Abstract
The nuclear receptor vitamin D receptor (VDR) is known to associate with two vitamin D response element (VDRE) containing chromatin regions of the insulin-like growth factor binding protein 3 (IGFBP3) gene. In non-malignant MCF-10A human mammary cells, we show that the natural VDR ligand 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3) causes cyclical IGFBP3 mRNA accumulation with a periodicity of 60 min, while in the presence of the potent VDR agonist Gemini the mRNA is continuously accumulated. Accordingly, VDR also showed cyclical ligand-dependent association with the chromatin regions of both VDREs. Histone deacetylases (HDACs) play an important role both in VDR signalling and in transcriptional cycling. From the 11 HDAC gene family members, only HDAC4 and HDAC6 are up-regulated in a cyclical fashion in response to 1α,25(OH)2D3, while even these two genes do not respond to Gemini. Interestingly, HDAC4 and HDAC6 proteins show cyclical VDR ligand-induced association with both VDRE regions of the IGFBP3 gene, which coincides with histone H4 deacetylation on these regions. Moreover, combined silencing of HDAC4 and HDAC6 abolishes the cycling of the IGFBP3 gene. We assume that due to more efficient VDR interaction, Gemini induces longer lasting chromatin activation and therefore no transcriptional cycling but monotonically increasing IGFBP3 mRNA. In conclusion, 1α,25(OH)2D3 regulates IGFBP3 transcription through short-term cyclical association of VDR, HDAC4 and HDAC6 to both VDRE-containing chromatin regions.
INTRODUCTION
The natural vitamin D receptor (VDR) ligand 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3) has an important role in the regulation of calcium and phosphate homoeostasis and bone mineralization (1). In addition to this classical role, there is both epidemiological and pre-clinical evidence that 1α,25(OH)2D3 is an anti-proliferative agent (2). The anti-proliferative effects of 1α,25(OH)2D3 include induction of a G1/G0 cell cycle arrest and stimulation of apoptosis, which are mediated by the up-regulation of tumour suppressors, such as the cyclin-dependent kinase inhibitory proteins p21 and p27 (3), and the down-regulation of oncogene products including Bcl-2 (4) and Myc (5). Mitogens, such as the insulin-like growth factors (IGFs), have also been reported to be down-regulated by 1α,25(OH)2D3 (6). In addition, also the up-regulation of factors that control the actions of mitogens, such as IGF binding proteins (IGFBPs), have important anti-cancer effects (7). Three members of the IFGBP gene family, IGFBP1, IGFBP3 and IGFBP5, respond to 1α,25(OH)2D3 (8), of which the IGFBP3 gene is the most prominent (9). This increases the impact of IGF-1 and the regulation of its circulating amounts by IGFBPs in models of the anti-proliferative action of 1α,25(OH)2D3 and its synthetic analogues (10). In addition, IGFBPs mediate IGF-independent actions, including the activation of the p21 gene, causing cell cycle arrest or cell death through induction of apoptosis (11). However, bound to cellular membranes, IGFBPs can have mitogenic, IGF-dependent effects on cellular growth (12,13).
As a member of the nuclear receptor superfamily, VDR acts as a transcription factor that binds to specific vitamin D response elements (VDREs) within the regulatory regions of its primary target genes (14). Most VDR target genes contain multiple VDREs (8,15–17). For example, the IGFBP3 gene has a tandem of two VDREs at position −400 and the other VDRE at position −3350 relative to the transcription start site (TSS) (8). In the absence of ligand, VDR associates via co-repressor proteins with histone deacetylases (HDACs) (18). HDACs can also inactivate directly non-histone proteins, such as p53, E2F or α-tubulin by deacetylation (19–21). Therefore, HDACs have multiple influences in cellular processes. At present 11 human HDACs are known (22). HDACs 1, 2, 3 and 8 belonging to Class I are ubiquitously expressed and seem to be involved more in general cellular processes. The Class II HDACs 4, 5, 6, 7, 9 and 10 have more tissue-specific functions and distributions, while HDAC11 forms its own class (23,24). All these HDACs are sensitive to the inhibitor trichostatin A (TSA) (25). In addition to the classical HDACs, on which we are focusing in this study, there is a family of functionally related HDACs, referred to as sirtuins (26). The seven members of this family are not sensitive to TSA but use NAD+ as an essential co-factor.
Recently, cyclical models have been proposed for the activation of transcription by nuclear receptors, including those for oestrogen receptor α on the trefoil factor-1 gene (27), for peroxisome proliferater-activated receptor δ on the pyruvate dehydrogenase kinase 4 gene (28) and for VDR on the genes 24-hydroxylase (CYP24) (29) and cyclin-dependent kinase inhibitor 1A (CDKN1A) (30). In these models, the ligand-dependent transcription is seen as a cyclical process, where alternating de-repressing, activating and initiation actions are required, providing means to stringently regulate the endurance and strength of the transcriptional response (28).
More than 3000 synthetic analogues of 1α,25(OH)2D3 are presently known and majority of them carry a modification in their aliphatic side chain (31). The 1α,25(OH)2D3 analogues have been developed with a goal to improve the biological profile of the natural hormone for therapeutic application either in hyper-proliferative diseases, such as psoriasis and different types of cancer, or in bone disorders, such as osteoporosis (32). Most of the analogues described to date are agonists, with a few having been identified as antagonists. An interesting exception is Gemini, which is the first 1α,25(OH)2D3 analogue that carries two side chains (33,34). Molecular dynamics simulations of the Gemini–VDR complex showed that the analogue can bind the VDR-LBD in two different conformations (35,36). In one of these conformations Gemini acts as an agonist with one side chain taking the same position as that of the natural hormone. In contrast, in its other conformation Gemini acts as an inverse agonist, since both of its side chains take alternative positions to that of 1α,25(OH)2D3 (37).
In this study, we performed detailed time course experiments and observed transcriptional cycling of IGFBP3 mRNA after 1α,25(OH)2D3 stimulation, but not in response to Gemini. This is reflected by ligand-dependent VDR association with both VDREs and histone 4 acetylation on the chromatin region of the more proximal VDRE of the IGFBP3 gene. The genes HDAC4 and HDAC6 are also up-regulated in a cyclical fashion in response to 1α,25(OH)2D3, whereas they do not respond to Gemini. Both HDACs are essential for the cycling of the IGFBP3 gene. Accordingly, HDAC4 and HDAC6 proteins show VDR ligand-induced association with both IGFBP3 VDREs. In conclusion, 1α,25(OH)2D3 regulates IGFBP3 transcription through cyclical association of HDAC4 and HDAC6 to its VDRE-containing chromatin regions.
