Abstract
MicroRNAs fine-tune the activity of hundreds of protein-coding genes. The identification of tissue-specific microRNAs and their promoters has been constrained by the limited sensitivity of prior microRNA quantification methods. Here, we determine the entire microRNAome of three endoderm-derived tissues, liver, jejunum and pancreas, using ultra-high throughput sequencing. Although many microRNA genes are expressed at comparable levels, 162 microRNAs exhibited striking tissue-specificity. After mapping the putative promoters for these microRNA genes using H3K4me3 histone occupancy, we analyzed the regulatory modules of 63 microRNAs differentially expressed between liver and jejunum or pancreas. We determined that the same transcriptional regulatory mechanisms govern tissue-specific gene expression of both mRNA and microRNA encoding genes in mammals.
INTRODUCTION
MicroRNAs are short non-coding RNAs of 21–23 nt that are present in multiple organisms and that are often evolutionarily conserved (1). MicroRNAs function by suppressing the expression of protein coding genes, with each microRNA targeting dozens or even hundreds of mRNAs. In mammals, microRNA function on a global level has been studied through mutational analysis of Dicer, an obligate enzyme in the processing of microRNA precursors. Thus, it was shown that microRNAs are required for ES self-renewal as well as development and function of tissues including liver (2,3), intestine (4) and heart (5).
There are more than 1000 microRNAs encoded in the mammalian genome, and these are derived from a complex series of processing steps. The primary transcript, or pri-microRNA, synthesized by RNA polymerase II or III is very labile, and quickly converted to ∼70 nt precursors, termed pre-microRNA (6). These pre-microRNAs exist as hairpins and are further processed through a series of endonuclease digestion steps to the final and functional microRNAs, which are loaded onto the so-called RNA inducing silencing complex (RISC) to exert their regulatory functions. Because of their very short sequence, quantification of microRNAs by array-based technologies has its limitations, as the hybridization conditions used cannot be optimized for all microRNA probes simultaneously. Previous tissue surveys used cloning and sequencing to determine the microRNA abundance in multiple tissues at low sequencing depth. While these assays could not capture the entire microRNAome, they nevertheless established that microRNAs are expressed in a tissue-specific manner (7). Recent studies have demonstrated that transcription factors can regulate microRNA expression; however, binding sites have been confirmed experimentally for only a small number of microRNA promoters, and little is known about the mechanisms that influence tissue-specific expression of microRNAs (8–10). In order to elucidate the regulatory networks that govern tissue-specific expression of microRNA genes, we determined their complete expression profile by ultra-high throughput sequencing in three endoderm-derived tissues. The greatly expanded number of differentially expressed microRNAs identified through this method provided sufficient sequence depth to determine the cis-regulatory modules that control the differentially expressed microRNA genes. Moreover, the results of our analysis established that microRNA genes are governed by the same transcription factor networks that also control protein-coding genes.
MATERIALS AND METHODS
Processing microRNA reads
Male CD-1 mice aged 8–12-week-old (Charles River Laboratories) underwent liver harvest between 8 AM and 12 PM. Small intestinal mucosa was isolated via mucosal scraping. Pancreata were harvested from the same mice and snap frozen. Chromatin was later prepared from a portion of the frozen tissue as described earlier (11) except that protease inhibitors were also included in the PBS and cell lysis buffers. H3K4Me3 ChIP was performed as described earlier (12). Total and small RNAs were extracted using the mirVana microRNA Isolation kit (Cat. # AM1561, Ambion, Austin, TX, USA). Small RNAs libraries were generated using the DGE-small RNA Sample prep kit (Cat. # FC-102-1009; Illumina, San Diego, CA, USA). Illumina sequencing libraries were prepared using the ‘long’ Illumina protocol according to the manufacturer’s directions. Purified PCR product was loaded on an Agilent Technologies 2100 Bioanalyzer to confirm sample quantity and integrity. Reads from six liver, five small intestinal mucosa, and two pancreas samples were sequenced on an Illumina GA-II following manufacturer’s instructions. The 3′-adapter was trimmed from the end of each read and the frequencies of the resulting oligos were tabulated for each lane and in total. The oligos were aligned to precursor hairpins (mirBase 14), RefSeq sequences, and the mouse genome (NCBI Build 36; mm8) using ELAND and allowing up to two mismatches. Alignments of reads in the length range 19–25 bp were assigned to the mature microRNA that they overlapped. When mature forms shared an oligo in this length range, they were merged into an ad hoc family for reporting read counts and for differential expression calculations. All high-throughput sequencing data are accessible from the NCBI Short Read Archive under accession number SRA023764.
Identifying differentially-expressed microRNAs
To identify differentially-expressed microRNAs we used read counts in reads per million (RPM) from six replicates from liver, five from small intestine, and two from pancreas. The RPM values were quantile normalized in R using the normalizeBetweenArrays function of the limma package. These values were then analyzed using SAMR, and microRNAs with an FDR ≤10%, a minimum of 1.5-fold change, and at least 100 RPM average expression (in the appropriate tissue) were selected as differentially expressed.
ChIP for histone modifications
Immunoprecipitations were performed as described earlier (11), except that 4 μg of chromatin and 4 μg of antibodies were used for each reaction. Chromatin was immunoprecipitated with antibodies for H3K4me3 (Millipore, Cat# CS200580). Immunoprecipitation was confirmed by calculating enrichment of control liver, jejunal mucosa and pancreas expressed genes using control intergenic regions, by comparing input DNA to ChIP DNA. The immunoprecipitated DNA was prepared for sequencing as per Illumina’s instructions (http://www.illumina.com) and previously described (13). High-throughput sequencing was performed on an Illumina GA-II following manufacturer’s instructions. The 36-bp reads were aligned to the mouse genome (NCBI Build 36; mm8) using ELAND and allowing up to two mismatches. Reads with a unique best alignment were included in further processing (liver H3K4me3 ChIP: 7 720 909, input: 12 056 786; small intestine H3K4me3 ChIP: 8 492 668, input: 11 961 006, pancreas H3K4me3 ChIP: 20 176 620).
