Abstract
Small nucleolar RNAs (snoRNAs) and microRNAs are two classes of non-protein-coding RNAs with distinct functions in RNA modification or post-transcriptional gene silencing. In this study, we introduce novel insights to RNA-induced gene activity adjustments in human cells by identifying numerous snoRNA-derived molecules with miRNA-like function, including H/ACA box snoRNAs and C/D box snoRNAs. In particular, we demonstrate that several C/D box snoRNAs give rise to gene regulatory RNAs, named sno-miRNAs here. Our data are complementing the increasing number of studies in the field of small RNAs with regulatory functions. In massively deep sequencing of small RNA fractions we identified high copy numbers of sub-sequences from >30 snoRNAs with lengths of ≥18 nt. RNA secondary structure prediction indicated for a majority of candidates a location in predicted stem regions. Experimental analysis revealed efficient gene silencing for 11 box C/D sno-miRNAs, indicating cytoplasmic processing and recruitment to the RNA silencing machinery. Assays in four different human cell lines indicated variations in both the snoRNA levels and their processing to active sno-miRNAs. In addition we show that box D elements are predominantly flanking at least one of the sno-miRNA strands, while the box C element locates within the sequence of the sno-miRNA guide strand.
INTRODUCTION
A variety of small non-protein coding RNAs (npcRNAs), including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs) are important mediators of gene regulation (1–3). piRNAs control retrotransposition (4,5) and are expressed in testes only (3). siRNAs and miRNAs are targeting mRNAs in a sequence-specific manner and guide translational inhibition, degradation or deadenylation of target-mRNAs (6–9). A role in transcriptional regulation for an endogenous miRNA was recently discovered (10,11) and previously shown for exogenously delivered siRNAs (12,13). Furthermore, a general transcriptional regulation mechanism was recently postulated for transcription start site (TSS) associated tiny RNAs, called tiRNAs (14,15). Components of the RNA induced silencing complex (RISC), in particular the Ago proteins, act in concert with small npcRNAs to regulate gene expression at the various levels mentioned earlier.
A new source of regulatory npcRNAs was recently discovered to originate from small nucleolar RNAs (snoRNAs) (14,16). SnoRNAs are ubiquitously expressed small npcRNAs (also named sRNAs here) that function in maturation and modification of other npcRNAs such as ribosomal RNAs (rRNAs) or small nuclear RNAs (snRNAs) (17). Box H/ACA snoRNAs trigger the site specific synthesis of pseudouridines in a broad variety of ribosomal and spliceosomal RNAs (18). In addition these snoRNAs play a role in nucleolytic processing of precursor rRNA (pre-rRNA) and also function in synthesis of telomeric DNA (19). Box C/D snoRNAs, which are the primary source of sno-miRNAs in this study, contain conserved Box C (UGAUGA) and Box D (CUGA) elements located closely to the 5′- and 3′-ends, respectively. Internal copies of these elements are termed Box C′ and Box D′ (20,21). An interaction between the 5′- and 3′-termini allows the formation of a stem bringing the Boxes C and D elements together to form a hairpin structure. Box C/D snoRNAs serve as guides for 2′-O-ribose methylation of ribosomal RNA. Pseudouridylation and 2′O-ribose methylation are important steps during maturation of ribosomal RNAs, thus playing a fundamental role in key processes of all cells. With exception of U3 all box C/D snoRNAs presented in this study are intron-encoded, as it is the general pathway for the biogenesis of this class of snoRNAs (22). However, apparently some snoRNAs have an additional function outside their normal ‘work-environment’. Ender et al. (16) demonstrated processing of the cajal body specific RNA ACA45 to cytoplasmic, Ago protein associated 20- to 25-nt RNAs with gene silencing activity. The identification of CDC2L6 as a target mRNA, which is down-regulated by the miRNA function of ACA45 underlined the physiological relevance of the dual function. Independence of nuclear micro-processing by Drosha/DGCR8 indicates that the mechanism is distinct from the classic miRNA pathway that was formulated in the early years of miRNA research (23) and processing factors and their regulations are constantly being refined (24,25). Today we know more about alternative processing of the primary miRNA transcripts (pri-miRNAs) in a Drosha-independent fashion (26,27) and regulation of the microprocessor complex itself (25,28,29). Recently, the possibility of processing box C/D snoRNAs to small RNA molecules with miRNA-like features and functions was postulated from intensive analysis of deep sequencing data and predicted for numerous candidates (30). The box C/D snoRNA MBII-52 was shown to give rise to shorter RNAs with a regulatory function in alternative splice site selection (31). We addressed the crucial task of providing further experimental evidences for regulatory snoRNA- mechanisms in human cells. In the ancient eukaryote Giardia lamblia a box C/D snoRNA was shown to be processed by Dicer and performed gene silencing in reporter gene assays (32) and similar snoRNA effects were postulated for humans (33).
