Abstract
Expression profiling experiments usually provide a static snapshot of messenger RNA (mRNA) levels. Improved understanding of the dynamics of mRNA synthesis and degradation will aid the development of sound bioinformatic models for control of gene expression. We studied mRNA stability in proliferating and differentiated myogenic cells using whole-genome exon arrays and reported the decay rates (half life) for ∼7000 mRNAs. We showed that the stability of many mRNAs strongly depends on the differentiation status and contributes to differences in abundance of these mRNAs. In addition, alternative splicing turns out to be coupled to mRNA degradation. Although different splice forms may be produced at comparable levels, their relative abundance is partly determined by their different stabilities in proliferating and differentiated cells. Where the 3′-untranslated region (3′-UTR) was previously thought to contain most RNA stabilizing and destabilizing elements, we showed that this also holds for transcript isoforms sharing the same 3′-UTR. There are two splice variants in Itga7, of which the isoform with an extra internal exon is highly stable in differentiated cells but preferentially degraded in the cytoplasm of proliferating cells. In conclusion, control of stability and degradation emerge as important determinants for differential expression of mRNA transcripts and splice variants.
INTRODUCTION
Although the abundance of messenger RNAs (mRNAs) in the cell is the net result of mRNA synthesis and degradation, expression profiling experiments usually take a snapshot of the mRNA levels in the cell and do not capture the dynamics of mRNA synthesis and breakdown. This lack of information on the dynamic component of the regulation of mRNA levels limits biological investigations in general and proper modeling of transcriptional networks in systems biology approaches in particular. When mRNA synthesis is shut down completely, the effect may be immediate for very short-lived mRNAs but only manifest after hours or days for mRNAs with long half lives. The current study fills in part of this gap and surveys mRNA degradation rates during muscle differentiation on a genome-wide scale. With the availability of these data, the genetic network orchestrating muscle differentiation can be revised and improved.
From the results of previous genome-wide analyses of mRNA degradation rates, we know that there are large differences between the stabilities of individual mRNAs, and that mRNA stability is strongly related to the molecular function of the encoded protein (1–8). However, the mechanisms underlying differential stability of transcripts remain poorly understood. MicroRNA (miRNA)-mediated mRNA degradation emerged as an important controlling factor for mRNA half life. Earlier research also characterized a plethora of RNA binding proteins that may affect mRNA stability (9). Recent studies in differentiating embryonic stem cells (7) and T-cells (6) demonstrated that mRNA stability may be affected by the cell’s biological state. Furthermore, an important subgroup of transcripts use alternative polyadenylation sites depending on the differentiation status (10). This results in 3′-untranslated regions (3′-UTRs) with variable lengths and probably contributes to the differential stability of these transcripts (10). The deciding factors determining the choice for shorter or longer 3′-UTRs are currently unknown.
We were interested in the control of decay rates during myogenic differentiation and studied proliferating and differentiated myogenic C2C12 cells, a well-established system to study differentiation of muscle cells. Cells were treated with actinomycin D to arrest de novo RNA synthesis, with sampling at short time intervals. We used Affymetrix whole-genome exon arrays covering all known and predicted exons. Where previous genome-wide studies determined expression levels and stability of the 3′-ends of transcripts, these arrays allowed us to assess expression level per exon and thereby, for the first time, the effect of alternative splicing on mRNA decay rates. Our results demonstrated that changes in mRNA abundance during differentiation of muscle cells are correlated with changes in decay rates and that the ratios of specific splice variants are controlled at the level of mRNA stability.
MATERIALS AND METHODS
Cell culture, RNA isolation, microarray hybridization
C2C12 cells were grown in proliferation medium [Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 2% Glutamax, 1% glucose and penicillin/streptomycin]. Myogenic differentiation was induced by shifting to differentiation medium (DMEM with 2% FBS, 2% Glutamax, 1% glucose and penicillin/streptomycin). C2C12 cells were maintained in differentiation medium for 8 days, when long and multinucleated myotubes had formed. Cultures of proliferating and differentiated C2C12 muscle cells in 6-well dishes containing 3-ml medium were treated with 5 µg/ml actinomycin D to arrest the transcription machinery. At seven different time points after addition of actinomycin D (0, 10, 20, 30, 60, 150, 480 min), cells were lysed and RNA was isolated with the Machery Nagel Nucleospin RNAII Kit. There was no notable decrease in cell viability within this time period. The experiment was performed in duplicate. After depletion of ribosomal RNAs, mRNA was reverse transcribed with random primers containing a T7 tag, and labeled with biotin during the T7 RNA amplification (which retains strand specificity), according to standard Affymetrix protocols. Equal amounts of amplified RNA were hybridized to 28 Affymetrix Mouse Exon v1 arrays [7 time points × 2 replicates × 2 conditions (proliferating/differentiated)].
Affymetrix exon arrays
Affymetrix Mouse Exon v1 arrays contain 1 236 087 non-control probe sets. Most probe sets consist of four probes and the intensity value of each probe set is the robust mean of the individual probe sets. Most exons are covered by at least one probe set. There is a subset of exons, called core exons, which represent all the exons for which there is substantial experimental proof for their existence. Next to the core set, the array covers a huge set of computationally predicted exons. The intensities of probe sets in most of these latter exons (in our experiment 91 versus 51% for core exons) are not above background, and these have not been considered in the analysis described here. Data from these arrays can be analyzed at the exon or the gene level. The presented gene level summarization, giving only one expression value per gene, considers the intensities of probes in core exons only.