EXPERIMENTAL PROCEDURES
Cell culture
MCF-10A cells (38) were cultured in a mixture of DMEM and Ham’s F12 medium (1:1) with 20 ng/ml of epidermal growth factor, 100 ng/ml of cholera toxin, 10 µg/ml insulin, 500 ng/ml hydrocortisone, 0.1 mg/ml streptomycin, 100 U/ml penicillin and 5% horse serum in a humidified 95% air/5% CO2 incubator. Twenty-four hours prior to the treatment, the cells were seeded into medium with 5% charcoal-treated fetal bovine serum (FBS) instead of horse serum. RWPE-1 adherent human prostate epithelial cells are derived from the peripheral zone of a histologically normal healthy 54-year-old male’s prostate (39). The cells were cultured in Keratinocyte serum free medium (SFM) containing l-glutamine, 2.5 µg human recombinant epidermal growth factor, 25 mg bovine pituitary extract, 0.1 mg/ml streptomycin and 100 U/ml penicillin. For mRNA extractions, the cells were seeded in the culture medium and grown to a density of 45–60%. For stimulation, the medium of cell supernatant was supplemented with stock solutions of the compounds but not changed. 1α,25(OH)2D3 and its two side chain analogue Gemini (33) (kindly provided by Dr Milan Uskokovic, BioXell Inc., Nutley, NJ, USA) were used at a final concentration of 10 nM. The ligand stocks were diluted first in ethanol to 100 µM and then in DMEM medium to 1 µM.
RNA extraction and real-time PCR
Total RNA was extracted using the RNA Isolation kit (Roche) and cDNA synthesis was performed for 30 min at 55°C using 1 µg of total RNA as a template and 100 pmol oligodT18 primers (Roche). Real-time quantitative polymerase chain reaction (PCR) was performed using a LightCycler® 480 System (Roche). The reactions were performed using 4 pmol of reverse and forward primers, 4 µl cDNA template and the Maxima™ SYBR Green/Fluorescein qPCR Master Mix (Fermentas, Vilnius, Lithuania) in a total volume of 10 µl. In the PCR reaction the DNA templates were pre-denaturated for 10 min at 95°C, followed by amplification steps cycles of 20 s denaturation at 95°C, 20 s annealing at primer-specific temperatures (Supplementary Table S1), 20 s elongation at 72°C and a final elongation for 10 min at 72°C.
Fold inductions were calculated using the formula 2−(ΔΔCt), where ΔΔCt is ΔCt(stimulus)−ΔCt(solvent), ΔCt is Ct (target gene) − Ct(contol gene) and the Ct is the cycle, at which the threshold is crossed. Basal expression levels were calculated using the formula 2−(ΔCt). The sequences of gene-specific primer pairs are given in Supplementary Table S1. PCR product quality was monitored using post-PCR melt curve analysis.
Chromatin immunoprecipitation assays
Nuclear proteins were cross-linked to DNA by adding formaldehyde directly to the medium to a final concentration of 1% for 5 min at room temperature. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M and incubating for 5 min at room temperature on a rocking platform. The medium was removed and the cells were washed twice with ice-cold phosphate buffered saline (PBS). The cells were then collected in ice-cold PBS and cell pellets were resuspended in lysis buffer (1% SDS, 10 mM EDTA, protease inhibitors, 50 mM Tris–HCl, pH 8.1) and the lysates were sonicated by a Bioruptor UCD-200 (Diagenode, Liege, Belgium) to result in DNA fragments of 300–1000 bp in length (Supplementary Figure S8). Cellular debris was removed by centrifugation and the lysates were diluted 1:10 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, protease inhibitors, 16.7 mM Tris–HCl, pH 8.1). Chromatin solutions were incubated at 4°C with rotation with 5 µl of antibodies against VDR (sc-1008), HDAC4 (sc-11418) and HDAC6 (sc-11420) (all from Santa Cruz Biotechnologies) or with 1 µl of anti-acetylated histone H4 (anti-acH4) antibody or control IgG (both from Upstate Biotechnology, Lake Placid, NY, USA). The immuno-complexes were collected with 60 µl of protein A agarose slurry (Upstate Biotechnology) for 1 h at 4°C with rotation. The beads were precipitated by centrifugation for 1 min at room temperature with 100g and washed sequentially for 4 min by rotation with 1 ml of the following buffers: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 8.1), high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris–HCl, pH 8.1) and LiCl wash buffer (0.25 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1). Finally, the beads were washed twice with 1 ml TE buffer (1 mM EDTA, 10 mM Tris–HCl, pH 8.1). The immuno-complexes were then eluted by adding 500 µl of elution buffer (25 mM Tris–HCl, pH 7.5, 10 mM EDTA, 0.5% SDS) and incubating for 30 min at 65°C. The cross-linking was reversed and the remaining proteins were digested by adding 2 µl of proteinase K (Fermentas) to a final concentration of 80 µg/ml and incubating overnight at 64°C. The DNA was recovered by phenol/chloroform/isoamyl alcohol (25:24:1) extractions and precipitated with 0.1 volume of 3 M sodium acetate, pH 5.2 and 2 volumes of ethanol using glycogen as carrier. Immuno-precipitated chromatin DNA was then used as a template for real-time quantitative PCR.
PCR of chromatin templates
For both of the VDRE-carrying regions of the IGFBP3 gene, genomic primers were designed (Supplementary Table S2) and for their quantification 6-carboxyfluorescein (6-FAM) and Black Hole Quencher 1 (BHQ-1)-modified hydrolysis probes were used (Supplementary Table S3). All oligonucleotides used in this study were obtained from Eurogentec (Liege, Belgium). Real-time quantitative PCR was performed with the Maxima Probe qPCR master mix (Fermentas) on a LightCycler® 480 System (Roche). The PCR cycling conditions were: pre-incubation for 10 min at 95°C, 50 cycles of 20 s at 95°C, 60 s at 60°C. The PCR products were also resolved on 2% agarose gels to control correct product size. Relative association of chromatin-bound protein or histone modifications were calculated using the formula 2−(ΔCt), where ΔCt is Ct(output)−Ct(input), output is the immuno-precipitated DNA and input is the purified genomic DNA from starting material of the ChIP assay. Results were normalized with respect to input and non-specific IgG using the formula [2−(ΔCt) (specific antibody)−2−(ΔCt) (non-specific IgG)]×100, where ΔCt is the Ct(immunoprecipitated DNA)−Ct(input). The input was corrected by the respective dilution factors.
