a nick or a recessive end; rather, it depends on the existence of inosine (Figures 3 and 4B).
The more interesting kinetic property of the inosine-dependent 3′-exonuclease activity is revealed from the time course analysis using the H214D mutant that has minimal non-specific nuclease activity (Figure 5). The ladders generated by H214D have a very distinct feature, i.e. the laddering patterns show little change over time. For example, the pattern at 60 min is quite similar to that at 7 min (Figure 5B). If the ladders are generated by non-specific 3′-exonuclease activity, one would expect to see an initial accumulation of high molecular weight products. Later on, the high molecular weight products would gradually disappear, which is accompanied by accumulation of low molecular weight products. This will be similar to the laddering pattern observed by the non-specific 5′-exonuclease activity shown in Figure 8A. However, this kind of kinetic behavior did not occur. As such, these data indicate that the 3′-exonuclease action only occurs once on an inosine-containing DNA. After the enzyme removes the inosine lesion and dissociates from the DNA, it has little chance to re-bind to the 3′-side to reinitiate a non-specific 3′-exonulcease action. As a result, an even laddering pattern is generated over time. Thus, the kinetic pattern reinforces the notion that Tma endo V contains inosine-dependent 3′-exonuclease activity.
An alternative explanation for the even laddering pattern throughout the time course is that it is caused by non-specific endonuclease action that allows the enzyme to randomly nick the DNA. We found this explanation implausible. First, H214D mutant did not show any laddering pattern on the non-inosine substrates (Figure 4). Second, if it had been caused by non-specific endonuclease action, we could have seen the ladder generated by H214D mutant extend below the 14-mer mark in the internally labeled T/I substrate due to the multiple cuts around the Fam position (Figure 6). Instead, the ladder stopped at the 14-mer position (Figure 6). Finally, if it had been caused by non-specific endonuclease, the substrates should have been degraded to low molecular weight products over time (Figure 5).
Does the wt enzyme possess non-specific 3′-exonuclease activity then? Although the data presented here may not give a definitive answer, we tend to believe the answer is no, based on the following two reasons. First, no obvious ladders were observed with 5′-labeled non-specific substrates (Figure 4). Second, the time course data from the 3′-labeled substrate does not show any low molecular weight fragments at the bottom of the gel (Figure 8A), which would indicate non-specific 3′-exonulcease activity.
5′-exonuclease activity
It has been proposed that Tma or E. coli endo V enzymes contain 5′-exonuclease activity (16–18). We measured the 5′-exonuclease activity using 3′-labeled substrates. A ladder formed by some mutants such as Q20A and R205K is a good indication of 5′-exonuclease activity (Figure 7). The 5′-exonuclease activity is non-specific since it does not depend on the existence of an inosine site (Figure 7). In addition, the generation of low molecular weight products as shown in Figure 4 is also indicative of non-specific 5′-exonuclease activity. A more definitive proof of the 5′-exonuclease activity comes from the time course analysis of the 3′-labeled substrate (Figure 8A). At the initial time points, the 5′-exonuclease action produces a ladder consisting of higher molecular weight fragments (Figure 8A, 5–20 s). At the later time points, the continuous 5′-exonuclease action results in the disappearance of the high molecular weight fragments with concurrent accumulation of low molecular weight fragments (Figure 8A, 5–20 min). On the other hand, a prominent non-specific endonuclease would result in generation of fragments of all sizes initially and later accumulation of low molecular weight fragments.