Data analysis—normalization
In our experimental set-up, normalization is not straightforward, since an important assumption underlying standard normalization procedures, equality of the total amount of mRNA under all conditions studied, is violated. Despite the decrease in the total amount of mRNA in the cell at later time points, equal amounts of RNA have been hybridized to the chips. This means that the samples from the later time points will be enriched for stable transcripts and depleted for fast decaying transcripts. We tried to use spike-in RNAs to correct for the differences in input RNA but this was not successful due to the low number of spike-in RNAs used. Thus the only alternative was to use a standard normalization method, with the intensities of more stable transcripts increasing at later time points.
The median polish summarization method [as implemented in RMA (11)] was applied to summarize the intensities of probes in a probe set and probes in a gene. VSN [variance stabilization and normalization (12)] was used for normalization. VSN and RMA normalization generated similar values in the high intensity range. In the lower intensity range, the variance stabilizing properties of the VSN algorithm corrects for noise-induced artificially inflated differences between arrays in near-background reads. As an additional advantage, the VSN data are on a natural logarithm scale compatible with our exponential decay model, whereas the RMA data are on a log2 scale. Since the arrays contain many negative and positive control spots, the effect of removing those before the normalization was investigated. This had no effect on the normalization of the non-control spots. As a threshold for background intensity levels, we used the mean + 2 SD of around 4000 negative control probes on the array.
All normalization and subsequent data analysis was performed in the statistical programming language R (13).
Raw and normalized data have been submitted to GEO and are available under series GSE14387.
Data analysis—model fit
To model decay rates, a standard linear model (1) was fitted with normalized intensity Y of each probe set j as dependent variable and time as independent, continuous variable.
| (1) |
where time is a vector (0, 10, 20, 30, 60, 150, 480); αj represents the intercept (estimated expression level at t = 0); βj represents the slope (negative of the decay constant).
The appropriateness of this model is discussed in the Supplementary Methods and in the caption of Supplementary Figure S1. Replicate time points were independent data points in the model. Two separate models were fit for the proliferating and differentiated cells. Decay constants were said to be significantly different between proliferating and differentiated cells when there was no overlap between their 95% confidence intervals (as determined from the standard error on β, and given the t-distribution and 12 degrees of freedom). Decay constants for entire transcripts were calculated using the exact same model, but after summarizing core probe sets in a transcript into a single expression value by RMA. The half life is given by –ln(2)/β.
For identification of probe sets with a behavior different from the other probe sets in the gene (possible alternative splicing events), we considered the mixed effect model (2) (nlme package; lme function in R), where time is a continuous variable and treated as a fixed effect, where the probe sets were considered as random effects and where we included an interaction term between the probe sets and the differentiation status. In formula form (14):
| (2) |
where bj ∼ N (0, σ12), bjk ∼ N (0, σ22), εijk ∼ N (0, σ2) and where Yijk represents normalized intensity of probe set j (j = 3, … , n probe sets in the gene) with differentiation status k (k = 1, 2) in gene i; time represents time vector (0, 10, 20, 30, 60, 150, 480); β represents fixed effect; bj represents random intercept; bjk represents the interaction between the probe set and differentiation status and εijk represents error term.
Data analysis—annotation and visualization
The R package Xmap (15), interfacing to a local Ensembl install (mouse release 47; NCBI genome build 37) was used for annotation and visualization of the exon-level data.
Quantitative PCR analysis
Quantitative polymerase chain reaction (qPCR) analysis with SYBR-Green detection was performed as described earlier (16). Bgn was used in the RNA decay studies as a reference gene to correct for differences in input cDNA. Bgn was found to be extremely stable and did not change significantly in Ct value over the time period analyzed. Gapdh was used as a reference gene for the nuclear versus cytoplasmic comparison.
3′-RACE
To determine the polyadenylation sites in the Itga7 transcript, we performed 3′-RACE (rapid amplification of cDNA ends). We performed a reverse transcription (RevertAid RNaseH-M-MuLV, MBI-Fermentas) on 500 ng of total RNA starting at the polyA site with an anchored primer oligo-dT primer (sequence: GCTCGCGAGCGCGTTTAAACGCGCACGCGTTTTTTTTTTTTTTTTTTVN). Subsequently, we performed two rounds of PCR [30 cycles, Faststart Taq (Roche)]. The forward primer was designed in exon 22 (sequence: CTGTTAGTCCTGGCCTTGCT); the sequence of the reverse primer in the first round was GCTCGCGAGCGCGTTTAAAC, whereas the sequence of the reverse primer in the second round of nested PCR was GCGTTTAAACGCGCACGCGT. Bands were excised from agarose gel and sequenced on an ABI-3700 sequencer.
Isolation of nuclei
Cells were scraped in phosphate buffered saline and pelleted by centrifugation for 5 min at 1500g. The cells were resuspended in cold lysis buffer (10 mM Tris–HCl pH 7.4, 3 mM MgCl2, 10 mM NaCl, 1mM EDTA) and pelleted by centrifugation for 5 min at 1500g. After resuspending in lysis buffer including 0.5% (v/v) Triton X-100, cells were disrupted in a Dounce homogenizer with 4 × 10 strokes of the pestle. Nuclei were pelleted by centrifugation for 2 × 5 min at 1500g and washed with lysis buffer (without Triton X-100). Nuclei were purified by sucrose gradient centrifugation. To this end, a centrifuge tube was filled with 4 ml of high sucrose buffer (2 M sucrose, 5 mM MgAc, 0.1 mM EDTA, 10 mM Tris–HCl pH 8.0, 1 mM DTT) and nuclei were added after mixing with 4.4 ml of low sucrose buffer [0.32 M sucrose, 3 mM CaCl2, 2 mM MgAc, 0.1 mM EDTA, 10 mM Tris–HCl pH 8.0, 1 mM DTT, 0.5% (v/v) Triton X-100], with subsequent centrifugation for 30 min at 30 000g in an SW41 rotor. Pellet was resuspended in 300 µl of storage buffer [50 mM Tris–HCl pH 8.3, 40% (v/v) glycerol, 2 mM MgCl2, 0.1 mM EDTA], aliquotted and snap frozen in liquid nitrogen.