By second generation sequencing of small RNA fractions (up to 40 nt) from human T lymphocytes we identified 22 fragments of C/D box snoRNAs by bioinformatics analysis. These may serve as substrates for Dicer processing as described in ref. (27) and may enter the RNA silencing machinery as it is usually observed for (pre-)miRNAs (34). For these candidate sequences we functionally characterized the putative miRNA-like activity by in vitro experiments. Two small RNAs derived from C/D box snoRNA HBII-99b and SNORD126 are reported as miRNAs in the latest release of the miRNA database (miRBase).
Furthermore, we identified miRNAs in miRBase, which de facto are box H/ACA snoRNA-derived RNAs (sdRNAs). These snoRNAs, i.e. ACA34, ACA36b and HBI-61, share common features in terms of predicted structures, processing and silencing capacity as was described for the small cajal body specific RNA (scaRNA) ACA45 (16) and additional box H/ACA sdRNAs (35). Based on the striking similarity shared by the two RNA species the theory was introduced that some miRNAs may have evolved from ancient box H/ACA snoRNAs (35). In their study Scott et al. (35) provide a list of 14 snoRNAs, which give rise of sdRNAs, including the three miRBase listed candidates (miR-1291/ACA34, miR-1248/HBI-61, miR-664/ACA36b). ACA36b was already predicted to have a miRNA-like activity that may compare to the miRNA-activity of ACA45 (16) and we provide experimental data for this function in three human cell lines. In this study we present an expanded range of candidate npcRNAs with putative miRNA-features, including evidence for the gene silencing activity of human box C/D sno-miRNAs. In addition, we show functional data for three previously discovered box H/ACA sno-miRNAs. Moreover, we provide experimental evidence for cell type specific activities of sno-miRNAs, suggesting a general and controlled participation of snoRNAs in gene regulation in a cell type dependent manner.
In a detailed analysis of snoRNA-derived sequences from our small RNA libraries we identified an interesting feature. In 15 out of 17 box C/D snoRNAs at least one of the sequenced (and functionally tested) sno-miRNA candidates was flanked by the box D element (CUGA) at the 3′-end and/or a box D′ element close to the 5′-end. The box C element (UGAUGA) was identified in 12 snoRNA sequences and located within the sequence of 11 sno-miRNAs. This pattern could be confirmed more frequently for 107 snoRNAs that express sequences from our sRNA libraries. Our findings indicate a relation of the functional snoRNA elements to the processing and activity of sno-miRNAs.
MATERIALS AND METHODS
Small npcRNA cloning and sequencing
Small RNA cloning was carried out by Vertis Biotechnology (Weihenstephan, Germany) and has been described earlier (36). The small RNA species were isolated from total RNAs using the mirVana miRNA isolation kit (Ambion). The small RNAs were separated on denaturing 15% polyacrylamide (PAA) gels. Oligonucleotides of 18- to 40-nt size were poly(A)-tailed using poly(A) polymerase, and an adaptor was ligated to the 5′-phosphate of the miRNAs: 5′-end adaptor GCCTCCCTCGCGCCATCAGCTNNNNGACCTTGGCTGTCACTCA. Next, first-strand cDNA synthesis was performed using an oligo(dT)-linker primer and M-MLV-RNaseH reverse transcriptase 3′-end adaptor GCCGGGGCGATGTCTCGTCTGAGCGGGCTGGCAAGGC. The resulting cDNAs were PCR amplified in 16 cycles using the high-fidelity Phusion polymerase (Finnzymes). The 120–135-bp amplification products were confirmed by polyacrylamide gel electrophoresis (PAGE) analysis. sRNA library sequencing was performed in a Roche FLX System (Roche Diagnostics, Mannheim, Germany) and yielded 550 000 reads in total.
sRNA extraction from library
Each library sequence (read) comes with a unique identifier and is composed of a fixed-length 5′-adaptor, a transcribed sRNA sequence of maximum 40 nt, a variable-length poly(A) sequence A+NA+, and a 3′-adaptor (see above). The following protocol is used to extract the sRNA sequences from the library:
The start position is simply found by removing the fixed-length 5′-primer. Only in a few cases the precise position may be missed in this way due to single nucleotide deletions (or insertions) inside the adaptor as a result of sequencing errors.