Identifying regions of significant H3K4me3 modification
We used GLITR (13) to identify regions of significant enrichment of H3K4me3 as compared to input using a 1% FDR. Adjacent GLITR regions in each tissue were merged if they were within 1500 bp. The merged regions were considered to be candidate TSSs. We created an atlas of all H3K4me3 regions by merging overlapping regions from all three endodermal tissues. To quantify the strength of modification in each tissue, we computed the length-normalized rate of tissue-specific reads (reads per kilo basepair), then applied quantile normalization to correct for differences in total read count and ChIP efficiency. The closest H3K4me3 peak with a normalized intensity of at least 25 reads and within 200 000-bp upstream of a pre-miRNA was considered its most likely transcriptional start site. Only microRNAs with a normalized intensity of at least 32 RPM were considered.
Identifying enriched predicted transcription factor binding sites
We selected putative TSS for liver-expressed microRNA genes. We extracted sequence covering ±2 kb from the middle of the putative TSS and masked out poorly-conserved regions by removing areas with a phastCons score <0.15 in the UCSC 17-way vertebrate conservation track. The phastCons score ranges between 0 (not conserved) and 1 (well conserved). Regions with similar dimensions anchored at the TSS of protein coding genes were selected as a background set. The background set was chosen so that it had the same joint distribution of conserved sequence and base composition as the microRNA promoters. Receiver-operating characteristic (ROC) curves were computed for each vertebrate PWM in TRANSFAC (v2009.2) by varying the scoring threshold. P-values were computed for each score threshold that yielded a hit in an additional microRNA TSS using a chi-squared distribution for each threshold that yielded hits >5 microRNA or background TSSs. The best chi-squared P-value was tracked for each PWM. The best P-value was corrected for multiple testing using a Bonferroni factor equal to the number of tests, which we took to be the number of positive regions microRNA TSS regions (between 25 and 32 depending on the comparison), multiplied by the number of PWMs (644 vertebrate PWMs). This is a conservative correction as many of the TRANSFAC PWMs are similar and therefore do not constitute independent trials which the Bonferroni correction assumes. We also tested HNF4α sites using the SVM (support vector machine) prediction method developed by Sladek and colleagues (14). We submitted all sequences to the website and tabulated the best score for each masked sequence. The chi-squared P-value was 0.00013 which we corrected to 0.00013 * 39 = 0.0052.
Associating Foxa2-binding sites with TSS regions
We assumed that a Foxa2-binding event is likely to regulate the gene or microRNA associated with the closest H3K4me3 region to the Foxa2 site. However, in cases where a binding site was located in the common promoter of divergently transcribed genes, in an array with overlapping genes, or in an intergenic region adjacent to multiple genes, identifying the target gene was less straightforward. We assigned experimentally-defined Foxa2-binding events to the gene with a H3K4me3 region that was closest to the Foxa2 site, as well as any additional genes with a H3K4me3 region that was no >50% further than the closest region.
Visualizing genomic data
The positions of expressed miRNAs, genes, H3K4me3 regions and profiles, miRNA-TSS associations from this and previous work were visualized using the TessLA system which consists of a genome browser and a data analysis tool kit (unpublished data).
RESULTS AND DISCUSSION
The microRNAome of liver, jejunum and pancreas
We isolated small RNA fractions from six livers, six samples of small intestinal mucosa (from the jejunum) and two pancreata, converted them into libraries, and obtained 26 574 536, 55 885 851 and 38 613 301 sequence reads for liver, jejunal mucosa and pancreas, respectively. The resulting sequence reads were aligned to known microRNA precursor genes, obtained from miRBase v14 (15), in order to assess the abundance of each mature microRNA. Next, we verified that our sequence reads represented microRNAs and not degraded mRNAs by aligning them to the RefSeq database as well. As shown in Supplementary Figure S1, <10% of the reads in the microRNA size range (21–23 nt) aligned to mRNAs, while >80% matched to precursor microRNAs, indicating that our small RNA preparation was highly enriched for true microRNAs. In total, we generated 19 754 019, 45 949 823 and 13 487 288 trimmed reads in the range of 19–25 nt that aligned to precursor microRNAs for liver, jejunum and pancreas, respectively. Using these reads, we found evidence for expression of 769 of the 1094 (69.9%) known or predicted mature microRNAs, corresponding to 459 of 547 (83.9%) pre-microRNAs. (Supplementary Table S1). We confirmed high-level expression of previously known abundant microRNAs in the respective tissue, such as mir-122 and mir-192 in the liver, miR-215 and miR-192 in the intestine, and miR-375 and miR-152 in the pancreas (Supplementary Table S1) (16–20). The let-7 family was highly expressed in all tissues. The extraordinary dynamic range (about six orders of magnitude) of the technology used allowed us to detect and quantify microRNAs present in a few copies per million as well as those that contribute up to ∼44% of the total microRNA pool, i.e. miR-122 in the liver. Because of technical limitations of prior efforts, many of the microRNAs identified here had been missed in previous studies (7,21).