RESULTS
Experimental setting
Cultures of proliferating C2C12 muscle precursor cells and cultures of C2C12 cells differentiated to elongated and fused myotubes were treated with actinomycin D to arrest the transcription machinery. At seven different time points after addition of actinomycin D (0, 10, 20, 30, 60, 150, 480 min), RNA was isolated and hybridized to Affymetrix Mouse Exon v1 arrays (in duplicate, 28 arrays in total). With the selected time intervals, it is possible to accurately estimate the degradation rates of relatively short-lived transcripts. For very stable transcripts (t1/2 > 480 min) the estimate becomes inaccurate, but longer incubation times resulted in a significant decrease in cell viability severely complicating the analysis.
To model mRNA degradation, we consider an exponential decay model. This model becomes linear on the logarithmic scale used to report the normalized exon-array signals (model 1; see Supplementary Methods and Supplementary Figure S1 for details on the model and the goodness-of-fit). The slope in this linear model is the (negative of the) decay constant, whereas the intercept reflects the expression level before addition of actinomycin D.
Estimation of decay rates
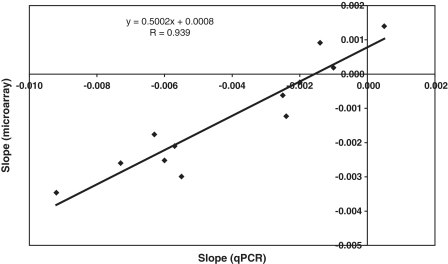
All mRNAs decay over time. The normalization method used (see ‘Materials and Methods’ section), centers the average slopes around zero (Figure 1A). Thus, stable transcripts appear to have positive slopes relative to the average, whereas transcripts with fast degradation will have negative slopes. The relative differences in decay rates (slow decaying versus fast decaying) can be well estimated using this model (Figure 1B, Supplementary Figure S1 and Supplementary Methods). The maximum slope for expressed genes is 0.0028 intensity units/minute, the mean is −0.00042 (median −0.00020), and the minimum −0.0073. If we assume that the slowest decaying transcript is stable over the period analyzed, all slopes would need to be corrected with −0.0028. The half life is calculated as −ln(2)/slope and the shortest half life we observed is then 69 min, the median half life of all expressed transcripts 231 min. This is in a similar range as reported before for proliferating C2C12 cells (8). The decay rates determined on the microarrays correlate well with those measured by quantitative PCR [Figure 2, Pearson’s correlation coefficient: 0.94 (P = 3.2 × 10−6)], in spite of the array data appearing to overestimate the half life of short-lived transcripts and to underestimate those of stable transcripts. Relative decay rates for all genes with detectable expression (6852 in total) are reported in Supplementary Table S1.
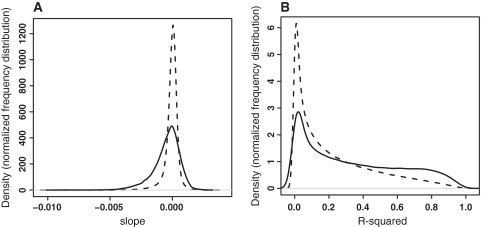
Figure 1.
Estimated relative decay rates measured on microarrays. Distribution of the slopes (negative of degradation rate constants) (A) and R2 values of the linear model (B) for all probe sets (dashed line) and for probe sets in genes expressed above background (solid line). The majority of probe sets do not give detectable signals and have very flat (slope near zero) and noisy (low R2 values) decay curves. In contrast, expressed genes frequently demonstrate more positive or negative slopes, and their decay rate constants can be reliably measured (high R2 values). This is expected since background signals remain constant over time, thus generating a slope close to zero, whereas true signals tend to increase or decrease, generating a slope away from zero.
Figure 2.
Correlation of microarray- and quantitative PCR-based degradation rates. For both microarray and qPCR assays, the natural logarithm of the expression level was plotted against the time after start of actinomycin D treatment and a linear curve was fitted. The slope of this curve is the negative of the decay constant. The slopes for the Bmp4, Ccng2, Cebpg, Pde4dip-short, Pde4dip-long and Itga7-exon27 genes in proliferating and differentiated myoblasts measured with qPCR with primers designed in the 3′-UTR are plotted on the x-axis, and the slopes measured with microarray probe sets in the same regions are plotted on the y-axis. The correlation coefficient is the Pearson correlation coefficient.
Factors determining mRNA half life
It has been noted that mRNA stability appears to correlate with the cellular function of the encoded proteins (1–8). To study this correlation in our data we functionally classified the most stable and unstable mRNA transcripts in proliferating and differentiated cells (Table 1). We see a similar correlation. Stable RNAs code for proteins involved in cell maintenance and structure, short-lived RNAs code for proteins involved in regulation of gene expression, including histones. RNAs coding for structural and contractile muscle proteins, which are only expressed in differentiated cells, appear to be very stable. Transcripts in the category ‘muscle development’ are either very short- or very long-lived, because this category contains both regulatory and structural proteins.