Small inhibitory RNA transfection
MCF-10A cells (350 000) were reverse transfected with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions using a mixture of three double-stranded small inhibitory RNA (siRNA) oligonucleotides per gene (Eurogentec, 200 pmol of each siRNA, Supplementary Table S4). Cells were treated 24 h after plating and RNA extraction, real-time quantitative PCR was carried out as described above. Silencing at the protein level was verified by western blotting using 25 µg of whole cell extract and the same antibodies as used for ChIP and anti-β-actin antibody (Sigma). Cellular proteins were separated on 12% SDS polyacrylamide gels. The blotted proteins were detected using IR800 fluorescence labelled secondary antibodies (Thermo Scientific, Rockford, IL, USA) and an Odyssey reader (Li-Cor Biotechnology, Nebraska, USA).
RESULTS
Cyclical induction of IGFBP3 mRNA expression by 1α,25(OH)2D3 but not by Gemini
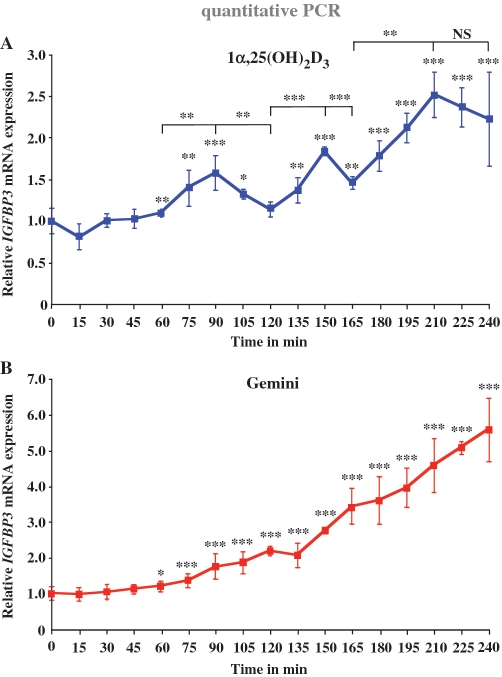
The IGFBP3 gene has initially been shown to respond to 1α,25(OH)2D3 in prostate cancer cells (8,9). Since (i) IGFBP3 has also important functions in the growth regulation of mammary cells (40) and (ii) we are mainly interested in the cancer-preventive actions of 1α,25(OH)2D3, we used the human non-tumourigenic epithelial cell line MCF-10A (38) as a cellular model. In order to elucidate the dynamics of IGFBP3 induction, we performed real-time quantitative PCR analysis of IGFBP3 mRNA expression after stimulation with 1α,25(OH)2D3 and Gemini (10 nM in all experiments of this study) in a detailed time course of 240 min with 15-min intervals (Figure 1). In response to 1α,25(OH)2D3, the first significant peak of IGFBP3 mRNA expression appeared after 90 min (1.6-fold induction), the second at time point 150 min (1.8-fold induction) followed by a third peak at time point 210 min (2.6-fold induction), i.e. the peaks showed a periodicity of 60 min (Figure 1A). After each peak, the accumulation of IGFBP3 mRNA ceased resulting in a decrease of IGFBP3 mRNA levels. It has to be noted that the cells had not been synchronized. Thus, 1α,25(OH)2D3 itself seems to be sufficient for the induction of cyclicity in IGFBP3 transcription. In contrast, in response to Gemini the IGFBP3 mRNA expression continuously increased without any signs of cycling to a 5.5-fold induction after 240 min (Figure 1B). As a control for the potency of Gemini versus 1α,25(OH)2D3, we measured with the same samples CYP24 induction (Supplementary Figure S1). Furthermore, with this gene, Gemini induced a continuous accumulation of mRNA [up to 25 000-fold after 240 min, (Supplementary Figure S1B)], whereas the response to 1α,25(OH)2D3 showed a more staircase-like profile (Supplementary Figure S1A). As a further control, we measured IGFBP3 mRNA accumulation of in RWPE-1 non-malignant human prostate cells in response to 1α,25(OH)2D3 (Supplementary Figure S2A) and Gemini (Supplementary Figure S2B). Interestingly, in this cellular model IGFBP3 mRNA increased steadily without any sign of cycling also after stimulation with 1α,25(OH)2D3. It has to be noted that the basal expression of IGFBP3 mRNA in RWPE-1 prostate cells is significantly higher as in MCF10A breast cells (data not shown).
Figure 1.
Cyclical induction of IGFBP3 transcription by 1α,25(OH)2D3. Real-time quantitative PCR was performed in order to measure the time-dependent mRNA expression of the IGFBP3 gene in MCF-10A cells after treatment with 10 nM 1α,25(OH)2D3 (A) or 10 nM Gemini (B). The data were normalized to the expression of the housekeeping gene ribosomal protein large P0 (RPLP0) and fold inductions were calculated in reference to vehicle control. Data points indicate the means of at least three independent cell treatments and the bars represent standard deviations. A two-tailed Student’s t-test was performed to determine the significance of the stimulation in reference to vehicle-treated control and for the comparison of the peaks to the minima (*P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant).
Taken together, the natural VDR ligand 1α,25(OH)2D3 induces IGFBP3 transcription only for short periods at selected time points, while the strong synthetic VDR agonist Gemini seems to continuously activate the gene.