Tissue-specific microRNA expression
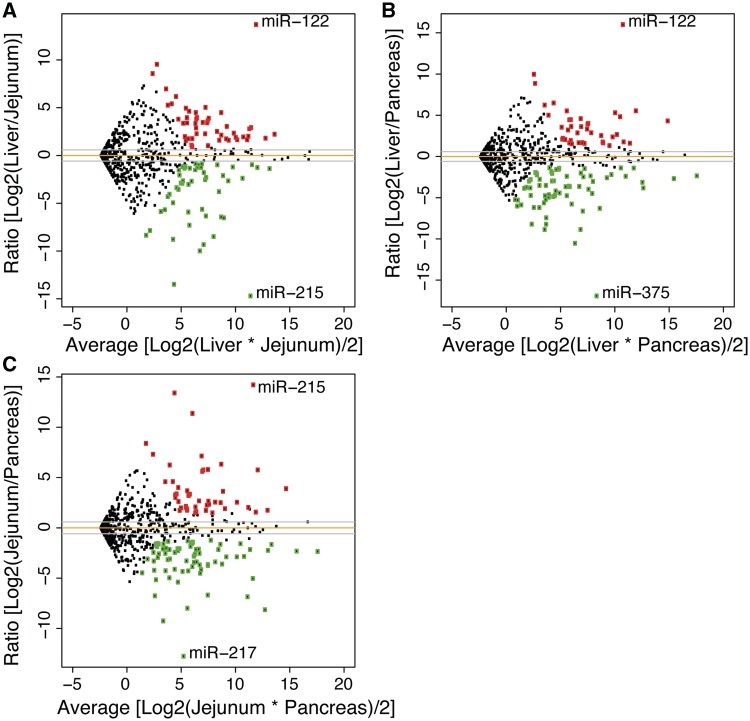
Next, we determined the differential microRNA gene expression between the three tissues. To this end, we employed computational tools established previously for the analysis of microarray expression profiling (for details, see ‘Materials and Methods’ section). After quantile normalization, expression levels of the independent samples for each tissue were compared. As shown in Figure 1, most microRNA genes are expressed at similar levels between any two tissues, suggesting that organs of related developmental origin such as liver and intestinal mucosa co-express many microRNA genes, just as they co-express many protein-coding genes. However, 162 microRNA genes exhibited statistically significant enrichment in either liver (63), small intestine (65) or pancreas (96), with differential expression of up to 120 000-fold. The top 30 microRNAs enriched in each organ versus the other two are listed in Tables 1–3. Full lists are available in Supplementary Tables S2–S4; see also Supplementary Figure S2.
Figure 1.
Differential expression of microRNAs in three endoderm-derived tissues. microRNAs were identified as differentially expressed in (A) liver versus small intestinal epithelium, (B) liver versus pancreas and (C) small intestine versus pancreas using an FDR of 10% and a minimum fold change of 1.5×. Several abundant microRNAs such as mmu-miR-192 are expressed at similar levels in both tissues (dots near the orange line). Each tissue has one highly-expressed, highly-differential microRNA as well as ∼30 other microRNAs that exhibit significant differential expression, which are highlighted in green (down) and red (up).
Table 1.
Top 20 microRNAs enriched in liver versus jejunum and/or Pancreas
| miRNA | Liver [RPM] | Ratio [log2] | Jejunum [RPM] | Ratio [log2] | Pancreas [RPM] |
|---|---|---|---|---|---|
| mmu-miR-122 | 447350.96 | 13.69 | 33.75 | 15.99 | 6.86 |
| mmu-miR-122-3p | 188.71 | 9.53 | 0.25 | 9.96 | 0.19 |
| mmu-miR-1948 | 138.99 | 6.96 | 1.12 | 8.86 | 0.3 |
| mmu-miR-485* | 102.56 | 8.57 | 0.27 | 6.22 | 1.37 |
| mmu-miR-455 | 199.55 | 6.16 | 2.78 | 6.49 | 2.23 |
| mmu-miR-21 | 27 344.49 | – | – | 5.53 | 591.84 |
| mmu-miR-340-5p | 332.16 | 2.26 | 69.11 | 5.53 | 7.21 |
| mmu-miR-193 | 116.31 | 5.4 | 2.76 | – | – |
| mmu-miR-101b | 6708.91 | 2.79 | 968.95 | 5.29 | 171.02 |
| mmu-miR-1937a | 86.26 | 5.26 | 2.25 | 5 | 2.69 |
| mmu-miR-30a* | 881.83 | 5.03 | 26.96 | 1.64 | 282.37 |
| mmu-miR-30c-2* | 244.84 | 4.95 | 7.94 | – | – |
| mmu-miR-107 | 7795.86 | 2.92 | 1032.13 | 4.82 | 276.37 |
| mmu-miR-22* | 269.87 | 4.62 | 10.96 | 4.45 | 12.38 |
| mmu-miR-31 | 578.92 | – | – | 4.58 | 24.21 |
| mmu-miR-99a | 394.09 | 4.48 | 17.63 | – | – |
| mmu-miR-221 | 1992.34 | 4.48 | 89.2 | – | – |
| mmu-miR-29b-2 | 294.92 | – | – | 4.46 | 13.43 |
| mmu-miR-192 | 136 451.85 | – | – | 4.31 | 6889.72 |
| mmu-miR-98 | 770.18 | 2.51 | 135.44 | 4 | 48.17 |
| mmu-miR-127 | 352.69 | 3.96 | 22.7 | – | – |
| mmu-miR-486 | 185.32 | 3.95 | 11.98 | 1.66 | 58.51 |
| mmu-miR-194-2 | 719.8 | – | – | 3.93 | 47.28 |
| mmu-miR-451 | 730.88 | 3.89 | 49.29 | – | – |
| mmu-miR-125b-1-5p | 320.5 | 3.8 | 23.05 | – | – |
| mmu-miR-541 | 101.72 | 3.8 | 7.32 | 1.63 | 32.79 |
| mmu-miR-142-5p | 228.22 | – | – | 3.63 | 18.38 |
| mmu-miR-125a-5p | 627.32 | 3.63 | 50.61 | – | – |
| mmu-miR-130a | 429.6 | 3.45 | 39.31 | 3.61 | 35.08 |
| mmu-miR-126-5p | 120.17 | 3.37 | 11.66 | 3.54 | 10.33 |
All expression levels are expressed as reads per million. Selection was based on a fold change of at least 1.5 and a false discovery rate of 10%.