Table 1.
Functional classification of fast and slow decaying transcripts in proliferating and differentiated cells
| Fast | P-value | Slow | P-value | |
|---|---|---|---|---|
| Proliferating | Regulation of cellular processes | 3.7 × 10−8 | DNA-dependent DNA replication | 3.6 × 10−4 |
| Establishment and/or maintenance of chromatin architecture | 7.1 × 10−4 | |||
| Differentiated | Regulation of cellular procesess | 4.6 × 10−10 | Muscle development | 1.1 × 10−4 |
| Regulation of transcription | 8.3 × 10−5 | Muscle contraction | 7.6 × 10−28 | |
| Negative regulation of protein kinase activity | 3.1 × 10−7 | |||
| Cell cycle | 5.0 × 10−5 | |||
| Development | 2.1 × 10−7 | |||
| Muscle development | 1.3 × 10−4 | |||
Gene annotation was derived from Gene Ontology. The biological processes significantly overrepresented in the 100 fastest and slowest decaying transcripts in proliferating and differentiated cells are displayed, together with the P-value from the hypergeometric test used for testing overrepresentation. Given multiple testing issues, P < 1 × 10−4 was considered to be significant.
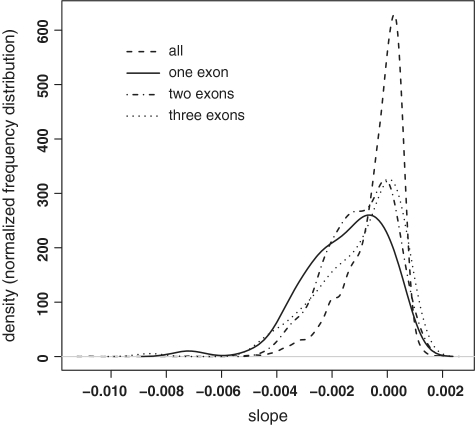
Short mRNAs tend to be less stable than average (Supplementary Figure S2). The median half lives of mRNAs consisting of one, two, three exons in proliferating cells are 172, 189 and 199 min, respectively. This is significantly shorter (P ≤ 1 × 10−5 Kolmogorov–Smirnov test) than the average half life in proliferating cells (237 min). Similar results were obtained for differentiated cells. Figure 3 visualizes the statistically significant shift to shorter half lives in mRNAs with three, two or one exons.
Figure 3.
One, two and three exon mRNAs have shorter half lives. Distribution of slopes (negative of degradation rate constants) for all mRNAs (dashed line), and mRNAs consisting of one (solid line), two (dash-dotted line) or three (dotted line) exons. The latter three distributions demonstrate statistically significant shifts to the left (shorter half lives), with Kolmogorov–Smirnov P-values of 4.3 × 10−9, 1.3 × 10−7 and 1.2 × 10−5, for one, two and three exons, respectively. mRNAs with 4 exons were only marginally different from the overall distribution (P = 0.015). The plot is for proliferating cells only, but the plot and results for differentiated cells are highly similar.
There is no correlation between 3′-UTR length and transcript stability (Supplementary Figure S3).
mRNA stability is dependent on the cellular state
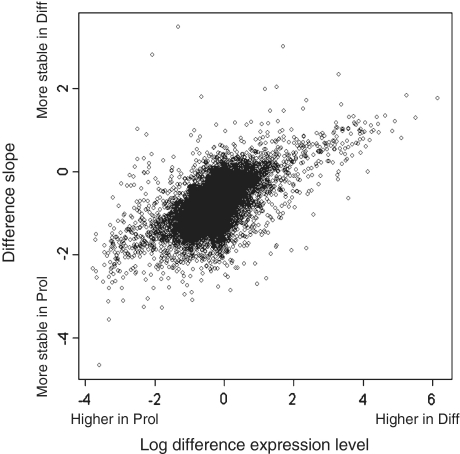
We calculated the differences in decay rates between differentiated and proliferating cells. There were 170 mRNAs displaying significant differences in stability between the two states; 99 of those were more stable in proliferating cells and 71 were more stable in differentiated cells (Supplementary Table S1). Interestingly, many of the mRNAs with differences in stability were also differentially expressed between the two conditions. Table 2 summarizes some of the results for transcripts playing an established role in muscle development. Slower degradation in differentiated than in proliferating cells goes with higher expression levels in differentiated cells, and vice versa. Myog and Cdkn1a (p21), two important regulators of differentiation previously shown to have increased expression and stability in differentiated cells (17–19) are amongst the genes with clearly different mRNA degradation rates: The half life of the Myog mRNA increased from 165 min in proliferating cells to 268 min in differentiated cells; the half life of the Cdkn1a mRNA increased from 138 to 371 min. The generality of these observations is illustrated by the strong positive correlation obtained after plotting the differences in gene expression against the differences in decay constants (Figure 4, Pearson’s correlation coefficient: 0.57; P < 2.2 × 10−16). Thus, control of mRNA stability and degradation emerges as an important determinant for the regulation of steady-state mRNA levels.
Table 2.
Half lives of myogenesis-related genes change during differentiation
| Gene | Fold-change differentiated versus proliferating cells | Half life proliferating cells (min) | Half life differentiated cells (min) |
|---|---|---|---|
| Tnc | −3.86 | 444 | 229 |
| Mbnl3 | −2.72 | 342 | 264 |
| Musk | 2.82 | 246 | 438 |
| Lrp4 | 3.29 | 236 | 416 |
| Cdkn1a | 3.73 | 138 | 371 |
| Tnnt2 | 4.75 | 242 | 500 |
| Tmod1 | 7.23 | 201 | 455 |
| Acta1 | 7.75 | 322 | 555 |
| Myog | 9.09 | 165 | 268 |
| Cacna1s | 17.01 | 229 | 798 |
Only genes annotated with ‘muscle development’ in Gene Ontology and a minimum absolute fold change of two are shown.