Cyclical enrichment of VDR on 1α,25(OH)2D3-responsive regions
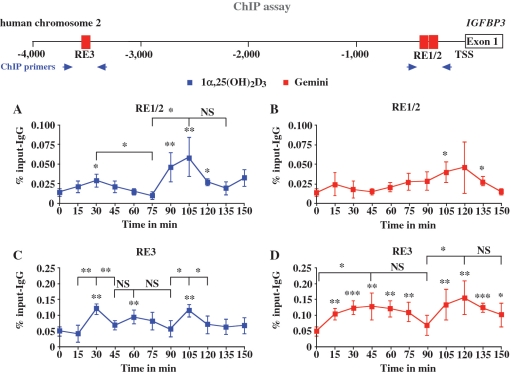
In order to study whether the cyclical induction of IGFBP3 mRNA in response to 1α,25(OH)2D3 is based on parallel cyclical association of VDR with the regulatory regions of the IGFBP3 gene, we performed in 1α,25(OH)2D3- and Gemini-treated MCF-10A cells ChIP assays with antibodies against VDR (Figure 2). We analysed the time period of 0–150 min with 15-min intervals, in which the IGFBP3 mRNA peaked two times (Figure 1). On the chromatin templates, we determined by real-time quantitative PCR using TaqMan probes the VDR ligand-induced enrichment of the two previously identified VDR-associated regions RE1/2 and RE3 (8) in comparison to that in untreated cells. After 1α,25(OH)2D3 treatment, VDR showed cyclical association both with RE1/2 (Figure 2A) and with RE3 (Figure 2C) showing peaks at 30 and 105 min. However, in the first cycle (0–75 min), the VDR association with RE1/2 is far lower than in the second cycle. In contrast, in response to Gemini, we did not observe any statistically significant increase of VDR binding to the region of RE1/2 and no signs of cycling (Figure 2B). Interestingly, Gemini could induce VDR association with RE3 in a cyclical fashion with maximal levels at time points 45 and 120 min and minima at 90 and 150 min (Figure 2D).
Figure 2.
Recruitment of VDR to the IGFBP3 promoter. Chromatin was extracted from MCF-10A cells that had been treated for indicated time points with 10 nM 1α,25(OH)2D3 (A and C) or 10 nM Gemini (B and D). ChIP experiments were performed using antibody against VDR. Real-time quantitative PCR using TaqMan probes was performed with primers specific for the VDRE containing regions RE1/2 (A and B) and RE3 (C and D) of the IGFBP3 promoter (schematically depicted on top). PCR conducted on chromatin input template served as a positive control and that on IgG-precipitated template as specificity control. Data points indicate the means of at least three independent cell treatments and the bars represent standard deviations. A two-tailed Student’s t-test was performed to determine the significance of the stimulation in reference to vehicle-treated control and for the comparison of the peaks to the minima (*P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant).
In summary, the association of VDR with both VDRE-containing regions of the IGFBP3 gene shows a cyclical behaviour after 1α,25(OH)2D3 treatment with peaks at 30 and 105 min, while in response to Gemini VDR cycles were observed only on RE3 with peaks at 45 and 120 min.
HDAC4 and HDAC6 are targets of 1α,25(OH)2D3 but not of Gemini
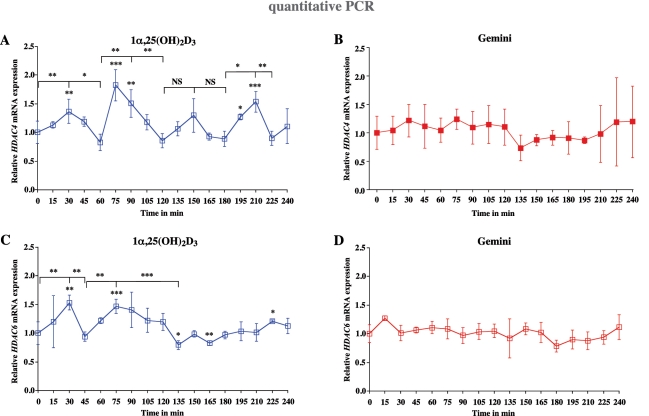
In a previous project, we studied the effects of 1α,25(OH)2D3 and HDAC inhibitors on the growth of mammary cells (41). Therefore, we screened in MCF-10A cells whether any of the 11 HDAC gene family members are direct targets of 1α,25(OH)2D3 or Gemini. While the basal mRNA expression of HDAC1, HDAC2 and HDAC3 is very high, HDAC4, HDAC5, HDAC10 and HDAC11 are expressed more than 100 times lower (Supplementary Figure S3). The basal expression of HDAC6, HDAC7 and HDAC8 is between these two groups and HDAC9 expression is not detectable by real-time quantitative PCR. In response to 1α,25(OH)2D3, only HDAC4 (Figure 3A) and HDAC6 (Figure 3C) showed to be primary targets. Within the measuring period of 240 min HDAC4 mRNA showed four maxima at 30 min (1.4-fold induction), 75 min (1.8-fold induction), 150 min (1.4-fold induction, although statistically not significant) and 210 min (1.6-fold induction) (Figure 3A), while significant peaks were observed for HDAC6 mRNA only at time points 30 min (1.6-fold induction) and 75 min (1.5-fold induction) (Figure 3C). This indicates that HDAC4 and HDAC6 are early 1α,25(OH)2D3 responding genes. Accordingly, an in silico screening for VDREs in both genes (Supplementary Figure S4) showed that within the first 10 kB upstream of the HDAC4 TSS two VDREs are located and the comparable region of the HDA6 gene carries even three potential VDREs. In contrast, none of the remaining eight HDAC gene family members, which were expressed in MCF-10A cells, was significantly regulated by 1α,25(OH)2D3 (Supplementary Figure S5). In contrast, neither HDAC4 (Figure 3B), HDAC6 (Figure 3D) nor any other HDAC gene family members (Supplementary Figure S6) was significantly regulated by Gemini.
Figure 3.
Cyclical induction of HDAC4 and HDAC6 transcription by 1α,25(OH)2D3. Real-time quantitative PCR was performed in order to measure the time-dependent mRNA expression of the genes HDAC4 (A and B) and HDAC6 (C and D) in MCF-10A cells after treatment with 10 nM 1α,25(OH)2D3 (A and C) or 10 nM Gemini (B and D). The data were normalized to the expression of the housekeeping gene RPLP0 and fold inductions were calculated in reference to vehicle control. Data points indicate the means of at least three independent cell treatments and the bars represent standard deviations. A two-tailed Student’s t-test was performed to determine the significance of the stimulation in reference to vehicle-treated control and for the comparison of the peaks to the minima (*P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant).
