Abstract
RNA-Seq has emerged as a revolutionary technology for transcriptome analysis. In this article, we report a systematic comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. On a panel of human/chimpanzee/rhesus cerebellum RNA samples previously examined by the high-density human exon junction array (HJAY) and real-time qPCR, we generated 48.68 million RNA-Seq reads. Our results indicate that RNA-Seq has significantly improved gene coverage and increased sensitivity for differentially expressed genes compared with the high-density HJAY array. Meanwhile, we observed a systematic increase in the RNA-Seq error rate for lowly expressed genes. Specifically, between-species DEGs detected by array/qPCR but missed by RNA-Seq were characterized by relatively low expression levels, as indicated by lower RNA-Seq read counts, lower HJAY array expression indices and higher qPCR raw cycle threshold values. Furthermore, this issue was not unique to between-species comparisons of gene expression. In the RNA-Seq analysis of MicroArray Quality Control human reference RNA samples with extensive qPCR data, we also observed an increase in both the false-negative rate and the false-positive rate for lowly expressed genes. These findings have important implications for the design and data interpretation of RNA-Seq studies on gene expression differences between and within species.
INTRODUCTION
In the past decade, there has been great interest in using genomic tools to identify gene expression differences between closely related species (1–3). A series of studies have used the DNA microarray technology to globally compare expression levels of orthologous genes between humans and non-human primates (4–10). These studies offer important insight into human evolution and diseases (1,3). For example, it has been reported that the expression level of energy-metabolism genes has an overall increase during human evolution (11–13). This finding is attributed to the important role of increased energy production in the evolution of the human brain size (14). A popular approach in microarray-based interrogation of primate transcriptomes is to hybridize primate tissue samples to a microarray platform designed for analysis of human genes (2,15,16). Based on the fluorescent intensities of individual oligonucleotide probes that perfectly match orthologous transcripts, one can estimate and compare gene expression levels in multiple species. Indeed, with the rapid increase in probe density on common microarray platforms, this strategy of ‘cross-species microarray hybridization’ (15) is able to reliably detect a large number of genes with differential expression between humans and non-human primates. For example, we recently reported the use of a high-density Affymetrix Human Exon Junction Array (HJAY array) to profile gene expression in chimpanzees and rhesus macaques (17). This array contains eight probes per exon for ∼315 000 exons in the human genome (17–19). By exploiting the high probe density and the large number of HJAY probes that perfectly match orthologous transcripts of all three species, we detected differences in gene expression levels between species with a validation rate of >95% by real-time qPCR (i.e. 32 validated out of 33 tested) (17).
However, despite these successes, microarray-based comparison of gene expression between closely related species has its inherent limitations. Most importantly, microarray studies are restricted by probes designed to target the genes in a given species’ genome. In cross-species comparison of gene expression, since sequence divergence between species could affect microarray probe hybridization, it is important to select oligonucleotide probes that perfectly match orthologous transcripts for accurate estimation of expression levels (10). However, this necessary probe-selection step leads to a significant reduction in gene coverage especially in the analysis of more distant species (16). For example, on the extremely probe-rich HJAY array, with a threshold of 11 perfect-match probes for orthologous transcripts we were able to compare expression levels of 12 481 orthologous gene groups from human, chimpanzee, orangutan and rhesus genomes, out of 16 774 RefSeq human genes initially interrogated by the array design (17). Thus, a significant fraction of genes, particularly those with a higher rate of mRNA sequence divergence, cannot be compared across species by the microarray approach.
Recently, RNA-Seq has emerged as a powerful new technology for transcriptome analysis (20–24). By mapping millions of RNA-Seq reads to individual genes’ transcripts, one can estimate the overall mRNA abundance and detect differentially expressed genes (DEGs) (21,25). In particular, since RNA-Seq is not constrained by any prior platform design and can be used to analyze any transcriptome of interest, it provides an attractive approach for globally comparing gene expression between species (26). However, the accuracy of RNA-Seq in detecting DEGs between species and a systematic comparison of RNA-Seq with other technologies (such as high-density arrays and real-time qPCR) for this application have not been reported.
In this article, we report an RNA-Seq analysis of differential gene expression between humans and non-human primates. On a panel of human/chimpanzee/rhesus cerebellum RNA samples previously examined by the high-density HJAY array and real-time qPCR (17), we generated a total of 48.68 million 36-bp RNA-Seq reads for these three species. By comparing RNA-Seq data with results obtained by the high-density HJAY array and real-time qPCR, we systematically assessed how accurately RNA-Seq can reveal gene expression differences between closely related species.
MATERIALS AND METHODS
RNA-Seq, HJAY array and real-time qPCR data of human, chimpanzee and rhesus macaque cerebellum tissues
We performed single-end, 36-bp RNA-Seq of a panel of three cerebellum RNA samples from humans, chimpanzees and rhesus macaques. RNA-Seq libraries were prepared by the Iowa State University DNA facility, and sequencing was conducted on the Illumina Genome Analyzer II following the manufacturer’s standard protocol. The human cerebellum RNA sample was a pool of 24 male and female donors purchased from Clontech (Mountain View, CA, USA). The chimpanzee and rhesus cerebellum RNA samples were both pools of cerebellum tissues of three animals, which were generously provided by the Southwest National Primate Research Center (San Antonio, TX, USA). These RNA samples were previously profiled by microarray and real-time qPCR for gene expression differences between species (17). We performed two lanes of RNA-Seq per sample for the human and rhesus cerebellum samples, and one lane of RNA-Seq for the chimpanzee cerebellum sample. This produced a total of 48.68 million RNA-Seq reads (Supplementary Table S1). The resulting sequencing reads have been deposited to the National Center for Biotechnology Information (NCBI) short-read archive under the accession number SRA023554.1.