The identification of the end position is more challenging due to the poly(A) string that precedes the 3′-primer. To cope with its variable length and patterns, we used Perl regular expression matching. In >99% of the sequences at least one of the subexpressions AA+NA+, A+NAA+, AAA+N, NAAA+ or AAAAAA+ matches. Here, we have to accept that at least for sRNAs ending on A+, cutting at the matching position is imprecise. The non-matching reads are discarded, together with all extracted sRNA sequences that are empty or just too short (<15 nt) for further processing (<5%).
sno-miRNA identification
To discover both (i) expressed/transcribed fragments of known snoRNAs in the short RNA library and (ii) novel candidates for snoRNA-derived RNAs (sno-miRNAs), we aligned all extracted sRNA sequences against all 402 known snoRNAs from snoRNABase (37) using the offline version of NCBI BLAST. Only perfect alignments (without gaps or mismatches) are accepted which cover at least 17 nt. This is the minimum length found for mature human miRNAs. From the BLAST analysis a snoRNA-expression profile was created counting the frequencies of matching sequences from the library (∼9500 in total). From this profile we selected sufficiently expressed snoRNAs which include a subsequence that (i) is minimum 25-nt long; (ii) is covered by a significant number of expressed snoRNA fragments; and (iii) whose most conserved part (≥20 nt) is located in a hairpin stem of the snoRNA folding. Secondary structure folding was calculated with RNAfold (rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) and sRNA sequences were aligned with ClustalW2 (38). Table 1 lists all C/D box snoRNAs, which were identified in this way for further functional (miRNA) analysis. To test whether some known miRNAs may actually be derived from snoRNAs all 718 mature human miRNAs from miRBase (39) (release 14.0, www.mirbase.org) are matched against the filtered snoRNA fragments. The five matches found are also listed in Table 1.
Table 1.
snoRNA derived sRNAs (sno-miRs), positions on original snoRNA sequences and miRNA-like activity
| snoRNA | HGNC symbol (host gene) | Type | Sno-miR pos. (major) | Sno-miR *pos. (minor) | miRNA activity (H/J/R) | References |
|---|---|---|---|---|---|---|
| ACA45 | SCARNA15 (Hs.513091) | H/ACA | 64–85 | 102–127 | +/+/O | (16) |
| snR39b | SNORD2 (EIF4A2) | C/D | 2–29 | 36–63 | +/+/− | |
| U3 | X14945 | C/D | 193–214 | (93–127) | +/O/+ | |
| U3-3 | SNORD3 | C/D | 45–66 | −/O/n | ||
| U3-4 | SNORD3 | C/D | 75–96 | O/+/+ | ||
| U78 | SNORD78 (GAS5) | C/D | 32–61 | 1–24 | +/+/O | |
| HBII-336 | SNORD93 (EST cluster) | C/D | 40–62 | 2–23 | −/+/+ | This study |
| HBI-43 | SNORD17 (SNX5) | C/D | 207–233 | 2–29 | O/–/n | |
| HBII-142 | SNORD66 (EIF4G1) | C/D | 47–70 | 1–23 | O/+/n | |
| U48 | SNORD48 (C6orf48) | C/D | 1–25 | 40–61 | −/O/n | |
| U51 | SNORD51 (EEF1B2) | C/D | 41–65 | 2–39 | −/O/n | |
| U21 | SNORD21 (RPL5) | C/D | 2–27 | 52–86 | −/−/n | |
| U27 | SNORD27 (U22 Host Gene, UHG) | C/D | 43–67 | 2–24 | −/+/− | |
| U44 | SNORD44 (AL110141) | C/D | 2–26 | 28–57 | −/O/n | |
| HBII-429 | SNORD100 (RPS12) | C/D | 4–36 | 37–70 | +/O/n | |
| U59b | SNORD59b (ATP5B) | C/D | 44–67 | 6–25 | −/O/n | |
| U83a | SNORD83a (RPL3) | C/D | 4–26 | n.d. | −/+/+ | |
| U15a | SNORD15a (RPS3) | C/D | 2–27 | 114–137 | +/+/n | |
| U74 | SNORD74 (GAS5) | C/D | 40–68 | 3–30 | −/+/n | |
| U45AC | SNORD45aSNORD45c (RABGGTB) | C/D | 57–73 | 1–29 | O/−/n | |
| miRbase listed snoRNAs | Identified | |||||