Table 2.
microRNAs enriched in Small Intestine versus Liver and/or Pancreas
| miRNA | Jejunum [RPM] | Ratio [log2] | Liver [RPM] | Ratio [log2] | Pancreas [RPM] |
|---|---|---|---|---|---|
| mmu-miR-215 | 447 350.96 | 14.71 | 16.73 | 14.21 | 23.66 |
| mmu-miR-215-3p | 2221.8 | 13.49 | 0.19 | 13.4 | 0.21 |
| mmu-miR-194-1 | 3448.07 | 9.99 | 3.38 | 11.36 | 1.31 |
| mmu-miR-200c | 3495.91 | 9.33 | 5.41 | – | – |
| mmu-miR-375 | 408.84 | 8.78 | 0.93 | – | – |
| mmu-miR-145 | 4903.86 | 8.49 | 13.6 | – | – |
| mmu-miR-194-1-3p | 63.24 | 8.36 | 0.19 | 8.39 | 0.19 |
| mmu-miR-141 | 68 | 7.86 | 0.29 | 2.61 | 11.12 |
| mmu-miR-363-5p | 68.34 | 5.88 | 1.16 | 7.31 | 0.43 |
| mmu-miR-429 | 693.71 | 7.3 | 4.4 | 2.55 | 118.23 |
| mmu-miR-1-1 | 1429.3 | 6.34 | 17.67 | 7.13 | 10.18 |
| mmu-miR-200a | 4802.45 | 6.53 | 51.99 | 2.03 | 1179.74 |
| mmu-miR-200b | 3905.74 | 6.43 | 45.17 | – | – |
| mmu-miR-194-2 | 3769.81 | 2.39 | 719.8 | 6.32 | 47.28 |
| mmu-miR-33 | 136.43 | 1.53 | 47.12 | 6.23 | 1.81 |
| mmu-miR-200b* | 401.44 | 5.93 | 6.6 | 1.48 | 143.63 |
| mmu-miR-31 | 1343.17 | 1.21 | 578.92 | 5.79 | 24.21 |
| mmu-miR-21 | 31 885.58 | – | – | 5.75 | 591.84 |
| mmu-miR-142-5p | 986.36 | 2.11 | 228.22 | 5.75 | 18.38 |
| mmu-miR-130b | 82.83 | 5.68 | 1.61 | – | – |
| mmu-miR-1-2 | 880.44 | 5.64 | 17.67 | 5.61 | 18.04 |
| mmu-miR-142-3p | 108.57 | 4.96 | 3.49 | 4 | 6.78 |
| mmu-miR-20a | 94.27 | 3.56 | 8.02 | 4.6 | 3.89 |
| mmu-miR-872 | 58 | – | – | 4.58 | 2.42 |
| mmu-miR-1274a | 61.77 | 4.31 | 3.12 | – | – |
| mmu-miR-182 | 63.73 | 4.07 | 3.8 | – | – |
| mmu-miR-192 | 102 132.57 | – | – | 3.89 | 6889.72 |
| mmu-miR-143 | 1357.07 | 3.79 | 98.12 | – | – |
| mmu-miR-29b-2 | 172.11 | – | – | 3.68 | 13.43 |
| mmu-miR-203 | 1660.92 | 2.66 | 262.14 | 3.63 | 134.5 |
All expression levels are in RPM. Selection was based on a fold change of at least 1.5 and a false discovery rate of 10%.
Table 3.
microRNAs enriched in pancreas versus liver and/or small intestine
| miRNA | Pancreas [RPM] | Ratio [log2] | Liver [RPM] | Ratio [log2] | Jejunum [RPM] |
|---|---|---|---|---|---|
| mmu-miR-375 | 115 969.49 | 16.93 | 0.93 | 8.15 | 408.84 |
| mmu-miR-217 | 3177.41 | 10.54 | 2.13 | 12.77 | 0.46 |
| mmu-miR-216b | 257.11 | 8.88 | 0.55 | 9.26 | 0.42 |
| mmu-miR-200c | 2528.08 | 8.87 | 5.41 | – | – |
| mmu-miR-672 | 226.77 | 8.23 | 0.75 | 4.29 | 11.62 |
| mmu-miR-138-1 | 89.86 | 8.22 | 0.3 | 4.12 | 5.16 |
| mmu-miR-676 | 779.03 | 3.98 | 49.21 | 8 | 3.05 |
| mmu-miR-184 | 173.65 | 7.44 | 1 | 5.4 | 4.1 |
| mmu-miR-148a | 24 381.18 | 3.82 | 1721.49 | 6.86 | 209.89 |
| mmu-miR-148a* | 63.48 | 4.77 | 2.33 | 6.76 | 0.58 |
| mmu-miR-99a | 1814.3 | 2.2 | 394.09 | 6.69 | 17.63 |
| mmu-miR-145 | 1353.92 | 6.64 | 13.6 | – | – |
| mmu-miR-200b | 3572.31 | 6.31 | 45.17 | – | – |
| mmu-miR-183 | 72.59 | 6.21 | 0.98 | – | – |
| mmu-miR-10b | 50.92 | 6.2 | 0.69 | 2.46 | 9.27 |
| mmu-miR-7b | 16.55 | 6.02 | 0.25 | ||
| mmu-miR-155 | 20.46 | 5.88 | 0.35 | 2.95 | 2.65 |
| mmu-miR-2143-3 | 494.87 | 5.78 | 9 | 4.17 | 27.48 |
| mmu-miR-130b | 79.92 | 5.63 | 1.61 | – | – |
| mmu-miR-224 | 13.32 | 5.3 | 0.34 | – | – |
| mmu-miR-141 | 11.12 | 5.25 | 0.29 | – | – |
| mmu-miR-802 | 78.2 | 5.25 | 2.06 | 1.3 | 31.67 |
| mmu-miR-676* | 39.2 | 2.06 | 9.4 | 5.17 | 1.09 |
| mmu-miR-152 | 18 175.19 | 3.15 | 2053.99 | 5.04 | 552.97 |
| mmu-miR-2133-2-3p | 86.25 | 4.96 | 2.78 | 4.97 | 2.75 |
| mmu-miR-802-3p | 112.04 | 4.88 | 3.81 | 1.55 | 38.37 |
| mmu-miR-1195-3p | 254.2 | 4.77 | 9.35 | 3.86 | 17.51 |
| mmu-miR-429 | 118.23 | 4.75 | 4.4 | – | – |
| mmu-miR-34a | 25.21 | 4.74 | 0.95 | 4.22 | 1.35 |
| mmu-miR-1274a | 78.33 | 4.65 | 3.12 | – | – |
All expression levels are in RPM. Selection was based on a fold change of at least 1.5 and a false discovery rate of 10%.