Taken together, from all members of the HDAC gene family only HDAC4 and HDAC6 were responsive to 1α,25(OH)2D3, but interestingly also in a cyclical fashion. In contrast, none of the HDAC genes was significantly regulated by Gemini.
Association of HDAC4 and HDAC6 and chromatin activation in response to 1α,25(OH)2D3 and Gemini on IGFBP3 VDRE regions
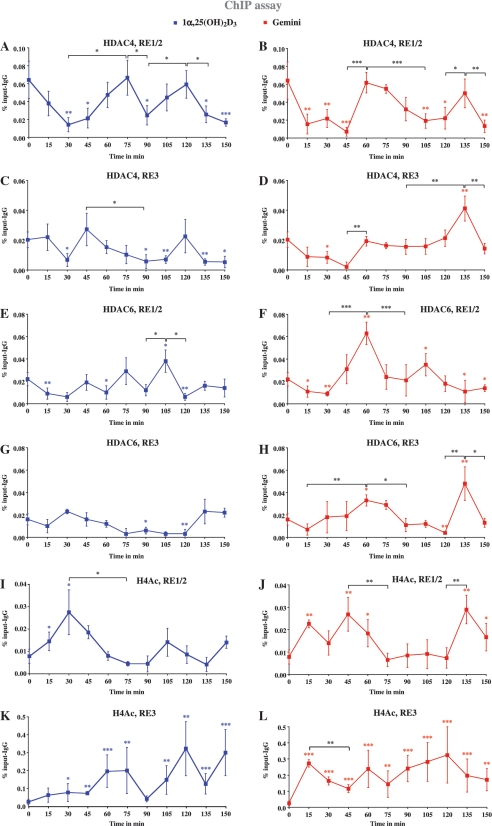
In order to test whether HDAC4 and HDAC6 show a physical interaction with the two VDRE regions of the IGFBP3 promoter and to monitor general chromatin activation of these regions, we performed ChIP assays with MCF-10A cells under the same conditions as in Figure 2, i.e. stimulation with 1α,25(OH)2D3 or Gemini and measurements every 15 min for a time period of 150 min, but now using antibodies against HDAC4, HDAC6 and AcH4 (Figure 4). The region of RE1/2 showed a high basal association with HDAC4, which after 1α,25(OH)2D3 stimulation significantly reduces within 30 min, restores at 75 min, diminishes again at 90 min and shows another maximum at 120 min, i.e. there is obvious cycling of HDAC4 on RE1/2 (Figure 4A). The basal HDAC4 binding to RE3 is less prominent than that to RE1/2, and also reduces in response to 1α,25(OH)2D3 with a minimum at 30 min, a maximum at 45 min, another minimum at 90–105 min and a last (non-significant) maximum at 120 min (Figure 4C). In addition, Gemini is able to reduce the association of HDAC4 with the regions of RE1/2 and RE3 for the time frame of 15–45 min (Figure 4B and D). On both VDRE regions HDAC4 binding restores at 60 min and shows a second maximum at 135 min.
Figure 4.
Recruitment of HDAC4 and HDAC6 to and histone acetylation of VDRE regions of the IGFBP3 promoter. Chromatin was extracted from MCF-10A cells that had been treated for indicated time points with 10 nM 1α,25(OH)2D3 (A, C, E, G, I and K) or 10 nM Gemini (B, D, F, H, J and L). ChIP experiments were performed using antibodies against HDAC4 (A–D), HDAC6 (E–H) and AcH4 (I–L). Real-time quantitative PCR using Taqman probes was performed with primers specific for the VDRE containing regions RE1/2 (A, B, E, F, I and J) and RE3 (C, D, G, H, K and L) of the IGFBP3 promoter. PCR conducted on chromatin input template served as a positive control and that on IgG-precipitated template as specificity control. Data points indicate the means of three to six independent cell treatments and the bars represent standard deviations. A two-tailed Student’s t-test was performed to determine the significance of the stimulation in reference to vehicle-treated control and for the comparison of the peaks to the minima (*P < 0.05, **P < 0.01, ***P < 0.001).
There is also basal association of HDAC6 with the regions of both RE1/2 and RE3. Treatment with 1α,25(OH)2D3 reduces HDAC6 binding at time points 15–30, 60, 90 and 120 min on RE1/2 leaving maxima at time points 45, 75 and 105 min (Figure 4E). On RE3 HDAC6 shows a short maximum at 30 min and long minimum at 75–120 min (Figure 4G). Also Gemini is able to reduce the basal HDAC6 association levels on both VDRE regions, but with a different profile from 1α,25(OH)2D3. HDAC6 binding to RE1/2 abolishes after 15–30 min, restores to higher than basal association levels at time point 60 min, is low again at 90 and 135 min and peaks in between at 105 min (Figure 4F). On RE3 HDAC6 levels are reduced after 15 min, peak at 60 min, are down at 90–120 min and show a last peak at 135 min (Figure 4H).
On RE1/2, we observed in response to 1α,25(OH)2D3 significant increase of chromatin activation only at time points 15 and 30 with a maximum at 30 min (Figure 4I), which coincides with a minimum of both HDAC4 and HDAC6 association to this region, while a second maximum at 105 min was not statistically significant (Figure 4I). After Gemini treatment significant chromatin activation could be measured at time points 15, 45 and 135 min, where it was also stronger than the induction by 1α,25(OH)2D3 (Figure 4J). Interestingly, on RE3 1α,25(OH)2D3 showed maxima at 60–75 min and 120 min (Figure 4K), while Gemini induced a rather high rate of chromatin activation, which was at nearly all time points higher than that induced by 1α,25(OH)2D3 (Figure 4L).
In summary, HDAC4 and HDAC6 both associate with the two VDRE regions of the IGFBP3 gene. Moreover, both 1α,25(OH)2D3 and Gemini reduce these basal levels after 15–45 min and induce rather individual profiles of association and dissociation of both HDACs. Interestingly, on RE1/2 both agonists show a similar acetylation pattern but with shifted maxima, while the chromatin on the region of RE3 is nearly constantly activated in response to Gemini. This suggests that the cycling of acetylation levels that were induced by Gemini cannot be related to HDAC expression levels (Figure 3).