| ACA34 hsa-miR-1291 | SNORA34 (FLJ20436) | H/ACA | 2–25 | (39–63) | +/+/+ | (35) |
| HBI-61 hsa-miR-1248 | SNORA81 (EIF4A2) | H/ACA | 1–27 | n.d. | +/+/+ | |
| ACA36b hsa-miR-664 | SNORA36b | H/ACA | 106–128 | 68–91 | +/O/− | |
| HBII-99b hsa-miR-1259 | SNORD12b (C20orf199) | C/D | 16–36 | n.d. | n.d. | This study |
| SNORD126 hsa-miR-1201 | SNORD126 (CCNB1IP1) | C/D | 52–75 | n.d. | n.d. | |
Host gene, pre-mRNA source of intron-encoded snoRNA; sno-miR, small RNA originating from snoRNAs; major, dominant sRNA, found in high copy numbers by deep sequencing; minor, sRNA found in significantly lower copy numbers; var., various minor sRNAs; n.d., not detected; miRNA activity: HeLa, Jurkat, RPMI8866 +, <60% remaining Renilla luciferase activity; O, 60–80%; −, no reduction.
Cell culture
Human CD4+ T cells (Jurkat) and B cells (RPMI8866) were maintained in RPMI 1640 Medium supplemented with 10% FCS and 1% antibiotics solution (Pen/Strep). Human HeLa and MCF7 cells were maintained in DMEM (Dulbecco’s modified Eagle’s Medium) supplemented with 10% FCS and antibiotics. RNA preparation was performed with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s guidelines.
Reporter gene assays
Silencing activity of snoRNA derived sRNA was measured in the dual luciferase reporter gene system psiCHECK-2 (Promega, Madison, WI, USA). Fully complementary target sites for sRNAs were synthesized with flanking overhangs for NotI and XhoI restriction sites, annealed in isostoichiometric ratio in annealing buffer (10 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA) and ligated into the 3′-UTR of the Renilla luciferase gene in the XhoI/NotI-digested psiCHECK-2 vector. For a complete list of ligated target site oligonucleotides see the Supplementary data. Transient transfection of human cell lines was performed with either nucleofection (suspension cell lines Jurkat and RPMI8866) according to manufacturer’s protocols (Amaxa) or with lipofection (adherent HeLa and MCF7 cells) utilizing FuGENE HD transfection reagent (Roche Diagnostics, Mannheim, Germany). Luciferase activities were determined with either the Dual-Glo Luciferase Assay or the two individual assays for Renilla and Firefly luciferase (all Promega). psiCHECK-2 transfected cells were treated in accordance to manufacturer’s protocols and luminescence was assayed on a Victor2 microplate reader (Perkin Elmer). Luminescence intensities for the sno-miRNA-target associated Renilla luciferase were normalized against the simultaneously examined firefly luciferase values to minimize the risk of transfection and assay artefacts. Normalized R/F values were standardized with two negative controls, i.e. the empty dual luciferase vector and a scrambled, non-cognate target sequence.
Northern blot analysis
Total RNA was isolated from cell cultures by the TRIzol method (Invitrogen Life Technologies). An amount of 10 µg of total RNA were size-separated on denaturing 8% acrylamide/bisacrylamide (19:1; 7 M Urea) gels (Invitrogen), transferred onto nylon membranes (BrightStar-Plus, Ambion), UV- crosslinked (Stratagene crosslinker) and pre-hybridized for 1 h at 40°C in 1M sodium phosphate buffer (pH 6.2), 7% SDS. Hybridizations to 32P-ATP, end-labelled oligonucleotides complementary to the respective npcRNAs (Supplementary Data) were performed in 1 M sodium phosphate (pH 6.2), 7% SDS for 12 h at 38°C. Membranes were washed twice for 30 min at room temperature in 2× SSC, 0.05% SDS buffer and exposed to Kodak MS-1 films for 3 h–2 days.