Nuclease activities and inosine repair
Since endo V cuts at the 3′-side of the lesion, the inosine damage still remains in the DNA. Additional enzymatic actions are required for the removal of the damage. Based on the results obtained from an oligonucleotide-based study in E. coli, after nicking by the inosine-dependent endonuclease activity of E. coli endo V, 2-nt from 5′-side and 3 nt from 3′-side are removed to create a gap (12). The enzymes responsible for the removal of inosine and subsequent creation of the 5-nt gap in the repair process are as yet unidentified. However, the existence of inosine-dependent 3′-exonuclease and non-specific 5′-exonuclease activities raises the possibility that endo V, facilitated by other factor(s) than the Mn2+ used in the in vitro study, could potentially be involved in the creation of the short gap during the repair process. If this is the case, then endo V can latch onto DNA and search for an inosine site. Once the enzyme nicks at the 3′-side of an inosine site, it may recruit other factor(s) to allow it to use its inosine-dependent 3′-exonuclease and non-specific 5′-exonuclease to remove the inosine in the DNA. The inosine-dependent 3′-exonuclease activity in vivo may be limited to remove the inosine lesion from the first few nucleotide instead of extensive nucleotide degradation. The gap created by the multiple enzymatic actions can then be filled by a DNA polymerase and the resulting nick sealed by a DNA ligase. Interestingly, in the case of mismatch repair, human MutLα is found to be a latent endonuclease that can be activated in the presence of a mismatch, ATP, Mg2+ and multiple protein factors (23). This latent endonuclease activity from MutH-less species is activated when Mn2+ is used as a metal factor (23–26). Of course, it is also possible that the in vitro exonuclease activities are irrelevant to the in vivo process. In this scenario, independent nuclease(s) instead of endo V is involved in the post-inosine cleavage process. Regardless of whether the multiple nuclease activities are physiologically relevant, this work reveals a series of concerted enzymatic functions that may define the inner workings of endonuclease V.
FUNDING
Cooperative State Research, Education and Extension Service/US Department of Agriculture (SC-1700274, technical contribution No. 5767); Department of Defense (W911NF-05-1-0335, W911NF-07-1-0141, W81XWH-10-1-0385); National Institutes of Health (GM090141); Calhoun Honors College of Clemson University; Howard Hughes Medical Institute Undergraduate Fellowship. Funding for open access charge: Department of Defense.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Bernard Weiss and Dr Yoke Wah Kow at Emory University and members of the Cao laboratory for stimulating discussions.
REFERENCES
- 1.Feng H, Klutz AM, Cao W. Active Site Plasticity of Endonuclease V from Salmonella typhimurium. Biochemistry. 2005;44:675–683. doi: 10.1021/bi048752j. [DOI] [PubMed] [Google Scholar]
- 2.Hitchcock TM, Gao H, Cao W. Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V. Nucleic Acids Res. 2004;32:4071–4080. doi: 10.1093/nar/gkh747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang J, Lu J, Barany F, Cao W. Multiple Cleavage Activities of Endonuclease V from Thermotoga maritima: recognition and strand nicking mechanism. Biochemistry. 2001;40:8738–8748. doi: 10.1021/bi010183h. [DOI] [PubMed] [Google Scholar]
- 4.Yao M, Hatahet Z, Melamede RJ, Kow YW. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem. 1994;269:16260–16268. [PubMed] [Google Scholar]
- 5.Yao M, Hatahet Z, Melamede RJ, Kow YW. Deoxyinosine 3′ endonuclease, a novel deoxyinosine-specific endonuclease from Escherichia coli. Ann. NY. Acad. Sci. 1994;726:315–316. doi: 10.1111/j.1749-6632.1994.tb52837.x. [DOI] [PubMed] [Google Scholar]