Figure 4.
Correlation between differential expression and differences in decay rates. Correlation of the differences in slopes (per minute) between differentiated and proliferating cells (y-axis) and differences in normalized expression levels between differentiating and proliferating cells (x-axis) for gene level data (Pearson’s correlation: R = 0.57). Transcripts that are higher expressed under differentiating conditions (right side of the plot) seem to have less negative slopes (upper part of the plot) and thus to be more stable under differentiating conditions. Probe set level data give similar results but are more difficult to visualize since the plot contains many more data points.
Relative abundance of splice isoforms is controlled by mRNA stability
The observation that the cellular state determines the overall decay rates of transcripts triggered us to investigate the role of mRNA degradation in controlling the ratio between different splice isoforms. To this end, we used the mixed effect model (2) to identify genes where the decay constant of a particular probe set differed more than 2 SDs from the decay constant of all other probe sets in the gene. Sixty-nine probe sets met these criteria (Supplementary Table S2). After further filtering for probes that show a low intensity across all conditions, we were left with 39 probe sets in 36 genes meeting our very stringent criteria. The positions of these probe sets in the mRNAs were evaluated (Table 3). As expected, we found 10 probe sets in 3′-UTRs, the length of which is known to affect transcript stability (10). The mRNAs include Ankrd2, Myom2, Itgb1, Cacna1s, Pde4dip and Xirp1, with a known function in muscle development, highlighting the importance of the shift in 3′-UTR usage during myogenic differentiation. Another 13 probe sets were located in the 5′-UTR and one in the coding part of a first exon. Given the known variability in the use of transcription start sites (20), this implicates that the choice for a transcription start site and thus the length of the 5′-UTR may influence the stability of the transcript. A further 15 probe sets were located in internal exons, many of them known to be subjected to alternative splicing. This demonstrates for the first time that transcript isoforms that differ in just one or two internal exons may differ in stability. This category includes mRNAs coding for structural muscle proteins, like Myom1, Myom2, Itga7, Tnnt3 and Atp1a2.
Table 3.
Location of probe sets with aberrant slopes as compared with other probe sets in the gene
| Number of probe sets | Probe set location | Genes |
|---|---|---|
| 14 | 5′-UTR, alternative 5′-UTR, or coding part of first exon | Tbrg4, Foxm1, Casq1, Trim37, Ly75, Casq2, Ezh2, Atp2a1, Ncapd3, Serpine1, Osbp2, Mamdc2, Bub1b, Mybph |
| 10 | 3′-UTR or alternative 3′-UTR | Kif20a, Ankrd2, Myom2, Foxo6, Cp, Mcm5, Itgb1, Cacna1s, Pde4dip, Xirp1 |
| 9 | Internal exon subject to alternative splicing | Adssl1, Myom1a, Itga7a, Pygm, Rbm9, Sec24c, Tnnt3 |
| 6 | Internal exon not knownb to be subject to alternative splicing | Tcfap4, Atp1a2a, Cabc1, Myom2, Mmp15 |
aTwo probe sets in two different exons affected.
bAccording to Ensembl annotation.
The decay of the transcript isoforms of two mRNAs was investigated in more detail: Pde4dip representing one of the mRNAs with differential use of 3′-UTRs, and Itga7, representing one of the mRNAs subject to alternative splicing of internal exons.
Pde4dip
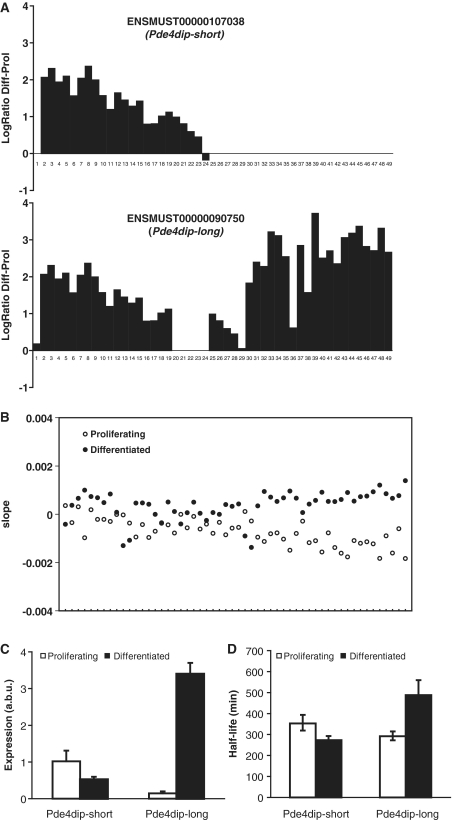
Pde4dip codes for the protein myomegalin, a protein associated with the Z-discs of skeletal muscles (21). In the ENSEMBL database, there are three annotated transcripts for this gene: a long transcript (indicated as Pde4dip-long) and a shorter transcript with an alternative 3′-UTR (indicated as Pde4dip-short) (Figure 5A). The expression levels of all exons are higher in differentiated than in proliferating cells (Figure 5A, note that exon expression levels reflect the cumulative expression of all transcripts containing that exon). Quantitative PCR analysis with primers specific for the 3′-UTRs of the long and short isoforms demonstrated that the short isoforms are expressed at similar levels in differentiated and proliferating cells, but that the long isoform is 25-fold higher expressed in differentiated compared with proliferating cells (Figure 5C). Figure 5B shows that the decay of all exons except for those unique to the short isoforms is slower in differentiated than in proliferating cells. In addition, the exons unique to the long isoform are significantly more stable in differentiating than in proliferating cells. qPCR experiments (Figure 5D) validate the longer half life of the long isoform in differentiated cells. Thus, the ratio between the long and the short isoform is regulated, at least in part, at the level of mRNA degradation, and depends on the differentiation status of the cells.