RESULTS
Pool of ncRNAs in human T lymphocytes
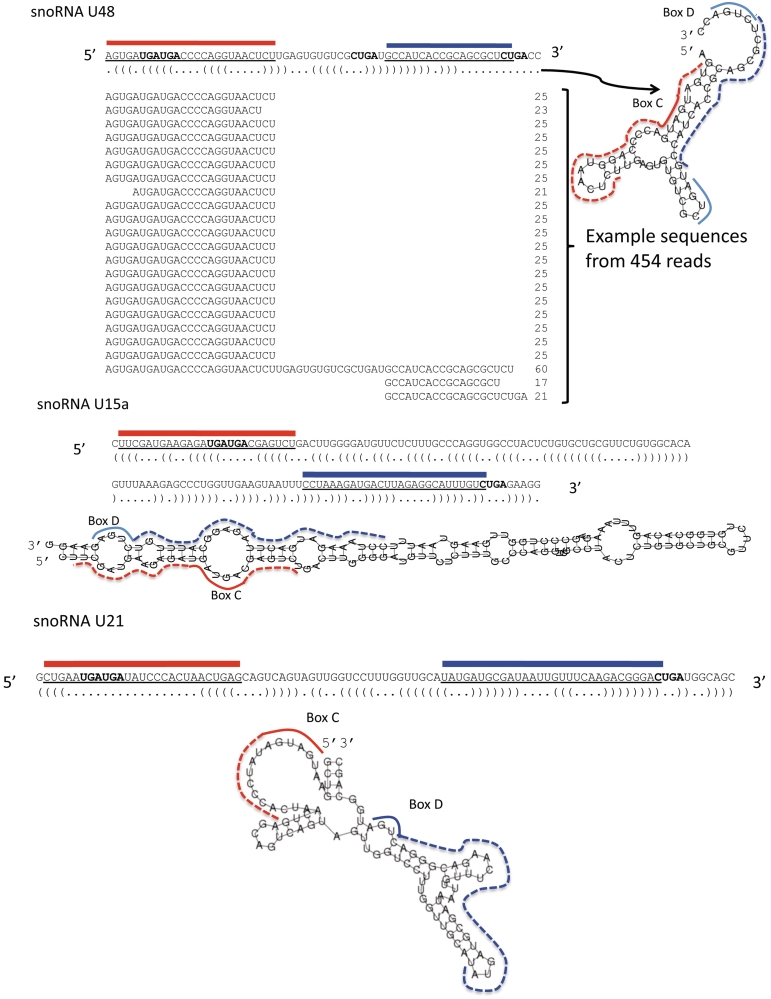
Small RNA fractions (up to 40 nt) were generated from RNA extracts of human CD4+ T lymphocytes and subjected to massive parallel pyrosequencing. We identified approximately 240 000 sequences originating from known human miRNAs, reflecting >60% of all small RNAs in the deeply sequenced library. In addition we identified more than 9000 RNAs of 18–28 nt in length that originate from bona fide snoRNAs. Among these sequences 20 box C/D snoRNAs were selected using the procedure described in ‘Materials and Methods’ section and are enriched in human Jurkat cells (Table 1), besides the previously characterized H/ACA box snoRNA (ACA45). Interestingly, the majority of sno-miRNAs with increased frequencies match perfectly in regions of ∼18-nt and display only minor differences in containing additional 1- to 5-nt in the 3′- or 5′-flanking region. When compared to secondary structure predictions we found most of the sno-miRNA sequences located in regions that are likely to form stem-loops when thermodynamics is the sole folding determinant. Three structurally distinct C/D box snoRNAs and their corresponding sno-miRNAs are shown in Figure 1. For U48 we include the alignment of sno-miRNA sequences, the dominant or guide strand is highlighted with a red bar, the aligned minority or passenger strand is marked with a blue bar. The location of sno-miRNAs on predicted snoRNA structures of U15a, U48 and U21 is indicated, red marks the major, blue the minor sno-miRNA-strand. The RNAfold-predicted structures of all 20 snoRNAs with embedded candidate sno-miRNAs are provided in Supplementary Figure S1. We frequently identified the sno-miRNA guide strand to contain the box C element of the snoRNA, while guides as well as the passenger strands that were only present in very low copy numbers in the small RNA libraries were in most cases flanked by the box D element in their 3′-end (compare Table 2).