Silencing HDAC4 and HDAC6 diminishes cycling of IGFBP3
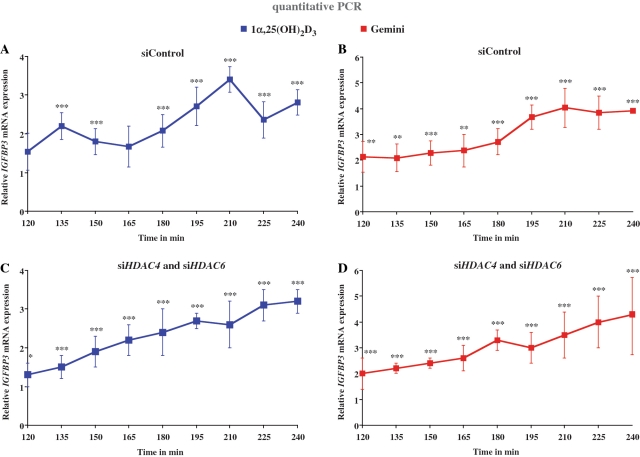
The observation that HDAC4 and HDAC6 are cycling target genes of 1α,25(OH)2D3 but not of Gemini led us to the hypothesis that the cycling phenomenon on the IGFBP3 gene may involve these two HDACs. Therefore, we silenced both HDACs simultaneously by gene-specific siRNA transfection of MCF-10A cells using a mixture of three gene-specific siRNAs (Figure 5). The efficiency of the silencing was 87.5% for HDAC4 and 83.4% for HDAC6 on mRNA level (Supplementary Figure S7A) and 62.1 and 87.9% on protein level (Supplementary Figure S7B). By real-time quantitative PCR, we monitored in the time period of 120–240 min every 15 min IGFBP3 mRNA accumulation in response to stimulation with 1α,25(OH)2D3 and Gemini. In control siRNA-transfected cells, we could repeat the results of Figure 1 that 1α,25(OH)2D3 induced peaks at approximately the same time points (Figure 5A), while Gemini stimulated continuous IGFBP3 mRNA accumulation (Figure 5B). Interestingly, while the knock-down of HDAC4 and HDAC6 did not significantly change the profile of the response of IGFBP3 mRNA accumulation to Gemini (Figure 5D), it diminished the cycling of the IGFBP3 gene in response to 1α,25(OH)2D3 (Figure 5C). This observation confirmed our hypothesis.
Figure 5.
Silencing of HDAC4 and HDAC6 diminishes transcriptional cycling of the IGFBP3 gene. MCF-10A cells were transfected for 24 h with 200 pmol unspecific control siRNA (A and B) or siRNAs against HDAC4 and HDAC6 (C and D). Then the cells were stimulated for indicated time points with 10 nM 1α,25(OH)2D3 (A and C) or 10 nM Gemini (B and D) and real-time quantitative PCR was performed to measure IGFBP3 gene expression. The data were normalized to the expression of the housekeeping gene RPLP0 and fold inductions were calculated in reference to vehicle control. Data points indicate the means of at least three independent cell treatments and the bars represent standard deviations. A two-tailed Student’s t-test was performed to determine the significance of the stimulation in reference to vehicle-treated control (*P < 0.05; **P < 0.01; ***P < 0.001).
Taken together, silencing HDAC4 and HDAC6 abolishes the cycling of the IGFBP3 gene in response to 1α,25(OH)2D3, but had no effect on the response of the gene to Gemini.
DISCUSSION
In this study, we used the well-known gene IGFBP3 as a model to describe how a simple signal, such as a stimulation with the natural VDR ligand 1α,25(OH)2D3 or its potent synthetic analogue Gemini, can lead to a rather different result in mRNA accumulation. While 1α,25(OH)2D3 induces in MCF-10A cells cyclical mRNA accumulation with phases of RNA synthesis and degradation of a periodicity of 60 min, a stimulation with Gemini results in a steady accumulation of IGFBP3 mRNA. Therefore, after 240 min the IGFBP3 gene is already 5.5-fold induced by treatment with Gemini, while in the same time period 1α,25(OH)2D3 stimulation leads only to a 2.5-fold mRNA accumulation. Possibly due to the cell-specific stability of IGFBP3 mRNA, transcriptional cycling is observed only in cell lines such as MCF-10A, where the mRNA has a short half-life, but not in others such as RWPE-1, where a longer half-life can be assumed. Due to a continuous mRNA accumulation in the latter type of cells 1α,25(OH)2D3 stimulation leads to similar inductions as a treatment with Gemini.
Transcriptional cycling is a rather recently discovered process of nuclear receptor target genes (27–30), and has also been observed with NF-κB target genes (42). mRNA cycling is reflected by cyclical association of transcription factors, RNA polymerase II and their co-factors. In this study, we show cyclical association of VDR, HDAC4 and HDAC6 and chromatin activation on both VDRE regions. The natural ligand 1α,25(OH)2D3 seems to have more prominent effects via the more proximal region RE1/2 and induces cycling, while the analogue Gemini appears to work preferentially via RE3. Importantly, high association levels of VDR coincide with low levels in HDAC4 binding. This indicates that both proteins belong to different phases of the cycling process (discussed below). Moreover, chromatin activation at region RE1/2 correlates well with mRNA accumulation. This suggests that rather the VDREs in RE1/2 mediate the 1α,25(OH)2D3-dependent cycling of the IGFBP3 gene and that the region of RE3 may have only a minor contribution.
Transcriptional cycling is subdivided into three phases. In the first phase, the deactivation phase, co-repressors and HDACs bind to the regulating chromatin regions containing REs and the TSS. In the second activation phase they are replaced by transcription factors and their associated co-activators and in the third initiation phase RNA polymerase II and mediator proteins are associated with the regulatory regions (28,43). Therefore, an up-regulation of HDACs is enhancing and probably prolonging the deactivation phase in which mRNA degradation but no new RNA synthesis occurs.
The 1α,25(OH)2D3 analogue Gemini differs in a number of interesting functional properties from the natural VDR ligand: (i). Gemini induces more efficient association of VDR with RE3 than 1α,25(OH)2D3, which results in a prolonged phase of low levels of HDACs and suggest that there is more mRNA synthesis than degradation, i.e. no occurrence of mRNA cycling, (ii). The genes HDAC4 and HDAC6 are early targets of 1α,25(OH)2D3 but not of Gemini. Due to the negative feedback control mechanism of HDACs on primary VDR target genes, such as IGFBP3, Gemini activates the latter more efficiently than 1α,25(OH)2D3, (iii) In previous studies (36,44), we have shown that Gemini is the more efficient in stabilizing of the active conformation of the VDR than the natural ligand. This leads to a prolonged stability of the VDR–ligand complex and prolongs the phase of active mRNA synthesis.