Mapping transcriptional start sites of microRNAs
The analysis of cis-regulatory elements requires knowledge of the promoter used for the microRNA gene in question in the liver, small intestine or pancreas. To this end, we took advantage of the recent discovery that transcriptional start sites in the mammalian genome are marked by trimethylated histone H3 (H3K4me3), often in a characteristic double peak pattern (22). We performed ChIP-Seq experiments for H3K4me3 in liver, small intestine and pancreas, identified areas of H3K4me3 enrichment, and then mapped these putative transcriptional start sites to expressed microRNAs by associating each microRNA with the nearest upstream region of H3K4me3 occupancy. To increase the chances that a microRNA would have an H3K4me3-marked TSS, we only processed microRNAs that had an expression level of at least 32 RPM, and required that the H3K4me3 enrichment levels reached at least 25 (see ‘Materials and Methods’ section for details.) Of a total of 17 505 H3K4me3 regions present in at least one organ, we identified 106 as putative TSS for a total of 128 pre-miRNAs (Supplementary Table S5). About 80% of the TSS were within 50 kb of the miRNA (Supplementary Figure S3). As found previously for the analysis of microRNA promoters in embryonic stem cells, between 73.3% (jejunum) and 77.6% (pancreas) (Table 4) of the H3K4me3 loci covering putative transcriptional start sites overlapped CpG islands, a DNA sequence feature frequently associated with promoters. We compared microRNA expression levels to the degree of H3K4me3 occupancy at their promoters, but did not observe any obvious correlation (data not shown).
Table 4.
Overlap between putative miRNA TSS and other regulatory features
| Organ | TSS Regions | CpG Island, % | ESC Promoters, % | Foxa2 Sites, % |
|---|---|---|---|---|
| Liver | 74 | 77.0 | 62.2 | 10.8 |
| Jejunum | 75 | 73.3 | 66.7 | 8.0 |
| Pancreas | 76 | 77.6 | 64.5 | 9.2 |
| All | 106 | 74.5 | 55.7 | 7.5 |
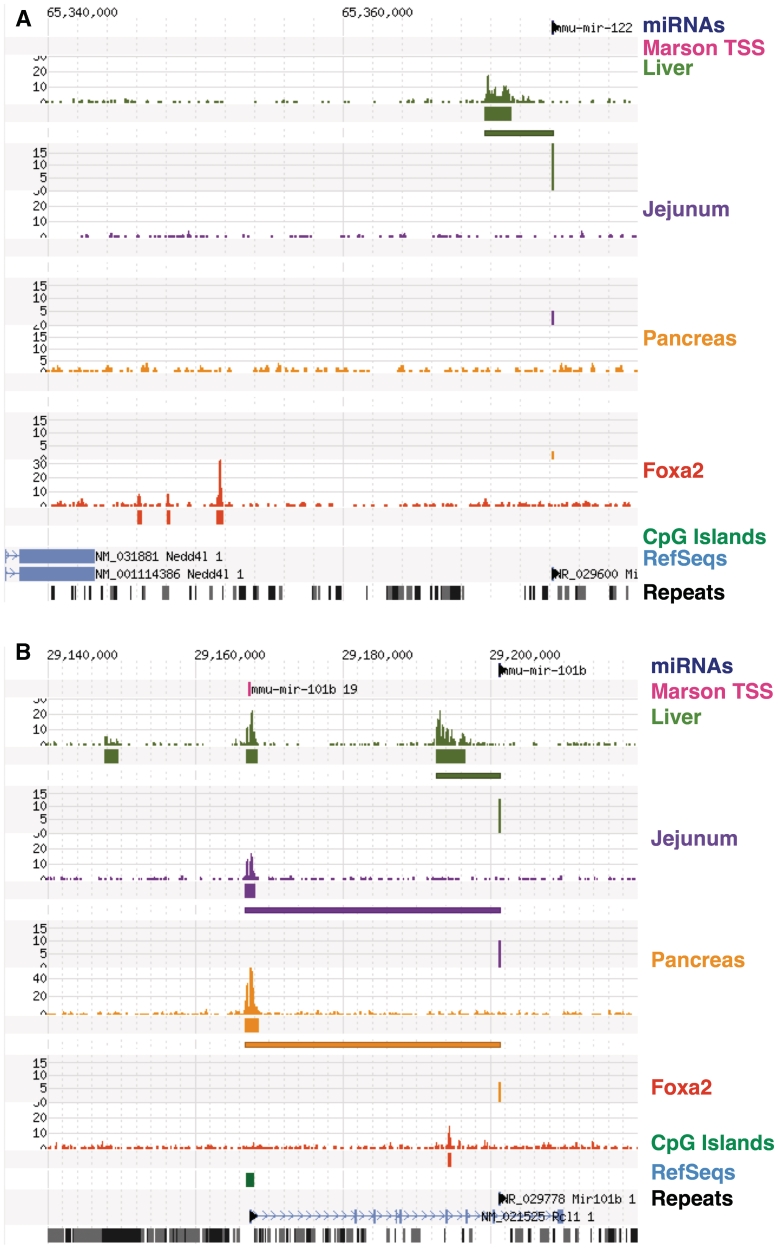
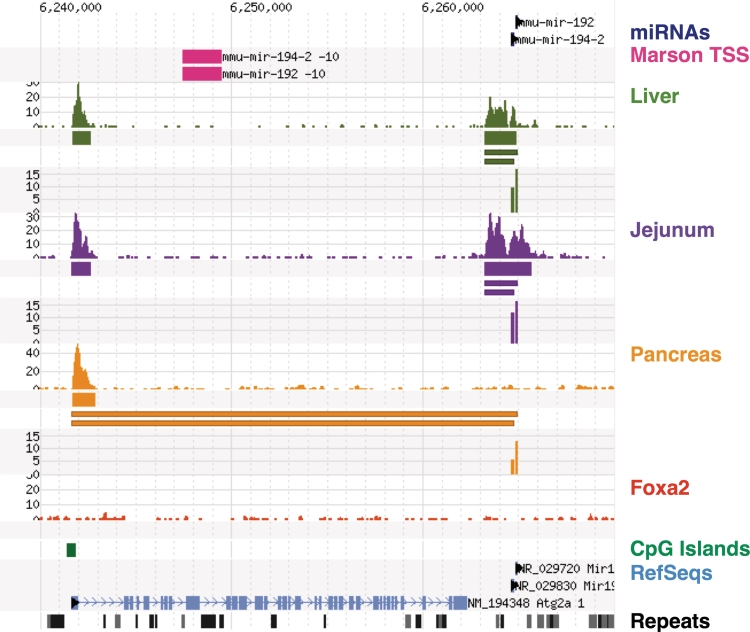
Comparing our results with previous efforts that had mapped transcriptional start sites in ES cells (23), we found agreement at 59 of 106 (55.7%) of our H3K4me3 loci. Reasons for this discrepancy might include the reliance on historic H3K4me3 mapping data in the previous study, and the use of different transcriptional start sites in liver and intestinal mucosa compared to embryonic stem