For comparison with RNA-Seq analysis, we also analyzed our existing microarray and real-time qPCR data on the same set of samples. In our previous study, we profiled gene expression in these cerebellum samples using the HJAY (NCBI GEO GSE15666) (17). This array contained eight probes per exon for ∼315 000 exons in the human genome (17–19). On HJAY probes that perfectly matched human/chimpanzee genomes or human/rhesus genomes at a single unique location, we used our iterative probe-selection algorithm developed for gene-level analysis of exon array data to estimate the expression levels of orthologous genes in human, chimpanzee and rhesus tissues (27,28). After quantile normalization of the calculated expression indices, we used the Bioconductor LIMMA package (29) to identify DEGs between humans and chimpanzees or humans and rhesus macaques, with a minimum fold-change of 2 and a false discovery rate (FDR) cutoff of 0.01. Additionally, among the DEGs detected from the HJAY array analysis, we previously selected 33 gene pairs and tested the relative expression levels between species by SYBR-green real-time qPCR using the housekeeping gene HPRT1 as the reference. Of the 33 DEGs tested, 32 were confirmed by qPCR as having at least 2-fold changes in expression levels. The between-species expression fold changes estimated by the HJAY array and by qPCR had a correlation of >0.85 (17). In the present work, these preexisting HJAY array and real-time qPCR data sets were compared against RNA-Seq data to assess how accurately RNA-Seq can detect differential gene expression between closely related species.
Estimation and comparison of overall gene expression levels in humans and non-human primates from RNA-Seq data
We used the short-read mapping tool SeqMap (30) to search the RNA-Seq reads against the genomes of human (hg18), chimpanzee (panTro2) and rhesus macaque (rheMac2), when appropriate. From the SeqMap results, we identified reads that matched a single unique location in the corresponding genome, allowing up to two mismatches.
In the context of RNA-Seq analysis, we define the overall gene expression level as the total abundance of all mRNAs transcribed from a single gene locus, including various forms of final transcripts arising from alternative promoter usage, alternative splicing and alternative polyadenylation. To derive an estimate of the overall gene expression level which was robust toward variations in RNA processing patterns, for each gene we counted the number of RNA-Seq reads that uniquely mapped to its constitutive exons, i.e. exons always incorporated into the final transcripts during splicing. To identify these constitutive exons, for each human gene we used transcript annotations in the UCSC KnownGenes database (31,32) to identify exons that were shared among all transcripts annotated for this gene. To avoid spurious transcript annotations in the KnownGenes database, we removed transcripts whose number of exons was less than half of the maximum number of exons in any transcript of this gene. We then identified the orthologous regions of all human constitutive exons in the chimpanzee and rhesus genomes using the UCSC pairwise genome alignments of the human genome (hg18) to the genomes of chimpanzee (panTro2) and rhesus macaque (rheMac2) (33). In comparison of overall gene expression levels between human and chimpanzee (or rhesus) tissues, for each pair of orthologous genes we counted the total number of RNA-Seq reads that uniquely mapped to the human constitutive exons in the human genome, and the total number of RNA-Seq reads that uniquely mapped to the orthologous exon regions in the chimpanzee (or rhesus) genome. We used a previously described Poisson model (25), which controlled for the total number of mapped reads in each lane, to identify the DEGs between humans and rhesus macaques or between humans and chimpanzees, with a minimum fold-change of 2 and a FDR cutoff of 0.01. The FDR was calculated using the approach of Benjamini and Hochberg (34).
Real-time qPCR and RNA-Seq data on the MicroArray Quality Control human reference RNA samples
We collected RNA-Seq and TaqMan qPCR data for two reference RNA samples in the MicroArray Quality Control (MAQC) project (35). The TaqMan qPCR data for human Universal Human Reference RNA (UHR) and Human Brain Reference RNA (brain) samples were downloaded from the NCBI GEO database (NCBI GEO GSE5350), with four replicate assays for each gene in each sample (35). The downloaded data were the normalized expression values in which the housekeeping gene POLR2A was used as the reference gene. For each replicate assay, the cycle threshold (Ct) value in the TaqMan qPCR assay was subtracted from the average POLR2A Ct to obtain a delta Ct value (POLR2A − gene of interest). A higher delta Ct value indicates a higher expression level of the gene of interest. The Illumina RNA-Seq data on the same UHR and brain samples were downloaded from the NCBI SRA database (SRA008403). SeqMap (30) was used to search the reads against the human genome (hg18). From the SeqMap results, we identified reads that matched a single unique location in the human genome, allowing up to two mismatches. For each gene, we counted the total number of reads that mapped to its constitutive exons in the UHR and brain samples. Similar to the TaqMan qPCR analysis, the read counts in the two samples were normalized to POLR2A.
RESULTS
Detection of DEGs between humans and non-human primates using RNA-Seq
To assess how reliably RNA-Seq can identify gene expression differences between closely related species, we performed single-end, 36-bp Illumina RNA-Seq on a panel of three cerebellum RNA samples from humans, chimpanzees and rhesus macaques. We performed a total of five lanes of RNA-Seq: two lanes per sample for the human and rhesus cerebellum RNAs, and one lane for the chimpanzee cerebellum RNA. Altogether, we generated ∼49 million RNA-Seq reads from these five lanes. Of these five lanes of data, we mapped 85.9–92.4% of reads in each lane to the respective genome allowing up to 2-bp mismatches. Between 73.4% and 78.7% of the RNA-Seq reads in each lane were mapped to a single unique location in the respective genome, with an reads per kilobase per million mapped reads (RPKM) read density of 10.7–12.6 within constitutive exon regions (Supplementary Table S1). The number of reads generated per sample (∼10–20 million) and the mapping statistics were comparable with those in published RNA-Seq studies of gene expression (21,25). The resulting gene-level RNA-Seq read counts were highly reproducible for replicate lanes of the same sample (i.e. between the two human replicate lanes, and between the two rhesus replicate lanes), as demonstrated by the MA plots and the QQ-plots comparing gene-level counts of replicate lanes (Supplementary Figures S1 and S2). The lane-by-lane variation was mostly observed for genes with low expression levels (e.g. with less than 32 RNA-Seq reads in both lanes).