Figure 5.
Association between RNA degradation and alternative splicing in Pde4dip. (A) Exon structure of the Pde4dip-short and the Pde4dip-long mRNA and the natural logarithm of the ratio (y-axis) of the expression levels of the different exons (x-axis) between differentiated and proliferating cells, as assayed before actinomycin D treatment. Note: using CAGE-seq (cap analysis gene of expression analyzed by high-throughput sequencing, investigating transcription start sites) data, we found that the annotation of the first exon from ENSMUST00000090750 in ENSEMBL is incorrect (20), explaining its aberrant behavior. (B) Plot of the decay rates in differentiated (closed symbols) and proliferating (open symbols) cells for the different probe sets in the Pde4dip gene, ordered from 5′ to the 3′ end of the gene. Less stable parts of the RNA have more negative slopes. (C) Relative abundance (arbitrary units; a.b.u) of the short and long transcripts of the Pde4dip gene, as assayed by qPCR with primers in the two different 3′-UTRs. (D) Estimated half lives of the short and long transcripts of the Pde4dip gene, as assayed by qPCR with primers in the two different 3′-UTRs. Error bars reflect standard error of the mean.
Itga7
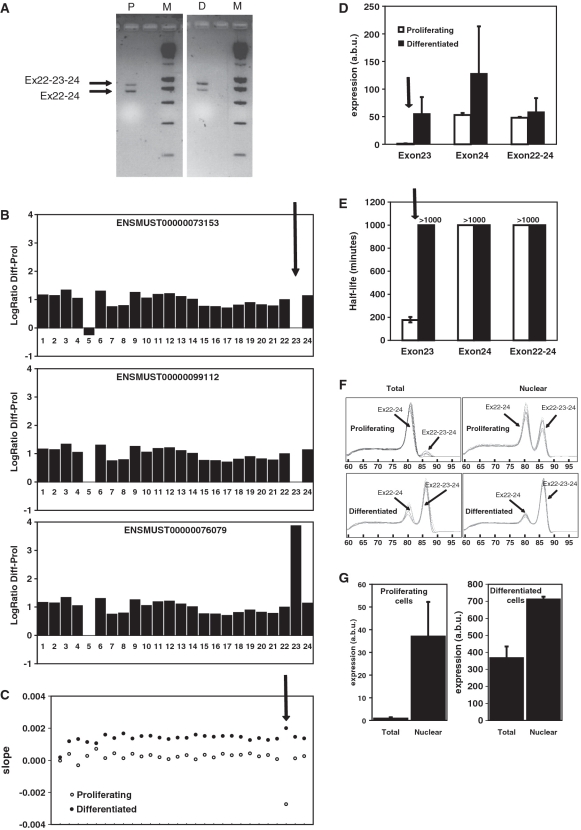
Itga7 is an example of a gene that codes for different transcript isoforms that share the same 3′-UTR but differ in stability. Itga7 codes for integrin α7, a transmembrane receptor important for maintaining the link between the extracellular matrix and the cytoskeleton of the muscle fiber (22,23). Itga7 is known to undergo alternative splicing, resulting in differences in both the extracellular (N-terminal) and cytoplasmic (C-terminal) domain. The splice isoform containing an additional exon (here indicated as 23, referred to in other publications as transcript α7A) is specific for skeletal muscle and expressed mainly during myogenic differentiation (24). Since this exon is out-of-frame, inclusion of this exon gives rise to a protein with an alternative C-terminal amino acid sequence, which may activate alternative signaling cascades. By 3′-RACE, with a primer in exon 22, we demonstrated that the transcripts with or without exon 23 indeed share the same 3′-UTR and use the same polyadenylation site (Figure 6A). As expected, transcripts containing exon 23 are almost exclusively present in differentiated cells (Figure 6B and D). Both exon-array (Figure 6C) and qPCR experiments (Figure 6E) with primers in exon 23 indicate that transcripts containing exon 23 are very unstable in proliferating cells. Moreover, analysis of the nuclear fraction of the mRNA shows that the splice variant that includes exon 23 is made in proliferating cells and is more abundant in the nucleus than in the cytoplasm (Figure 6F and G), and therefore preferentially degraded in the cytoplasm of the proliferating cells. This example shows that the stability of splice variants is actively controlled and dependent on the differentiation status.
Figure 6.