- 6.He B, Qing H, Kow YW. Deoxyxanthosine in DNA is repaired by Escherichia coli endonuclease V. Mutat Res. 2000;459:109–114. doi: 10.1016/s0921-8777(99)00063-4. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Lu J, Barany F, Cao W. Mutational analysis of endonuclease V from Thermotoga maritima. Biochemistry. 2002;41:8342–8350. doi: 10.1021/bi015960s. [DOI] [PubMed] [Google Scholar]
- 8.Yao M, Kow YW. Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem. 1995;270:28609–28616. doi: 10.1074/jbc.270.48.28609. [DOI] [PubMed] [Google Scholar]
- 9.Guo G, Weiss B. Endonuclease V (nfi) mutant of Escherichia coli K-12. J. Bacteriol. 1998;180:46–51. doi: 10.1128/jb.180.1.46-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss B. Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat Res. 2001;461:301–309. doi: 10.1016/s0921-8777(00)00062-8. [DOI] [PubMed] [Google Scholar]
- 11.Schouten KA, Weiss B. Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat Res. 1999;435:245–254. doi: 10.1016/s0921-8777(99)00049-x. [DOI] [PubMed] [Google Scholar]
- 12.Weiss B. Removal of deoxyinosine from the Escherichia coli chromosome as studied by oligonucleotide transformation. DNA Repair. 2008;7:205–212. doi: 10.1016/j.dnarep.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng H, Dong L, Klutz AM, Aghaebrahim N, Cao W. Defining amino acid residues involved in DNA-protein interactions and revelation of 3′-exonuclease activity in endonuclease V. Biochemistry. 2005;44:11486–11495. doi: 10.1021/bi050837c. [DOI] [PubMed] [Google Scholar]
- 14.Feng H, Dong L, Cao W. Catalytic mechanism of endonuclease V: a catalytic and regulatory two-metal model. Biochemistry. 2006;45:10251–10259. doi: 10.1021/bi060512b. [DOI] [PubMed] [Google Scholar]
- 15.Dalhus B, Arvai AS, Rosnes I, Olsen OE, Backe PH, Alseth I, Gao H, Cao W, Tainer JA, Bjoras M. Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nat. Struct. Mol. Biol. 2009;16:138–143. doi: 10.1038/nsmb.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao H, Huang J, Barany F, Cao W. Switching base preferences of mismatch cleavage in endonuclease V: an improved method for scanning point mutations. Nucleic Acids Res. 2007;35:e2. doi: 10.1093/nar/gkl916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pincas H, Pingle MR, Huang J, Lao K, Paty PB, Friedman AM, Barany F. High sensitivity EndoV mutation scanning through real-time ligase proofreading. Nucleic Acids Res. 2004;32:e148. doi: 10.1093/nar/gnh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao M, Kow YW. Cleavage of insertion/deletion mismatches, flap and pseudo-Y DNA structures by deoxyinosine 3′-endonuclease from Escherichia coli. J. Biol. Chem. 1996;271:30672–30676. doi: 10.1074/jbc.271.48.30672. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Gao H, Schallhorn KA, Harris RM, Cao W, Ke PC. Lesion recognition and cleavage by endonuclease V: a single-molecule study. Biochemistry. 2007;46:7132–7137. doi: 10.1021/bi6024534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barany F, Danzitz M, Zebala J, Mayer A. Cloning and sequencing of genes encoding the TthHB8I restriction and modification enzymes: comparison with the isoschizomeric TaqI enzymes. Gene. 1992;112:3–12. doi: 10.1016/0378-1119(92)90296-2. [DOI] [PubMed] [Google Scholar]
- 21.Yao M, Kow YW. Further characterization of Escherichia coli endonuclease V. J. Biol. Chem. 1997;272:30774–30779. doi: 10.1074/jbc.272.49.30774. [DOI] [PubMed] [Google Scholar]
- 22.Moe A, Ringvoll J, Nordstrand LM, Eide L, Bjoras M, Seeberg E, Rognes T, Klungland A. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 24.Fukui K, Nishida M, Nakagawa N, Masui R, Kuramitsu S. Bound nucleotide controls the endonuclease activity of mismatch repair enzyme MutL. J. Biol. Chem. 2008;283:12136–12145. doi: 10.1074/jbc.M800110200. [DOI] [PubMed] [Google Scholar]
- 25.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauris J, Evans TC. Adenosine triphosphate stimulates Aquifex aeolicus MutL endonuclease activity. PLoS One. 2009;4:e7175. doi: 10.1371/journal.pone.0007175. [DOI] [PMC free article] [PubMed] [Google Scholar]