To identify DEGs between human and rhesus cerebellum RNAs using RNA-Seq, we used a previously described Poisson model (25), which controlled for the total number of mapped reads in each lane. For all human genes in the NCBI Entrez Gene database, 16 769 genes had at least one mapped RNA-Seq read in at least one of the two sequencing lanes of either species. Of these genes, we identified 7244 DEGs between human and rhesus samples with a gene expression fold-change of at least 2 and an FDR of less than 0.01.
Comparison of RNA-Seq with a high-density exon junction array
We compared RNA-Seq detection of DEGs with results on the same RNA samples using an HJAY array. Previously, we hybridized these human, chimpanzee and rhesus RNA samples to the HJAY array. By exploiting the large number of HJAY probes that perfectly matched orthologous transcripts of all three species, we detected widespread differences in gene expression levels between species with a validation rate of >95% by real-time qPCR (i.e. 32 validated out of 33 tested) (17). Requiring at least six HJAY probes that perfectly matched both human and rhesus transcripts and passed our iterative probe-selection procedure (27,28), we were able to compare expression levels of 12 194 pairs of orthologous genes between humans and rhesus macaques using the HJAY array. Thus, on these two samples, RNA-Seq had substantially higher gene coverage for detecting DEGs between humans and rhesus macaques compared with the HJAY array (16 769 genes versus 12 194 genes).
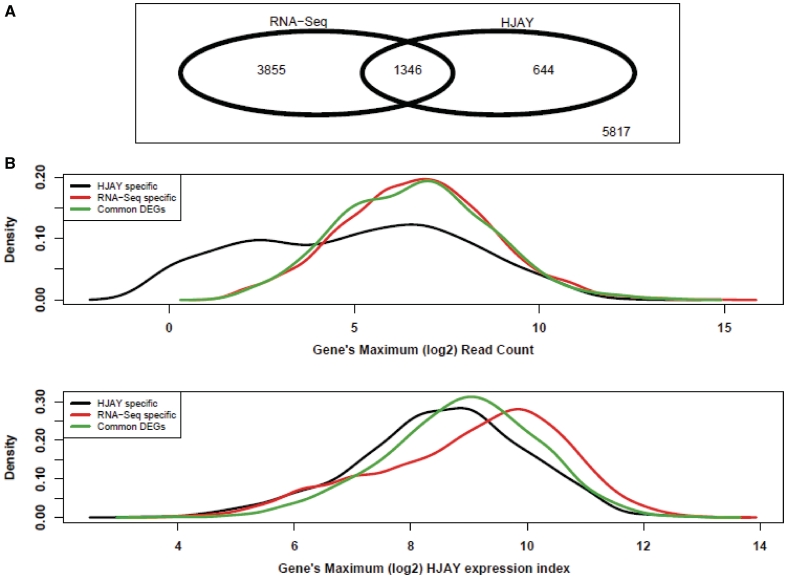
Next, we focused on the 11 662 human–rhesus gene pairs that can be analyzed by both platforms. We compared the log2 expression fold-change between human and rhesus tissues estimated by the HJAY array and by RNA-Seq. We observed a Spearman rank correlation of 0.61 (Supplementary Figure S3), indicating a fair degree of consistency between these two independent platforms. Among these 11 662 genes, we identified 5201 DEGs by RNA-Seq and 1990 DEGs by the HJAY array. A total of 1346 DEGs were identified by both RNA-Seq and the HJAY array (Figure 1A). We noted that, in general, RNA-Seq appeared to be much more sensitive than the HJAY array in detecting DEGs: 67.6% of DEGs identified by the HJAY array were also identified by RNA-Seq, while only 25.9% of DEGs identified by RNA-Seq were also identified by the HJAY array. These results are consistent with other studies reporting that RNA-Seq has a better dynamic range for gene expression levels and increased power to detect DEGs (20). Additionally, 5107 gene pairs analyzed by RNA-Seq did not have sufficient HJAY probes perfectly matching orthologous transcripts, and thus were completely missed by the HJAY analysis. Among these genes, RNA-Seq identified 2043 DEGs between human and rhesus cerebellum RNAs. This illustrates the advantage of the unbiased RNA-Seq analysis, which does not depend on any prior design and covers the entire transcriptome in the comparison of global expression profiles between species.
Figure 1.
A comparison of RNA-Seq and high-density HJAY array for detecting differential gene expression between human and rhesus cerebellum tissues. (A) The Venn diagram showing the overlap between differentially expressed genes (Human versus Rhesus Macaque, FDR <1% and FoldChange ≥2) detected by Illumina RNA-Seq and the Affymetrix HJAY array. Only genes that can be analyzed by both platforms are shown. (B and C) The density plot showing the distributions of gene-level RNA-Seq read counts and HJAY array expression indices for different sets of differentially expressed genes (DEGs) detected by the HJAY array only, or by RNA-Seq only, or by both (i.e. common DEGs). The x-axis is each gene’s maximum log2 RNA-Seq read count (B), or maximum HJAY log2 expression index (C) in human and rhesus samples.
Array-specific DEGs are characterized by low gene expression levels
Our comparison of the RNA-Seq and HJAY data sets also revealed DEGs that were identified by only one of the two platforms. For the 11 662 human–rhesus gene pairs that can be analyzed by both platforms, 3855 DEGs were identified by RNA-Seq only (RNA-Seq specific), and 644 DEGs were identified by HJAY only (HJAY specific). We investigated the representative features of RNA-Seq-specific, HJAY-specific and common DEGs shared by both platforms. Interestingly, we found that, compared with RNA-Seq-specific and common DEGs, HJAY-specific DEGs were marked by significantly lower gene expression levels (Figure 1B and C). For each DEG, we calculated its maximum RNA-Seq read count and maximum HJAY expression index in the human and rhesus samples. The median value of the maximum RNA-Seq read count of all HJAY-specific DEGs was significantly smaller than that of RNA-Seq-specific DEGs and that of common DEGs, respectively (Table 1). We observed a similar pattern when we used the HJAY expression index as the indicator of gene expression levels (Table 1). It should be noted that our previous study indicated a very low FDR (1/33 tested by real-time qPCR) among between-species DEGs identified by the HJAY array analysis (17). Thus, we expect that the vast majority of HJAY specific DEGs represent bona fide gene expression differences between these human and rhesus RNA samples. Together, these data suggest that while RNA-Seq generally has a higher sensitivity to detect between-species DEGs, it has a systematic false-negative problem to detect DEGs with relatively low expression levels in both species. It is possible that such DEGs could be identified by a high-density microarray, especially when there are high-affinity probes to detect signals of relatively low abundance transcripts. On the other hand, they could be missed by RNA-Seq due to insufficient transcript sampling of lowly expressed genes.