Association between RNA degradation and alternative splicing in Itga7. (A) Agarose gel displaying the products of 3′-RACE for Itga7 with forward primer in exon 22 in proliferating (P) and differentiated (D) cells and a 200-bp size marker (M). The longer fragment (more abundant in differentiated cells) contains exon 23, whereas the shorter fragment lacks exon 23. The size difference equals the size of exon 23 (113 bp). Subsequent sequencing confirmed this and mapped the polyadenylation site for both fragments at the previously identified and annotated polyA site (position 4018 in GenBank entry NM_008398.2). (B) Exon structure of three different transcript isoforms (indicated by their Ensembl transcript identifier) and the natural logarithm of the ratio (y-axis) of the expression levels of the different exons (x-axis) in the Itga7 gene, as assayed before actinomycin D treatment. The arrow indicates exon 23, which is preferentially incorporated in transcripts expressed in differentiated myoblasts. (C) Plot of the decay rates in differentiated (closed symbols) and proliferating (open symbols) cells for the different probe sets in the Itga7gene, ordered from 5′ to the 3′ end of the gene. Less stable parts of the RNA have more negative slopes. The arrow indicates the probe in exon 23. The transcript containing exon 23 is much more stable in differentiated than in proliferating myoblasts. (D) Relative abundance (arbitrary units; a.b.u) of the different transcripts of the Itga7 gene, as assayed by qPCR with primers in exon 23 (assaying only transcripts that include exon 23), exons 22 and 24 (a combination of transcripts that do and do not include exon 23), and exon 24 (assaying all transcripts), in proliferating (open bars) and differentiated (closed bars) cells. (E) Estimated half lives of the different transcripts of the Itga7 gene, as assayed by qPCR with the primers described under (D) in proliferating (open bars) and differentiated (closed bars) cells. Error bars reflect standard error of the mean. (F) Melting curves obtained after qPCR with with forward primer in exon 22 and reverse primer in exon 24 of the Itga7 gene in total (left) and nuclear (right) RNA of proliferating (top) and differentiated (bottom) cells. Two different PCR products were obtained. The product containing exon 23 melts at a higher temperature than the product without exon 23. The transcript containing exon 23 is present in the nuclei of proliferating myoblasts but barely detectable in total RNA (mainly cytoplasmatic). (G) Abundance of the Itga7 transcript containing exon 23 in total and nuclear RNA of proliferating (left) and differentiated (right cells), as determined by qPCR with forward primer in exon 22 and reverse primer in exon 23. Note the different scales for the left and right graphs.
DISCUSSION
We reported around 7000 mRNA half lives in proliferating and differentiated myoblast cells. Given the 480 min time interval analyzed, the estimates for transcripts with half lives shorter than 480 min are more precise than those for very stable transcripts. The reported half lives can guide the choice of evaluation time points in future studies where transcript levels are induced or repressed [with the obvious exception of small interfering RNA (siRNA) experiments influencing mRNA degradation itself]. Knowledge on mRNA half life can be used in the analysis of gene expression profiling studies to identify genes that are in fact completely shut down, whereas the data as such only indicate that expression is slightly reduced. In addition, the data can be used to flag transcripts or transcript isoforms that can not be valued since their mRNA half lives exceed the time points analyzed, or demand special attention due to significantly altered half lives.
Furthermore, we show that transcripts, including specific splice variants, with a role in myogenesis and differential expression between proliferating and differentiated myoblasts frequently demonstrate different stabilities in the two states. Transcripts with higher expression in differentiated than in proliferating cells are more rapidly degraded in proliferating cells and vice versa. A cell is in some cases reminiscent of a Dutch auction. When the clock is down to zero and there appears to be no demand, the commodities are to be destroyed.
Despite the characterization of a multitude of proteins and pathways involved in mRNA degradation (9), we are only beginning to understand the mechanisms that control the stability and degradation of individual transcripts. The 3′-UTR is very important for transcript stability given the presence of miRNA target sites, AU-rich elements (AREs) and binding sites for other RNA-binding proteins. Our observation that decay constants of first or last probe sets in a gene frequently differ (are higher or lower) from the other probe sets is consistent with the reported extensive use of differential 3′-UTRs (and alternative promoters) depending on the tissue or cellular state (10,25–27). In differentiating T-cells (10) and in cancer cells (27), differential expression of transcripts has been related to alternative 3′-UTR (polyadenylation site) use, resulting in differences in mRNA stability.
It is not yet clear which RNA binding proteins are responsible for the control of mRNA stability during myogenic differentiation. The HuR and KSRP can transiently affect the stability of transcripts containing AREs during myogenic differentiation (17–19,28). However, only a minority of the transcripts with variable decay rates contain AU-rich elements [ARE motifs, predicted by the ARED algorithm (29)] in their 3′-UTR. From our digital gene expression studies (20), two members of the tristetraprolin family of RNA binding protein emerge as other important candidate factors for differentiation state-dependent control of mRNA degradation: ZFP36L1, which has been shown to accelerate degradation of tumor necrosis factor-alpha (TNF-α), vascular endothelial growth factor (VEGF) and granulocyte macrophage-colony stimulating factor (GM-CSF) (30–32) and ZFP36L2. The former is 10-fold increased and 3-fold decreased in expression during myogenic differentiation. The muscle-specific RNA binding protein CUGBP has been shown to stabilize mRNAs by binding to GU-rich elements in the 3′-UTR (8,33). However, CUGBP shows little variation in expression between proliferating and differentiated cells and is therefore less likely to be responsible for the differences in decay rates observed in proliferating and differentiated myoblasts. In addition to RNA stabilizing and destabilizing proteins, also miRNAs and components of the nonsense-mediated decay machinery have been shown to have tissue-dependent expression levels and activities and may contribute to the observed differences in mRNA stability (34–38).