Figure 1.
Box C/D snoRNA derived sno-miRnas. Predicted secondary structures of the snoRNAs U48, U15a and U21 were calculated with RNAfold. Secondary structure sequences are given in dot-bracket notation with base pairings represented by two complementary parentheses ‘(’ and ‘)’ and non-pairing bases by dots ‘.’. A multiple sequence alignment of U48 derived sequences that were identified by 454 sequencing displays multiple copies of 23–25 nt of the 5′-region of U48, a minority of sequences was identified as 3′-fragments or full length U48. The positions of high frequency sequences from snoRNAs U15a, U48 and U21 are indicated with red bars, the corresponding low frequency strand positions are marked with blue bars. Box C and D elements are given in bold in the sequences.
Table 2.
Sequences of box C/D snoRNAs with indicated sno-miRNAs that were functionally tested
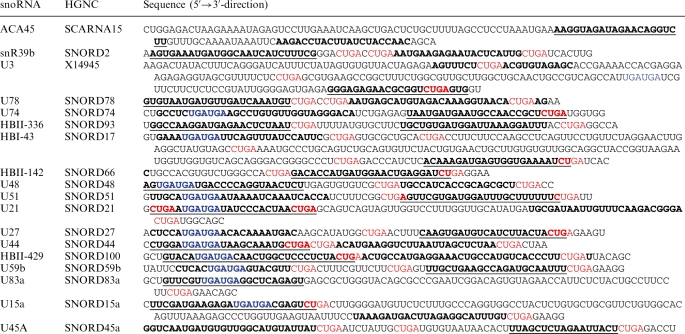 |
Sequences that were found in small RNA libraries are shown in bold, the putative guide strand (higher frequencies) is underlined. Blue indicates possible box C and C′ consensus sequences, red indicates consensus box D and D′ elements.
Functional characterization of candidate sno-miRNA sequences
Selected candidates were tested for their gene silencing capabilities in dual luciferase based reporter gene assays. Fully complementary target sites for putative sno-miRNA guide strands were cloned into the 3′-UTR of the Renilla luciferase gene of the psiCHECK-2 reporter vector. The second luciferase gene (Firefly, Photinus pyralis) remained unaffected and served as an internal control for subsequent normalization of the data obtained. Data variations due to different transfection efficiencies can be excluded, since the reporter gene and the control gene for normalization of the data are encoded on the same plasmid. Upon RISC incorporation of sno-miRNAs the transcripts were recognized and subjected to ago2 cleavage and subsequent degradation. A target for the previously described H/ACA box snoRNA ACA45 served as positive control, the unmodified (empty) psiCHECK-2 vector or insertion of a non-cognate target site as negative control.
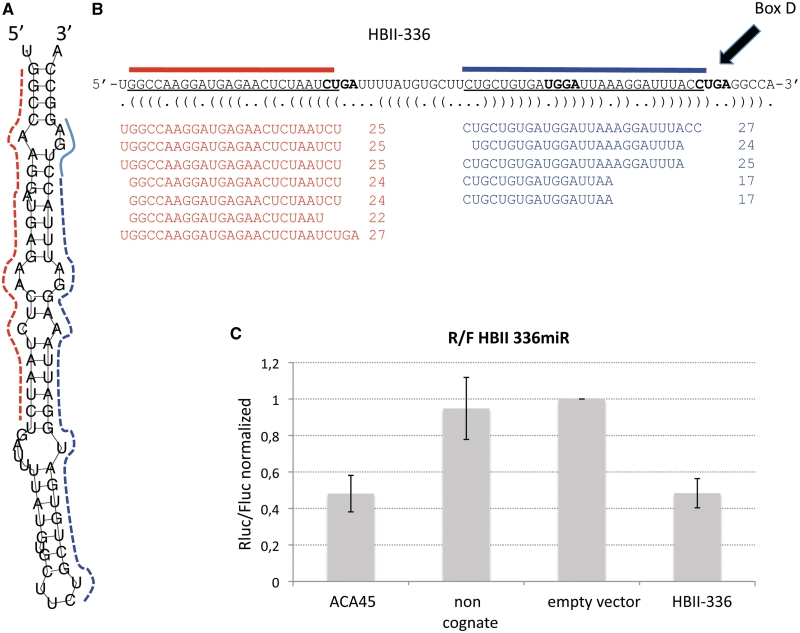
Figure 2 displays the general exemplar for the combined results of analysing the C/D box snoRNA HBII-336. The RNAfold-predicted secondary structure of HBII-336 is shown in Figure 2A. The major sno-miRNA strand, as was determined by frequencies in deep sequencing of small RNA libraries from human T cells is highlighted in red in (Figure 2B), nucleotides of the minority strand (sno-miRNA*) are shown in blue colour. Sequence alignments of deep-sequenced small RNAs are shown in Figure 2B, the 3′-flanking box D element is indicated with an arrow. The high degree of sequence stability taken together with the putative location in stable stem regions of the source-snoRNA and their abundance in small RNA libraries were selection criteria for functional analysis of the candidates. RISC incorporation and RNA silencing activity of the candidate sequences were determined in luciferase assays. The HBII-336 sno-miRNA was efficient in silencing the reporter gene upon transfection into Jurkat cells and displayed miRNA activity that was directly comparable to that of the ACA45 sno-miRNA (Figure 2C).