Via the feed-back loop control mechanism on transcriptional cycling HDACs seem to control overboarding responses of cells to natural nuclear receptor ligands. Only in cases, when reasonable amount of mRNA degradation occurs, such as after up-regulation of HDACs by 1α,25(OH)2D3, transcriptional cycling can be observed. This may be one explanation, why 1α,25(OH)2D3 but not Gemini is inducing transcriptional cycling of the IGFBP3 gene. As discussed above, the lack of HDAC up-regulation may shorten the de-activation phase. This suggests that potent VDR agonists, such as Gemini, may have an advantage over the natural ligand, because they bind the VDR so efficiently that the phases of induced RNA synthesis are longer than that of RNA degradation and no cycling is observed.
Loss of HDAC4 has already been shown to increase the basal expression of CDKN1A and to severely disturb its mRNA accumulation pattern upon ligand treatment (30). Interestingly, HDAC4 is also essential for optimal chromatin looping from VDREs to the TSS of the CDKN1A gene in response to the ligand. As the binding and substrate specificity as well as protein associations of HDAC4 are still largely unknown, the reason for its crucial importance in the 1α,25(OH)2D3 response of the genes CDKN1A and IGFBP3 remains unresolved.
In the previous studies (41,45), we could demonstrate that the level of HDAC expression varies significantly between non-malignant and malignant cells and this affects the interference of nuclear receptors and HDACs on the regulation of important cell cycle regulatory genes, such as CDKN1A and Cyclin C. So far it was assumed that nuclear receptor ligands, such as 1α,25(OH)2D3, interact primarily with the actions of HDAC inhibitors, such as TSA. This study now adds an additional level of complexity to the interference of nuclear receptor and HDAC signalling by suggesting that HDAC levels affect the cycling of nuclear receptor target genes. In this way, both the cell-specific basal levels of HDAC genes as well as their primary response to nuclear receptor ligands are important.
In conclusion, 1α,25(OH)2D3 induces a dynamic and orchestrated response of the IGFBP3 gene, where cyclical binding of VDR, HDAC4 and HDAC6 leads to repeated induction of IGFBP3 mRNA production. This feed-back loop mechanism is not used by Gemini, so that treatment with the compound does not lead to transcriptional cycling and consequently to stronger inductions of VDR target genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants from the Academy of Finland, the Juselius Foundation and the University of Luxembourg supported this research. Funding for open access charge: University of Luxembourg
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Milan Uskokovic for 1α,25(OH)2D3 and Gemini and Maija Hiltunen for skilled technical assistance.
REFERENCES
- 1.DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 2004;80:1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- 2.Ingraham BA, Bragdon B, Nohe A. Molecular basis of the potential of vitamin D to prevent cancer. Curr. Med. Res. Opin. 2008;24:139–149. doi: 10.1185/030079908x253519. [DOI] [PubMed] [Google Scholar]
- 3.Liu M, Lee M-H, Cohen M, Bommakanti M, Freedman LP. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 4.Xu HM, Tepper CG, Jones JB, Fernandez CE, Studzinski GP. 1,25-Dihydroxyvitamin D3 protects HL60 cells against apoptosis but down-regulates the expression of the bcl-2 gene. Exp. Cell Res. 1993;209:367–374. doi: 10.1006/excr.1993.1322. [DOI] [PubMed] [Google Scholar]
- 5.Pan Q, Simpson RU. c-myc intron element-binding proteins are required for 1,25-dihydroxyvitamin D3 regulation of c-myc during HL-60 cell differentiation and the involvement of HOXB4. J. Biol. Chem. 1999;274:8437–8444. doi: 10.1074/jbc.274.13.8437. [DOI] [PubMed] [Google Scholar]
- 6.Xie SP, Pirianov G, Colston KW. Vitamin D analogues suppress IGF-I signalling and promote apoptosis in breast cancer cells. Eur. J. Cancer. 1999;35:1717–1723. doi: 10.1016/s0959-8049(99)00200-2. [DOI] [PubMed] [Google Scholar]
- 7.Colston KW, Perks CM, Xie SP, Holly JM. Growth inhibition of both MCF-7 and Hs578T human breast cancer cell lines by vitamin D analogues is associated with increased expression of insulin-like growth factor binding protein-3. J. Mol. Endocrinol. 1998;20:157–162. doi: 10.1677/jme.0.0200157. [DOI] [PubMed] [Google Scholar]
- 8.Matilainen M, Malinen M, Saavalainen K, Carlberg C. Regulation of multiple insulin-like growth factor binding protein genes by 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2005;33:5521–5532. doi: 10.1093/nar/gki872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng L, Malloy PJ, Feldman D. Identification of a functional vitamin D response element in the human insulin-like growth factor binding protein-3 promoter. Mol. Endocrinol. 2004;18:1109–1119. doi: 10.1210/me.2003-0344. [DOI] [PubMed] [Google Scholar]
- 10.Rozen F, Pollak M. Inhibition of insulin-like growth factor I receptor signaling by the vitamin D analogue EB1089 in MCF-7 breast cancer cells: a role for insulin-like growth factor binding proteins. Int. J. Oncol. 1999;15:589–594. doi: 10.3892/ijo.15.3.589. [DOI] [PubMed] [Google Scholar]
- 11.Krishnan AV, Peehl DM, Feldman D. The role of vitamin D in prostate cancer. Recent Results Cancer Res. 2003;164:205–221. doi: 10.1007/978-3-642-55580-0_15. [DOI] [PubMed] [Google Scholar]
- 12.Kelley KM, Oh Y, Gargosky SE, Gucev Z, Matsumoto T, Hwa V, Ng L, Simpson DM, Rosenfeld RG. Insulin-like growth factor-binding proteins (IGFBPs) and their regulatory dynamics. Int. J. Biochem. Cell Biol. 1996;28:619–637. doi: 10.1016/1357-2725(96)00005-2. [DOI] [PubMed] [Google Scholar]