cells. We observe that the average concordance rate within a given organ (64.5%) is higher than the rate over all identified miRNA/TSS pairs, which is consistent with the idea that the TSS concordant between ES cells and a given organ or tissue correspond to ‘housekeeping’ miRNAs which are used in most tissues. As we determine TSS in different endoderm-derived tissues, we identify additional tissue-specific microRNA/TSS, while still re-identifying the same housekeeping microRNA/TSS, thus lowering the total concordance rate. A few examples of this latter category are discussed below. Figures 2 and 3 illustrate the advantage of identifying tissue-specific microRNA transcriptional start sites using H3K4me3 mapping in tissues that express the microRNA in question, and also illustrate the regulatory complexity of miRNAs. In Figure 4 we show strong local promoters active in liver and small intestine where miRNAs miR-192 and miR-194 are highly-expressed. In pancreas, the expression of these miRNAs is about 10× lower, though still strong, but the level of H3K4me3 has been reduced to background levels. Marson and colleagues identified a promoter region further upstream in the Atg2a gene, which is also at background levels in pancreas (23). However, the CpG-containing promoter for Atg2a is active, suggesting that it may be the source of miR-192 and miR-194 in the pancreas.
Figure 2.
Transcriptional start sites of microRNAs enriched in the liver. (A) Shows mmu-miR-122 that is highly expressed in liver and expressed about 1000–2000× lower in jejunal epithelium and pancreas. Each section (liver, jejunum, and pancreas) shows the profile of H3K4me3, regions of significant levels of H3K4me3, the link from the miRNA to the nearest H3K4me3 peak, and the log2 normalized expression level of miRNAs. Note the presence of H3K4me3-modified histones at the transcriptional start site in the liver (olive). No H3K4me3 is present in jejunum (purple) or pancreas (yellow). Additionally, this TSS was not identified in mouse embryonic stem cells [TSS from (23) in magenta]. This locus also includes an example of Foxa2 binding. In (B), mmu-miR-101b is located within the protein-coding Rcl1 gene, which is not highly expressed in liver, yet the microRNA TSS shows clear evidence of liver-specific H3K4me3. mmu-miR-101b is expressed at lower levels in jejunum and pancreas and may the use Rcl1 promoter.
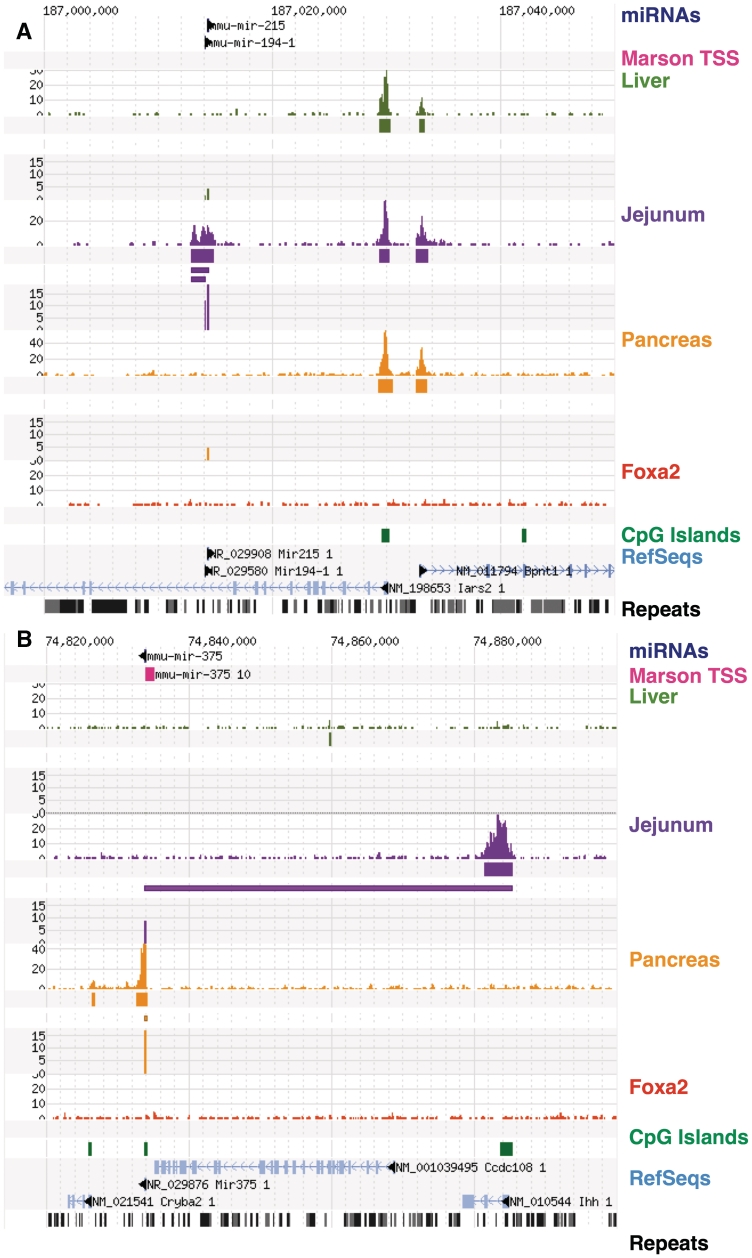
Figure 3.