One of the new aspects of the current study is the notion that also splice variants that share the same 3′-UTR but include alternative internal exons may differ in stability. It was previously thought that the splicing machinery determines the abundance of the different splice isoforms aided by tissue-specific splicing factors like FOX, CELF, PTB, MBNL, hnRNP A/B (26,39,40). From our study, it appears that the presence of stabilizing or destabilizing elements in the alternative exons or differences in structural characteristics of the synthesized transcripts may also contribute to their differential expression. We show this in detail for the Itga7 transcripts variants with or without exon 23. The transcript including exon 23 appears to be me far more stable in differentiated cells, whereas in proliferating cells it is subject to fast degradation. A comparison of the nuclear fraction of the RNA with total RNA (nuclear and cytoplasmic) shows that the degradation of the transcript containing exon 23 mainly occurs in the cytoplasm. This observation is in line with a recent publication showing major differences between transcript variants in their nucleocytoplasmic distribution, at least partly attributable to mechanistic differences between nuclear and cytoplasmic mRNA degradation (41). We investigated whether exon 23 contains any known destabilizing elements such as AREs or PUFs (7), but could not find any. It is still possible that exon 23 contains an miRNA binding site but prediction programs failed to predict a good candidate. Thus, the mechanism for its preferential cytoplasmic degradation in proliferating myoblast remains elusive.
We conclude that mRNA degradation is a key controlling factor in gene expression regulation, possibly as important as transcription factor-mediated induction of mRNA synthesis. The mechanisms of mRNA degradation are currently less well understood than the mechanisms behind transcription factor activity regulation. We have gained several mechanistic insights in the process of mRNA degradation, but many mechanistic aspects deserve more attention in future studies. Knowledge on mRNA degradation rates, as presented in this study, is important for the proper interpretation of many biological studies. Decay rates need to be taken into account when choosing a time point for evaluation of the perturbation of a biological system, aid the modeling of transcriptional networks, and should be incorporated as important parameters in systems biology approaches.
ACCESSION NUMBER
Gene Expression Omnibus GSE14387.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Centre for Medical Systems Biology within the framework of The Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO).
Conflict of interest statement. None declared.
REFERENCES
- 1.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC. Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2:RESEARCH0041. doi: 10.1186/gb-2001-2-10-research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLOS One. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl. 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 13.Ihaka R, Gentleman RC. R: a language for data analysis and graphics. Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- 14.Pinheiro JC, Bates DM. Mixed Effects Models in S and S-plus. New York: Springer; 2000. [Google Scholar]
- 15.Okoniewski MJ, Yates T, Dibben S, Miller CJ. An annotation infrastructure for the analysis and interpretation of Affymetrix exon array data. Genome Biol. 2007;8:R79. doi: 10.1186/gb-2007-8-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.'t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 18.van der Giessen K, Di Marco S, Clair E, Gallouzi IE. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J. Biol. Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- 19.Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hestand MS, Klingenhoff A, Scherf M, Ariyurek Y, Ramos Y, van WW, Suzuki M, Werner T, van Ommen GJ, den Dunnen JT, et al. Tissue-specific transcript annotation and expression profiling with complementary next-generation sequencing technologies. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq602. [Epub ahead of print, 7 July 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faul C, Dhume A, Schecter AD, Mundel P. Protein kinase A, Ca2+/calmodulin-dependent kinase II, and calcineurin regulate the intracellular trafficking of myopodin between the Z-disc and the nucleus of cardiac myocytes. Mol. Cell Biol. 2007;27:8215–8227. doi: 10.1128/MCB.00950-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul AC, Sheard PW, Kaufman SJ, Duxson MJ. Localization of alpha 7 integrins and dystrophin suggests potential for both lateral and longitudinal transmission of tension in large mammalian muscles. Cell Tissue Res. 2002;308:255–265. doi: 10.1007/s00441-002-0526-y. [DOI] [PubMed] [Google Scholar]
- 23.Postel R, Vakeel P, Topczewski J, Knoll R, Bakkers J. Zebrafish integrin-linked kinase is required in skeletal muscles for strengthening the integrin-ECM adhesion complex. Dev. Biol. 2008;318:92–101. doi: 10.1016/j.ydbio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Kaariainen M, Nissinen L, Kaufman S, Sonnenberg A, Jarvinen M, Heino J, Kalimo H. Expression of alpha7beta1 integrin splicing variants during skeletal muscle regeneration. Am. J. Pathol. 2002;161:1023–1031. doi: 10.1016/s0002-9440(10)64263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 26.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deschenes-Furry J, Belanger G, Mwanjewe J, Lunde JA, Parks RJ, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuR binds to acetylcholinesterase transcripts and regulates their expression in differentiating skeletal muscle cells. J. Biol. Chem. 2005;280:25361–25368. doi: 10.1074/jbc.M410929200. [DOI] [PubMed] [Google Scholar]
- 29.Halees AS, El Badrawi R, Khabar KS. ARED organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36:D137–D140. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciais D, Cherradi N, Bailly S, Grenier E, Berra E, Pouyssegur J, Lamarre J, Feige JJ. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 31.Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, Meyers EN, Mishina Y, Blackshear PJ. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol. Cell Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J. Biol. Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad Sci. USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X, Li X, Moriarty PM, Henics T, LaDuca JP, Maquat LE. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol. Biol. Cell. 2001;12:1009–1017. doi: 10.1091/mbc.12.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bateman JF, Freddi S, Nattrass G, Savarirayan R. Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum. Mol. Genet. 2003;12:217–225. doi: 10.1093/hmg/ddg054. [DOI] [PubMed] [Google Scholar]
- 38.Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl Acad. Sci. USA. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagnani M, Barash Y, Ip JY, Misquitta C, Pan Q, Saltzman AL, Shai O, Lee L, Rozenhek A, Mohammad N, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L. A global comparison between nuclear and cytosolic transcriptomes reveals differential compartmentalization of alternative transcript isoforms. Nucleic Acids Res. 2010;38:1086–1097. doi: 10.1093/nar/gkp1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.