Table 1.
HJAY-specific DEGs are characterized by significantly lower gene expression levels compared with RNA-Seq-specific and common DEGs
| Human versus rhesus |
Human versus chimpanzee |
|||
|---|---|---|---|---|
| Median of the maximum RNA-Seq read count | Median of the maximum HJAY expression index | Median of the maximum RNA-Seq read count | Median of the maximum HJAY expression index | |
| HJAY-specific DEGs | 38 (N/A) | 390 (N/A) | 33 (N/A) | 380 (N/A) |
| RNA-Seq-specific DEGs | 104 (P = 4.4e-42*) | 633 (3.0e-18) | 95 (2.5e-18) | 590 (2.5e-11) |
| Common DEGs | 99 (7.7e-31) | 493 (6.9e-7) | 81 (6.2e-12) | 537 (2.1e-6) |
*Compared with HJAY-specific DEGs (P-value of two-sided Wilcoxon rank sum test).
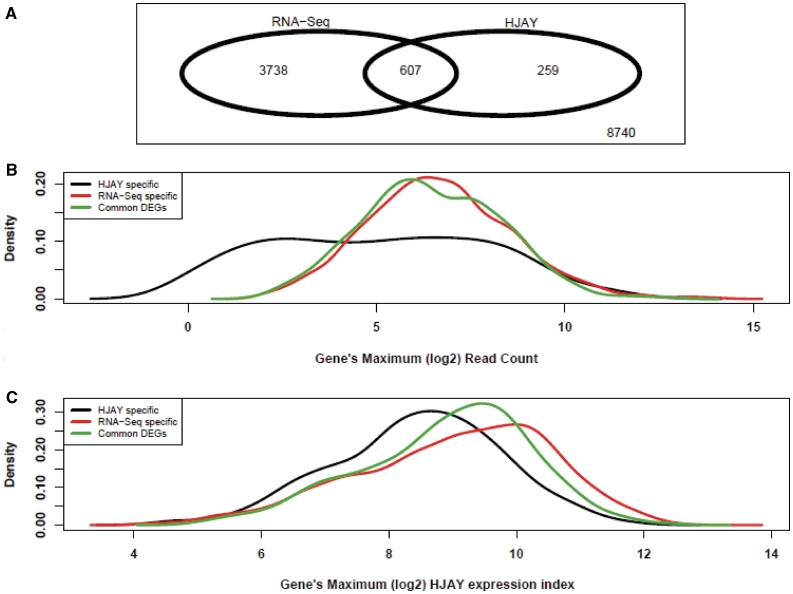
To assess if this false-negative issue for lowly expressed genes also affected RNA-Seq analysis of more closely related species, we performed one lane of RNA-Seq on the chimpanzee cerebellum RNA to produce ∼10.1 million reads. We compared the chimpanzee RNA-Seq data with the human RNA-Seq data to identify DEGs. Among the 13 344 genes that can be analyzed by both RNA-Seq and the HJAY array, we identified 4345 DEGs by RNA-Seq and 866 DEGs by the HJAY array. A total of 607 DEGs were shared by both platforms, accounting for 14.0% DEGs detected by RNA-Seq and 70.1% DEGs detected by the HJAY array (Figure 2A). There were 259 HJAY-specific and 3738 RNA-Seq-specific DEGs. Consistent with the trend observed in the human versus rhesus comparison, HJAY-specific DEGs were characterized by significantly lower expression levels compared with RNA-Seq-specific DEGs and common DEGs (Table 1 and Figure 2B and C).
Figure 2.
A comparison of RNA-Seq and high-density HJAY array for detecting differential gene expression between human and chimpanzee cerebellum tissues. (A) The Venn diagram showing the overlap between differentially expressed genes (Human versus Chimpanzee, FDR <1% and FoldChange ≥2) detected by Illumina RNA-Seq and the Affymetrix HJAY array. Only genes that can be analyzed by both platforms are shown. (B and C) The density plot showing the distributions of gene-level RNA-Seq read counts and HJAY array expression indices for different sets of differentially expressed genes (DEGs) detected by the HJAY array only, or by RNA-Seq only, or by both (i.e. common DEGs). The x-axis is each gene’s maximum log2 RNA-Seq read count (B) or maximum HJAY log2 expression index (C) in human and chimpanzee samples.
Real-time qPCR data confirm low expression levels of between-species DEGs missed by RNA-Seq
The above comparison of RNA-Seq and HJAY data suggest a systematic bias for RNA-Seq to miss between-species DEGs of genes with relatively low expression levels. To further confirm this finding, we examined the RNA-Seq data of 33 human-versus-chimpanzee or human-versus-rhesus orthologous gene pairs, for which we had measured their expression levels using SYBR-green real-time qPCR (17). These gene pairs were randomly selected in our previous HJAY array study of the same human and non-human primate samples for validation of between-species DEGs (17). Of the 33 candidate DEGs tested, 32 were validated by real-time qPCR (11/11 for human-versus-rhesus DEGs; 21/22 for human-versus-chimpanzee DEGs; Supplementary Tables S2 and S3). Among them, RNA-Seq correctly called between-species DEGs for 29 gene pairs, yielding a false-negative rate of 9.4% (i.e. 3/32). The only qPCR-confirmed false-positive DEG in our previous HJAY array study was NT5C. This gene was called by the HJAY array as having >2-fold reduction in expression level in humans as compared with chimpanzees. However, qPCR indicated that the human-versus-chimpanzee fold change was insignificant (1.3-fold reduction in humans; Supplementary Table S3). RNA-Seq called the expression difference of NT5C between humans and chimpanzees as insignificant, with an estimated fold reduction of 1.1, consistent with the qPCR data.