Transcriptional start sites of microRNAs enriched in the small intestinal epithelium or pancreas. (A) mmu-miR-215 and mmu-miR-194-1 are specific to jejunum and have a strong proximal jejunum-specific H3K4me3 region which is internal to the Iars2 gene. (B) mmu-miR-375, situated just downstream of Ccdc108 gene, has an adjacent region of H3K4me3 [also identified in ref. (23)] that is absent in liver and jejunum. mmu-miR-375 is expressed in jejunum, but the promoter identified is at the Ihh gene about 50-kb upstream.
Figure 4.
Transcriptional start sites for miR-192 and miR-194 which are expressed in all three tissues, but at decreased levels in pancreas where the H3K4me3 region at chr19:6263000 is nearly absent. The ES-cell promoter from (23) does not appear to be active in any of these tissues, but a CpG+ promoter for the Atg2 gene is active and may be the source of these miRNAs in the pancreas.
Cis-regulatory elements of differentially expressed microRNA genes
Next we employed positional weight matrices (PWMs) to identify potential binding sites of tissue-enriched transcription factors. We compared the occurrence of the best match to each PWM in the conserved regions surrounding the TSSs (for simplicity referred to as ‘promoters’ below) with their occurrence in randomly selected promoters from protein-coding genes. We used the sets of promoters that were associated with liver-, small intestine-, or pancreas-enriched microRNAs. Because we were interested in identifying motifs that may cover only a subset of microRNA promoters, we developed a novel enrichment statistic that emphasizes the enrichment of high-scoring motif matches in a group of promoters, but does not require strong motifs in all or even most of the tissue-specific promoters (see ‘Materials and Methods’ section for details).
In the set of liver-enriched microRNA genes, predicted sites for the factors MYB (M00183, pv = 3.1e–5), and SREBP (M01168, pv = 4.8e–3) were enriched within 1000 bp of H3K4me3 regions. Considering just the miRNA-only H3K4me3 regions, we found CREB (M00916 pv = 1.2e–4), AHR/ARNT (M00778 pv = 1.8e–4 or M00237 pv = 2.6e–3) and E2F (M00938 pv = 6.8e–3) to be enriched within 3000–8000 bp. All of these factors are known to be relevant to the function of the liver. Because of the major role played by the nuclear receptor HNF4α in gene regulation in hepatocytes, we assessed the potential for regulation of liver microRNAs by HNF4α using a recently developed support vector machine-based prediction method (14), and found the HNF4α motif significantly enriched (corrected P-value = 0.0052) among the liver-expressed microRNA promoters. In fact, eight microRNA genes expressed preferentially in the liver contain predicted HNF4α-binding sites above the recommended threshold. We note that HNF4α is a hepatocyte-specific transcription factor which is not expressed in all cell types in the liver; therefore, there may be a set of liver-specific miRNAs that are not expressed hepatocytes, for which HNF4α is irrelevant.
Next, we sought experimental evidence of binding of tissue-specific transcriptional regulators near microRNA TSSs. Foxa2 is an important transcriptional regulator of liver development and function (11,24–29) so we checked previously published experimental data for Foxa2 binding (13) and found that indeed seven of the putative liver-enriched microRNAs TSSs (associated with 10 microRNAs) were within 2KB of experimentally defined Foxa2 sites (Table 5). A further eight miRNA had a Foxa2 site within 15 kb of the TSS. Four of the miRNA TSS were not associated with a protein-coding gene and thus represent evidence of miRNA-specific regulation by Foxa2. In Figure 2A and B we illustrate two examples where we have identified a novel liver-specific microRNA TSS that is occupied by Foxa2 within a few hundred base pairs of the TSS.
Table 5.