- 13.Yin P, Xu Q, Duan C. Paradoxical actions of endogenous and exogenous insulin-like growth factor-binding protein-5 revealed by RNA interference analysis. J. Biol. Chem. 2004;279:32660–32666. doi: 10.1074/jbc.M401378200. [DOI] [PubMed] [Google Scholar]
- 14.Carlberg C, Polly P. Gene regulation by vitamin D3. Crit. Rev. Eukaryot. Gene Expr. 1998;8:19–42. doi: 10.1615/critreveukargeneexpr.v8.i1.20. [DOI] [PubMed] [Google Scholar]
- 15.Väisänen S, Dunlop TW, Sinkkonen L, Frank C, Carlberg C. Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1α,25-dihydroxyvitamin D3. J. Mol. Biol. 2005;350:65–77. doi: 10.1016/j.jmb.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 16.Sinkkonen L, Malinen M, Saavalainen K, Väisänen S, Carlberg C. Regulation of the human cyclin C gene via multiple vitamin D3-responsive regions in its promoter. Nucleic Acids Res. 2005;33:2440–2451. doi: 10.1093/nar/gki502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seuter S, Väisänen S, Radmark O, Carlberg C, Steinhilber D. Functional characterization of vitamin D responding regions in the human 5-lipoxygenase gene. Biochim. Biophys. Acta. 2007;1771:864–872. doi: 10.1016/j.bbalip.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Polly P, Herdick M, Moehren U, Baniahmad A, Heinzel T, Carlberg C. VDR-Alien: a novel, DNA-selective vitamin D3 receptor-corepressor partnership. FASEB J. 2000;14:1455–1463. doi: 10.1096/fj.14.10.1455. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace DM, Cotter TG. Histone deacetylase activity in conjunction with E2F-1 and p53 regulates Apaf-1 expression in 661W cells and the retina. J. Neurosci. Res. 2009;87:887–905. doi: 10.1002/jnr.21910. [DOI] [PubMed] [Google Scholar]
- 21.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 22.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 25.Monneret C. Histone deacetylase inhibitors. Eur. J. Med. Chem. 2005;40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol. Endocrinol. 2007;21:1745–1755. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 27.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 28.Degenhardt T, Rybakova KN, Tomaszewska A, Mone MJ, Westerhoff HV, Bruggeman FJ, Carlberg C. Population-level transcription cycles derive from stochastic timing of single-cell transcription. Cell. 2009;138:489–501. doi: 10.1016/j.cell.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J. Bone Miner. Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 30.Saramäki A, Diermeier S, Kellner R, Laitinen H, Väisänen S, Carlberg C. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1α,25-dihydroxyvitamin D3. J. Biol. Chem. 2009;284:8073–8082. doi: 10.1074/jbc.M808090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlberg C, Mouriño A. New vitamin D receptor ligands. Expert Opin. Ther. Pat. 2003;13:761–772. doi: 10.1517/13543776.2012.673590. [DOI] [PubMed] [Google Scholar]
- 32.Bouillon R, Okamura WH, Norman AW. Structure-function relationships in the vitamin D endocrine system. Endocr. Rev. 1995;16:200–257. doi: 10.1210/edrv-16-2-200. [DOI] [PubMed] [Google Scholar]
- 33.Herdick M, Bury Y, Quack M, Uskokovic M, Polly P, Carlberg C. Response element- and coactivator-mediated conformational change of the vitamin D3 receptor permits sensitive interaction with agonists. Mol. Pharmacol. 2000;57:1206–1217. [PubMed] [Google Scholar]
- 34.Norman AW, Manchand PS, Uskokovic MR, Okamura WH, Takeuchi JA, Bishop JE, Hisatake J-I, Koeffler HP, Peleg S. Characterization of a novel analog of 1α,25(OH)2-vitamin D3 with two side chains: interaction with its nuclear receptor and cellular actions. J. Med. Chem. 2000;43:2719–2730. doi: 10.1021/jm0000160. [DOI] [PubMed] [Google Scholar]
- 35.Väisänen S, Peräkylä M, Kärkkäinen JI, Uskokovic MR, Carlberg C. Structural evaluation of a vitamin D analogue with two side chains. Mol. Pharmacol. 2003;63:1230–1237. doi: 10.1124/mol.63.6.1230. [DOI] [PubMed] [Google Scholar]
- 36.Molnar F, Peräkylä M, Carlberg C. Vitamin D receptor agonists specifically modulate the volume of the ligand-binding pocket. J. Biol. Chem. 2006;281:10516–10526. doi: 10.1074/jbc.M513609200. [DOI] [PubMed] [Google Scholar]
- 37.Macias Gonzalez M, Samenfeld P, Peräkylä M, Carlberg C. Corepressor excess shifts the two-side chain vitamin D analog Gemini from an agonist to an inverse agonist of the vitamin D receptor. Mol. Endocrinol. 2003;17:2028–2038. doi: 10.1210/me.2003-0072. [DOI] [PubMed] [Google Scholar]
- 38.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 39.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 40.Renehan AG, Zwahlen M, Minder C, O'Dwyer ST, Shalet SM, Egger M. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet. 2004;363:1346–1353. doi: 10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 41.Malinen M, Saramäki A, Ropponen A, Degenhardt T, Väisänen S, Carlberg C. Distinct HDACs regulate the transcriptional response of human cyclin-dependent kinase inhibitor genes to Trichostatin A and 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2008;36:121–132. doi: 10.1093/nar/gkm913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, et al. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlberg C. The impact of transcriptional cycling on gene regulation. Transcription. 2010;1:1–4. doi: 10.4161/trns.1.1.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bury Y, Herdick M, Uskokovic MR, Carlberg C. Gene regulatory potential of 1α,25-dihydroxyvitamin D3 analogues with two side chains. J. Cell Biochem. 2001;81:179–190. doi: 10.1002/jcb.1082. [DOI] [PubMed] [Google Scholar]
- 45.Makkonen KM, Malinen M, Ropponen A, Väisanen S, Carlberg C. Cell cycle regulatory effects of retinoic acid and forskolin are mediated by the cyclin C gene. J. Mol. Biol. 2009;393:261–271. doi: 10.1016/j.jmb.2009.08.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.