Interestingly, the three ‘gold-standard’ between-species DEGs missed by RNA-Seq were characterized by relatively low expression levels in both the human and chimpanzee (or rhesus) samples. The first gene, EPHA6, was validated by qPCR as having >5-fold increase in humans compared with rhesus macaques (Supplementary Table S2). It has no more than four RNA-Seq reads in any of the human and rhesus macaque sequencing lanes. Although RNA-Seq correctly predicted the direction of expression differences (i.e. up in human), it failed to call this gene as a DEG. The second gene, LPXN, was validated by qPCR as having >2-fold increase in humans compared with chimpanzees (Supplementary Table S3). It was missed by RNA-Seq, with less than 34 reads in any of the human and chimpanzee sequencing lanes. For this gene, RNA-Seq failed to predict the significance of its fold change as well as the correct direction of fold change. The third gene missed by RNA-Seq was SNTG2 in the human-versus-chimpanzee comparison (Supplementary Table S3). Again, this gene has no more than 14 reads in either humans or chimpanzees. To definitely confirm that these three genes had relatively low expression levels, we examined the raw real-time qPCR Ct values of these genes in human and non-human primate samples, and compared them with the 29 ‘gold standard’ DEGs correctly predicted by RNA-Seq. For each DEG, we calculated the difference in Ct value of our reference gene HPRT1 to the Ct value of the gene of interest in both human and chimpanzee (or rhesus when appropriate) samples, and obtained the maximum delta Ct value in one of these two samples. DEGs whose maximum delta Ct values were low had relatively low expression levels in both samples used for identifying the DEGs. We found that the three DEGs missed by RNA-Seq (i.e. false negatives of RNA-Seq) had a median maximum delta Ct of −1.92, as compared with −0.25 for the other 29 DEGs correctly predicted by RNA-Seq (P = 0.07, one-sided Wilcoxon rank sum test). Thus, the gold-standard real-time qPCR data on this relatively small list of genes also indicates a systematic bias of RNA-Seq to have false negatives for DEGs of lowly expressed genes. This is consistent with our genome-scale comparison of RNA-Seq and HJAY array data.
MAQC reference RNA data sets confirm an increased error rate of RNA-Seq for detecting DEGs of lowly expressed genes
The above analysis of RNA-Seq, HJAY and real-time qPCR data on human and non-human primate tissues indicates that RNA-Seq tends to have an increased false-negative rate for genes with relatively low expression levels. However, two questions remained to be answered. First, is this issue unique to the detection of DEGs between closely related species, or does it also affect the detection of within-species DEGs? Second, our analysis so far focused on the false-negative issue of RNA-Seq. Does RNA-Seq also tend to have increased false positives for lowly expressed genes? To address these questions, we compared RNA-Seq and TaqMan qPCR data of two human reference RNA samples in the MAQC project (35). The MAQC project generated TaqMan qPCR data for 960 genes in two samples—human UHR and brain, with four replicate assays per gene in each sample. This provided extensive gold standard expression measurements for assessing the accuracy of RNA-Seq to detect DEGs.
We observed a strong overall concordance between TaqMan qPCR and RNA-Seq in the detection of DEGs. The fold-change estimates by qPCR and by RNA-Seq were strongly correlated (Pearson correlation coefficient was 0.95). We found that 12.7% of qPCR DEGs were missed by RNA-Seq, a false-negative rate comparable with what we observed (9.4%) in our comparison of RNA-Seq and qPCR data between multiple species. Meanwhile, 58 out of 323 non-DEGs according to qPCR data were predicted by RNA-Seq as DEGs, representing a false-positive rate of 18%.
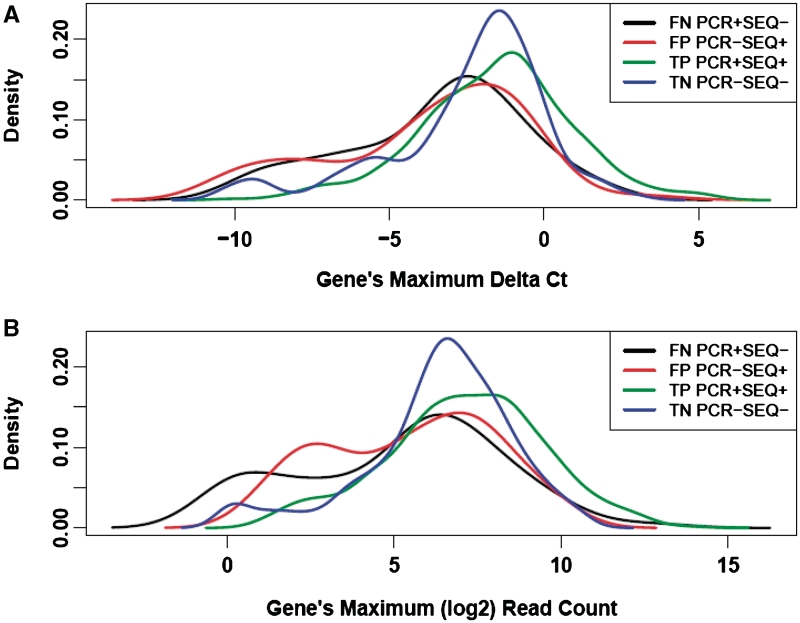
Based on the DEG selection by RNA-Seq, we further divided these genes into four subsets: the true positive (TP) set (qPCR DEGs correctly selected by RNA-Seq), the false positive (FP) set (qPCR non-DEGs incorrectly selected by RNA-Seq), the true negative (TN) set (qPCR non-DEGs correctly excluded by RNA-Seq) and the false negative (FN) set (qPCR DEGs incorrectly excluded by RNA-Seq). We found that both the FP and FN sets were characterized by significantly lower levels of gene expression compared with the TP and TN sets. For each gene, we obtained its maximum RNA-Seq read count and maximum qPCR delta Ct value (the reference gene POLR2A—gene of interest) in the UHR and brain samples. The median values of the maximum qPCR delta Ct and the maximum RNA-Seq gene count of the FN set were significantly smaller than those of the TP set (Table 2 and Figure 3). The median values of the maximum qPCR delta Ct and the maximum RNA-Seq gene count of the FP set were also significantly smaller than those of the TN set (Table 2 and Figure 3). Together, these results indicate that RNA-Seq has a reduced accuracy for detection of DEGs in genes with low expression levels, reflected by an increase in both the false-negative and the false-positive rates.