Location of Foxa2 sites within 10 kb of miRNA/TSS
| Foxa2 Location | TSS/miRNA Location | Distance [bp] | miRNA |
|---|---|---|---|
| chr11:86403625–86403809 | chr11:86400262–86403879 | 162 | mmu-mir-21 |
| chr6:31152438–31152697 | chr6:30992823–31151892 | 675 | mmu-mir-29a |
| chr1:196718801–196719044 | chr1:196717762–196737844 | 1160 | mmu-mir-29b-2 |
| chr1:196718801–196719044 | chr1:196717762–196738358 | 1160 | mmu-mir-29c |
| chr5:138400937–138401132 | chr5:138395109–138402675 | 1641 | mmu-mir-25 |
| chr5:138400937–138401132 | chr5:138395311–138402675 | 1641 | mmu-mir-93 |
| chr4:100854070–100854278 | chr4:100844877–100855891 | 1717 | mmu-mir-101a |
| chr19:29194307–29194636 | chr19:29192749–29201372 | 1722 | mmu-mir-101b |
| chr5:138400704–138400930 | chr5:138395109–138402675 | 1858 | mmu-mir-25 |
| chr5:138400704–138400930 | chr5:138395311–138402675 | 1858 | mmu-mir-93 |
| chr4:100853701–100853887 | chr4:100844877–100855891 | 2097 | mmu-mir-101a |
| chr16:18255150–18255448 | chr16:18240964–18257550 | 2251 | mmu-mir-185 |
| chr5:119931308–119931521 | chr5:119786217–119936556 | 5142 | mmu-mir-1959 |
| chr13:48564064–48564222 | chr13:48547949–48558966 | −5177 | mmu-let-7d |
| chr13:48564064–48564222 | chr13:48549766–48558966 | −5177 | mmu-let-7f-1 |
| chr13:48564064–48564222 | chr13:48550116–48558966 | −5177 | mmu-let-7a-1 |
| chr11:79470698–79470956 | chr11:79476766–79542706 | −5939 | mmu-mir-365-2 |
CONCLUSIONS
Here we have taken advantage of the substantial dynamic range of ultra-high throughput sequencing to detect and quantify microRNAs in three endoderm-derived tissues. Many of the microRNAs identified here had been missed in previous studies (21). However, despite the fact that we obtained >90 million sequence reads for small RNAs, no new microRNAs were discovered, strongly suggesting that mirBase covers all or nearly all existing microRNA genes.
As one might expect for developmentally related tissues such as liver, jejunum and pancreas, most of the microRNA genes are expressed at roughly similar levels in the three tissues. However, we also discovered tissue-specific expression between pairs of tissues, in some cases over several orders of magnitude. We utilized experimental mapping of transcriptional start sites in order to localize the promoters that direct this tissue-specific gene activation. Importantly, we found multiple cases where different transcriptional start sites are used by microRNA genes in endoderm-derived tissues as opposed to embryonic stem cells. These findings suggest that understanding microRNA gene promoters requires their experimental validation in each tissue of interest. We did not measure repressive chromatin marks, e.g. H3K27me3, in this work, so potentially active H3K4me3-marked TSS need to be evaluated for absence of repressive marks to further validate their activity.
Finally, we analyzed the cis-regulatory elements that contribute to the regulation of microRNA gene expression in liver, jejunum, and pancreas. We find that the same major transcription factors that regulate the tissue-specific expression of protein-coding genes also contribute to the regulation of microRNA genes, suggesting that both classes of genes utilize the same fundamental regulatory mechanisms. These results extend earlier findings by others. For example, SREBP-1c has been shown to indirectly regulate miRNAs in skeletal muscle in response to insulin signaling (30), so it is exciting to find evidence for a direct link in liver between SREBP-1c and microRNA regulation, as this is a tissue which also responds to insulin. Similarly, c-Myb has been shown to be both a target and a regulator of miRNA-15a in K562 myeloid leukemia cells (31).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (DK056669 to L.E.G., DK049210 and DK053839 to K.H.K.); American Liver Foundation Innovative Seed grant (to L.E.G.); Core services provided by the DERC at the University of Pennsylvania from a grant sponsored by National Institutes of Health (P30-DK19525). Funding for open access charge: DK049210 grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Alan Fox and Olga Smirnova for expert technical assistance
REFERENCES
- 1.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304–2315 e2301–e2304. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekine S, Ogawa R, McManus MT, Kanai Y, Hebrok M. Dicer is required for proper liver zonation. J. Pathol. 2009;219:365–372. doi: 10.1002/path.2606. [DOI] [PubMed] [Google Scholar]
- 4.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig N, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.07.040. doi:10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl Acad. Sci. USA. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grove CA, Walhout AJ. Transcription factor functionality and transcription regulatory networks. Mol. Biosyst. 2008;4:309–314. doi: 10.1039/b715909a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 10.Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuteja G, Jensen ST, White P, Kaestner KH. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–4157. doi: 10.1093/nar/gkn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhandare R, Schug J, Le Lay J, Fox A, Smirnova O, Liu C, Naji A, Kaestner KH. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 20:428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 17.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 18.Hino K, Fukao T, Watanabe M. Regulatory interaction of HNF1-alpha to microRNA-194 gene during intestinal epithelial cell differentiation. Nucleic Acids Symp. Ser. 2007:415–416. doi: 10.1093/nass/nrm208. [DOI] [PubMed] [Google Scholar]
- 19.Tzur G, Levy A, Meiri E, Barad O, Spector Y, Bentwich Z, Mizrahi L, Katzenellenbogen M, Ben-Shushan E, Reubinoff BE, et al. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS One. 2008;3:e3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl Acad. Sci. USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc. Natl Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat. Med. 2008;14:828–836. doi: 10.1038/nm.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochkis IM, Schug J, Rubins NE, Chopra AR, O'Malley BW, Kaestner KH. Foxa2-dependent hepatic gene regulatory networks depend on physiological state. Physiol. Genomics. 2009;38:186–195. doi: 10.1152/physiolgenomics.90376.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, White P, Tuteja G, Rubins N, Sackett S, Kaestner KH. Foxa1 and Foxa2 regulate bile duct development in mice. J. Clin. Invest. 2009;119:1537–1545. doi: 10.1172/JCI38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sund NJ, Ang SL, Sackett SD, Shen W, Daigle N, Magnuson MA, Kaestner KH. Hepatocyte nuclear factor 3beta (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol. Cell Biol. 2000;20:5175–5183. doi: 10.1128/mcb.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Rubins NE, Ahima RS, Greenbaum LE, Kaestner KH. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab. 2005;2:141–148. doi: 10.1016/j.cmet.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Granjon A, Gustin MP, Rieusset J, Lefai E, Meugnier E, Guller I, Cerutti C, Paultre C, Disse E, Rabasa-Lhoret R, et al. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes. 2009;58:2555–2564. doi: 10.2337/db09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113:505–516. doi: 10.1182/blood-2008-01-136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.