Table 2.
For DEG detection by RNA-Seq in the MAQC samples, both the false positive (FP) set and the false negative (FN) set are characterized by significantly lower gene expression levels compared with the true positive (TP) set and the true negative (TN) set
| Median of the maximum qPCR delta Ct value | Median of the maximum RNA-Seq read count | |
|---|---|---|
| FN set (PCR+SEQ-) | −2.83 (N/A) | 57 (N/A) |
| TP set (PCR+SEQ+) | −1.41 (8.8e-7*) | 145 (1.5e-6*) |
| FP set (PCR-SEQ+) | −3.20 (N/A) | 59 (N/A) |
| TN set (PCR-SEQ-) | −1.77 (2.7e-3**) | 98 (2.0e-2**) |
*Compared with the FN set (P-value of two-sided Wilcoxon rank sum test).
**Compared with the FP set (P-value of two-sided Wilcoxon rank sum test).
Figure 3.
The density plot showing the distribution of TaqMan qPCR delta Ct value (A) and gene-level RNA-Seq read counts (B) for different sets of genes: false negative set (black), false positive set (red), true positive set (green) and true negative set (blue) in the analysis of the MAQC reference RNA samples (UHR and brain). The x-axis is each gene’s maximum delta Ct value (POLR2A—gene of interest) (A), or maximum log2 RNA-Seq read count (B) in UHR and brain samples. The median TaqMan qPCR delta Ct of these four sets of gene is −2.83, −3.20, −1.41 and −1.77, respectively. The median RNA-Seq read count of these four sets of gene is 57, 59,145 and 98, respectively.
Recent work on RNA-Seq has revealed a potential detection bias toward differential expression of genes with long transcripts (36). To assess whether this issue confounded the trend that we observed in this study, we normalized the RNA-Seq gene count by the total length of constitutive exons and plotted the distribution of transcript-length normalized gene counts for different sets of genes. We observed the same trend (Supplementary Figure S4) that the accuracy of DEG detection by RNA-Seq was reduced for genes with low transcript-length normalized gene counts.
DEG detection by RNA-Seq: the influence of sequencing count and extent of expression change
The analysis of the MAQC RNA-Seq and TaqMan qPCR data confirmed a systematic bias for RNA-Seq in the analysis of genes with relatively low expression levels. This raised several interesting questions. How does the sequencing count affect the false-negative rate of RNA-Seq for detecting true DEGs? Does the false-negative rate also depend on the extent of expression change one wishes to study?
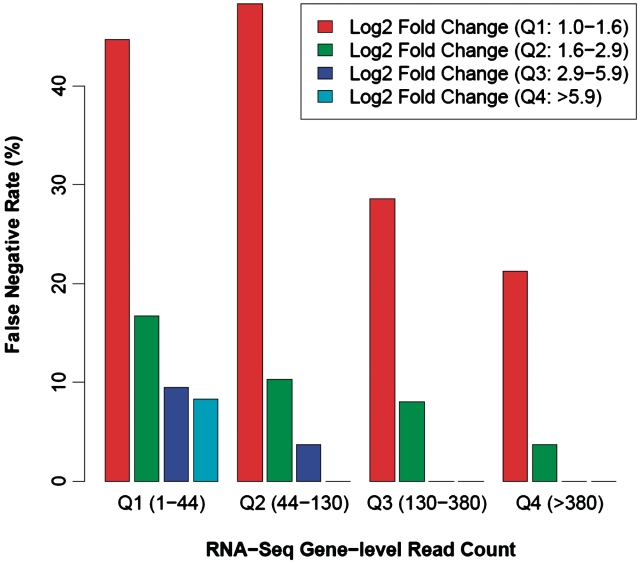
To address these questions, we again made use of the MAQC data to assess the influence of sequencing count and extent of expression change on DEG detection. Based on the TaqMan qPCR estimates of expression fold change (FC) between the MAQC human UHR and brain samples, we grouped the 510 qPCR DEGs into four quartiles: first quartile, log2 FC between 1 and 1.6; second quartile, 1.6–2.9; third quartile, 2.9–5.9 and fourth quartile, >5.9. Similarly, we grouped the 510 qPCR DEGs into four quartiles based on the maximum gene-level RNA-Seq read count in the human UHR and brain samples: first quartile, read count 1–44; second quartile, 44–130; third quartile, 130–380; and fourth quartile, >380. For each fold-change group, we calculated the proportion of TaqMan qPCR DEGs missed by RNA-Seq (i.e. RNA-Seq false-negative rate) in individual RNA-Seq read count groups. This analysis allowed us to investigate how the RNA-Seq false-negative rate could be affected by the extent of expression change and the number of sequences per gene.
Our results indicate that DEGs with larger extent of expression change require less sequencing count to identify the change (Table 3 and Figure 4). For DEGs with small expression fold-change (i.e. log2 FC between 1.0 and 1.6; first quartile based on the expression fold-change), only 55.3% of DEGs from the first RNA-Seq read-count quartile (i.e. <44 reads) can be detected by RNA-Seq, while 78.8% of DEGs from the fourth RNA-Seq read-count quartile (i.e. >380 reads) can be detected by RNA-Seq. In contrast, for DEGs with larger expression fold-change (i.e. log2 FC between 1.6 and 2.9; second quartile based on the expression fold-change), 83.3% of DEGs from the first RNA-Seq read-count quartile (i.e. <44 reads) can be detected by RNA-Seq, while 96.3% of DEGs from the fourth RNA-Seq read-count quartile (i.e. >380 reads) can be detected by RNA-Seq. Consistent with this trend, we obtained even lower false-negative rates (i.e. higher RNA-Seq detection rate) in DEGs from the third and fourth quartiles of expression-fold change (Table 3 and Figure 4). Together, our results confirm that the ability of RNA-Seq to identify significant DEGs is positively associated with both the number of sequences per gene and the extent of expression-fold change.
Table 3.
The proportions of MAQC TaqMan qPCR DEGs missed by RNA-Seq, stratified by the number of sequences per gene (i.e. gene-level RNA-Seq read counts) and the extent of expression change (i.e. log2 fold-change between brain and UHR)
| Q1a (1–44) (%) | Q2a (44–130) (%) | Q3a (130–380) (%) | Q4a (>380) (%) | |
|---|---|---|---|---|
| Q1b (1.0–1.6) | 44.7 | 48.3 | 28.6 | 21.2 |
| Q2b (1.6–2.9) | 16.7 | 10.3 | 8.0 | 3.7 |
| Q3b (2.9–5.9) | 9.5 | 3.7 | 0.0 | 0.0 |
| Q4b (>5.9) | 8.3 | 0.0 | 0.0 | 0.0 |
aThe four quartiles based on the gene-level RNA-Seq read counts.
bThe four quartiles based on the TaqMan qPCR estimates of expression fold change (log2 scale).
Figure 4.
The ability of RNA-Seq to identify significant DEGs is positively associated with both the number of sequences per gene and the extent of expression change. Based on the TaqMan qPCR estimates of expression fold-change (FC) between the MAQC human UHR and brain samples, we grouped the 510 qPCR DEGs into four quartiles: first quartile, log2 FC between 1 and 1.6; second quartile, 1.6–2.9; third quartile, 2.9–5.9 and fourth quartile, >5.9. Similarly, we grouped the 510 qPCR DEGs into four quartiles based on the maximum gene-level RNA-Seq read count in the human UHR and brain samples: first quartile, read count 1–44; second quartile, 44–130; third quartile, 130–380 and fourth quartile, >380. For each fold-change group, we calculated the proportion of TaqMan qPCR DEGs missed by RNA-Seq (i.e. RNA-Seq false-negative rate) in individual RNA-Seq read count groups, as shown in the barchart.
We also used the MAQC samples (8.1 million reads for UHR and 10.1 million reads for brain) to determine the overall distribution of sequence count per gene from a typical RNA-Seq data set of ∼10 million reads. At this sequencing depth, 34% of human genes had read counts >1 and <44 (first quartile in our analysis of the qPCR DEGs), 13% of human genes had read count >380 (fourth quartile) and 41% of human genes had intermediate read counts of between 44 and 380 (second and third quartiles) (Supplementary Figure S5).
DISCUSSION
RNA-Seq has emerged as a revolutionary technology for transcriptome analysis (20). Unlike gene expression microarray which relies on prior probe design and existing transcript annotations, RNA-Seq can be used to analyze any transcriptome. This is particularly useful for achieving complete gene coverage when comparing transcriptomes of multiple species.
In this work, we systematically assessed how accurately RNA-Seq can detect DEGs between closely related species, specifically between humans and non-human primates. We generated ∼49 million RNA-Seq reads on a panel of cerebellum RNA samples of humans, chimpanzees and rhesus macaques, which were previously profiled by the HJAY array and real-time qPCR. Although the HJAY array is a newly designed exon array with an unprecedented probe density (17–19), our results indicate that RNA-Seq has substantially better coverage for DEGs in cross-species comparison of gene expression. For example, in the human-versus-rhesus comparison of gene expression levels, 65.0% of DEGs identified by the HJAY array can be identified by RNA-Seq. In contrast, only 18.6% of DEGs identified by RNA-Seq can be identified by the HJAY array. In fact, 28.2% of RNA-Seq DEGs cannot even be analyzed by the HJAY array due to lack of probes perfectly matching orthologous transcripts of both species. It should be noted that with the same statistical criteria, we estimated a false-positive rate of ∼18% for RNA-Seq detection of DEGs in the MAQC human reference RNA samples. Thus, we expect that the vast majority of RNA-Seq-specific DEGs represent bona fide gene expression differences between these human and non-human primate samples. The significantly improved gene coverage and increased sensitivity for DEGs will provide a powerful tool that can greatly advance our understanding of transcriptome changes during human evolution.
Despite the strength of RNA-Seq for comparative studies of gene expression, we also observed a systematic bias for RNA-Seq in the analysis of genes with relatively low expression levels. Specifically, DEGs missed by RNA-Seq (i.e. false negatives) were characterized by low expression levels in samples being examined, as indicated by lower RNA-Seq read counts, lower HJAY expression indices and higher raw Ct values by real-time qPCR. Additionally, our analysis of the MAQC human data set showed that this issue was not restricted to the comparison of gene expression between species. In the comparison of two human reference RNA samples, we found an increase in both the false-negative rate and the false-positive rate for RNA-Seq detection of DEGs with relatively low expression levels.
Although our finding appears to contradict the general assumption that RNA-Seq is particularly suitable for analysis of lowly expressed genes as compared with microarrays (23,24), it is not entirely unexpected. For DEGs with low mRNA abundance, RNA-Seq at 10–20 million reads per sample may not achieve sufficient transcript sampling to accurately detect the changes in their mRNA concentrations. Meanwhile, for non-DEGs with low mRNA abundance, random sampling noise during RNA-Seq may have a more significant impact on their final read counts, thus causing an increase in false-positive predictions. It is reassuring that among the three RNA-Seq false negatives of qPCR-confirmed DEGs between humans and non-human-primates, RNA-Seq correctly predicted the direction of expression changes in two genes. It is possible that these DEGs can be eventually detected by even deeper RNA-Seq. It should be noted that the sequencing depth in this study (10–20 million reads per sample) is comparable with most published RNA-Seq studies of gene expression levels (21,25). Thus, our conclusions have important implications for the design and data interpretation of RNA-Seq studies on gene expression differences between and within species.
In our study, the RNA-Seq data were generated on pooled RNA samples from each species. Specifically, the human cerebellum RNA sample was a pool of 24 male and female donors, and the chimpanzee and rhesus cerebellum RNA samples were both pools of cerebellum tissues of three animals. It should be noted that RNA pooling is a common practice in many gene expression studies, and has been well justified based on statistical and practical considerations (37). Pooling is a desirable strategy when the cost to profile individual samples is high and the primary research goal is to identify differences in gene expression profiles between different biological classes. Studies on pooling have concluded that it does not adversely affect the inference for most genes (37). It should also be noted that because of the current high cost of RNA-Seq as well as the long running time and limited access of high-throughput sequencers (compared with arrays), it is common for RNA-Seq studies to adopt the pooling strategy to reduce the number of sequencing runs. Our experimental design and the number of reads generated (1–2 lanes of data per RNA pool) were representative of published RNA-Seq studies of gene expression. By designing our experiment in this way, we expect the conclusions drawn from our data set will be valuable to many investigators using the RNA-Seq technology in diverse settings. Moreover, in our previous HJAY array analysis of these samples, we found a strong correlation in expression profiles of biological replicates from individual species (17). For example, the three rhesus macaque cerebellum samples had a Pearson correlation coefficient of at least 0.98 in their expression profiles. This further justifies the use of the pooling strategy in our RNA-Seq study design.
Although the RNA-pooling strategy does not assess biological variations among individual samples from each species, it must be emphasized that the major finding of our study, i.e. there exist a systematic bias and a higher false-negative rate for RNA-Seq in the analysis of DEGs with relatively low expression levels, cannot be an artifact attributed to the pooling strategy. It is well known that the pooling strategy could potentially increase statistical power by reducing the biological variation among individual samples comprising the RNA pool (37,38). Thus, in a study without pooling we would expect a more pronounced false-negative problem for RNA-Seq to detect between-species DEGs with relatively low expression levels. Similarly, the MAQC data sets also do not contain information about biological variation. Nonetheless, the MAQC samples have been used extensively for assessing gene expression platforms and statistical methods of data analysis (35,39). The tremendous insights gained from these studies further demonstrate the validity and utility of evaluating gene expression technologies without addressing the issue of biological variation.
Another important issue in comparative studies of gene expression is to distinguish bona fide inter-species expression differences from intra-specific variations of gene expression, which can arise from a variety of confounding factors such as the age, gender and health conditions of individual samples. To address this issue, it is usually necessary to perform expression profiling and validation experiments on a large number of samples from each individual species. However, this issue is not a concern to the present study, as the goal of this work is to investigate the accuracy of RNA-Seq in between-species comparisons of gene expression levels. For this purpose, as long as there are genuine expression differences of orthologous genes in samples from different species, we can seek to assess various factors (such as gene expression level and extent of expression change) that affect the accuracy of RNA-Seq. The underlying cause of such expression differences does not confound our evaluation of RNA-Seq and the comparison to microarray technology. Nonetheless, it should be clarified that because of our limited sample size, the gene expression differences identified from the RNA-Seq data set are not necessarily equivalent to genuine species-specific expression patterns.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grants (R01HG004634 and R01GM088342, to Y.X.). Roswell Park Cancer Institute research startup fund (to S.L.). Funding for open access charge: National Institutes of Health grant (R01-HG004634).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Jerilyn Pecotte, Mary Jo Aivaliotis, Kelly Clark and Michael Baker for assistance. We thank David Eichmann, Ben Rogers and the University of Iowa Institute for Clinical and Translational Science (NIH grant UL1 RR024979) for computer support. This study used biological materials obtained from the Southwest National Primate Research Center, which is supported by NIH-NCRR grant P51 RR013986.
REFERENCES
- 1.Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 2.Gilad Y, Borevitz J. Using DNA microarrays to study natural variation. Curr. Opin. Genet. Dev. 2006;16:553–558. doi: 10.1016/j.gde.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Preuss TM, Caceres M, Oldham MC, Geschwind DH. Human brain evolution: insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- 4.Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 6.Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 7.Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 10.Gilad Y, Rifkin SA, Bertone P, Gerstein M, White KP. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 2005;15:674–680. doi: 10.1101/gr.3335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaitovich P, Lockstone HE, Wayland MT, Tsang TM, Jayatilaka SD, Guo AJ, Zhou J, Somel M, Harris LW, Holmes E, et al. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 2008;9:R124. doi: 10.1186/gb-2008-9-8-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat. Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- 13.Uddin M, Wildman DE, Liu G, Xu W, Johnson RM, Hof PR, Kapatos G, Grossman LI, Goodman M. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl Acad. Sci. USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varki A, Altheide TK. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 15.Gilad Y, Oshlack A, Rifkin SA. Natural selection on gene expression. Trends Genet. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Oshlack A, Chabot AE, Smyth GK, Gilad Y. Using DNA microarrays to study gene expression in closely related species. Bioinformatics. 2007;23:1235–1242. doi: 10.1093/bioinformatics/btm111. [DOI] [PubMed] [Google Scholar]
- 17.Lin L, Liu S, Brockway H, Seok J, Jiang P, Wong WH, Xing Y. Using high-density exon arrays to profile gene expression in closely related species. Nucleic Acids Res. 2009;37:e90. doi: 10.1093/nar/gkp420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto ML, Clark TA, Gee SL, Kang JA, Schweitzer AC, Wickrema A, Conboy JG. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood. 2009;113:3363–3370. doi: 10.1182/blood-2008-05-160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen S, Warzecha CC, Carstens RP, Xing Y. MADS+: discovery of differential splicing events from Affymetrix exon junction array data. Bioinformatics. 2010;26:268–269. doi: 10.1093/bioinformatics/btp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 24.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 25.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapur K, Xing Y, Ouyang Z, Wong WH. Exon arrays provide accurate assessments of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing Y, Kapur K, Wong WH. Probe selection and expression index computation of affymetrix exon arrays. PLoS ONE. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 30.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 33.Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 35.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol. Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc. Natl Acad. Sci. USA. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng X, Wood CL, Blalock EM, Chen KC, Landfield PW, Stromberg AJ. Statistical implications of pooling RNA samples for microarray experiments. BMC Bioinformatics. 2003;4:26. doi: 10.1186/1471-2105-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.