Figure 2.
Structural and functional analysis of the box C/D snoRNA HBII-336 sno-miRNA. (A) Secondary structure prediction for HBII-336 was calculated using RNAfold. A dashed red line marks the guide-sno-miRNA strand, the dashed blue line indicates the positions of the passenger strands. (B) The dominant sno-miRNA sequence found in deep sequencing of Jurkat small RNAs is highlighted in red, the minor sequence is marked with a blue bar. The sno-miRNA is located in a very stably base paired stem that can possibly be recognized and processed by Dicer. ClustalW alignments of the guide and passenger HBII-336- sno-miRNAs show homogenous distributions of the sequences found in small RNA libraries. The guide sno-miRNA (red) shows length variability of 21- to 27-nt, while the length distribution for the minor species (or the passenger-strand, blue) is from 17- to 27-nt. (C) Functional investigation of the HBII-336 sno-miRNA in human HeLa cells by the dual luciferase assay shows similar silencing effects when compared to the previously described ACA45 sno-miRNA. Insertion of a scrambled target site sequence into the Renilla luciferase gene did not lead to significant luciferase reduction when compared to the unmanipulated vector. Data represent normalized outcomes of three individual experiments.
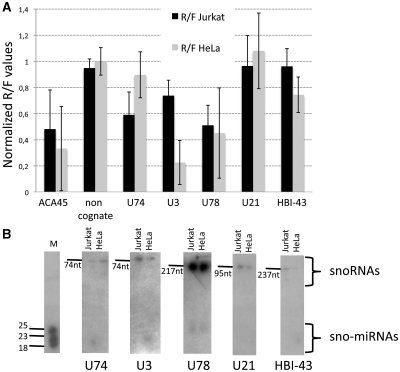
Total RNA extracts from human HeLa and Jurkat cells were analyzed by northern blots with probes against sno-miRNAs U74, U3, U78, U21 and HBI-43 (Figure 3). The snoRNAs were processed to sno-miRNAs in a cell type specific manner, e.g. U74 was processed only in Jurkat, but not in HeLa cells. Reporter gene assays for the sno-miRNA activities indicated a possible correlation between presence of the sno-miRNA and gene silencing activity (Figure 3A). The snoRNA U21 was not detected as a corresponding sno-miRNA and gene silencing was not observed in Jurkat and HeLa cells. A correlation of the expression level of the host snoRNA to the sno-miRNA activity could not be concluded from the results. U78 was the most abundant snoRNA that was present as a sno-miRNA but still the silencing efficiency was lower when compared to the U3 sno-miRNA.
Figure 3.
Sno-miRNA analysis in human HeLa and Jurkat cells. Five individual snoRNAs were probed for expression and processing via northern blots with total RNA extracts from HeLa and Jurkat cells as shown in (B). Processed sno-miRNAs can be identified in both cell types. In Hela extracts sno-miRNAs from U3, U78 and HBI-43 can be seen, in Jurkat cell extracts the sno-miRNAs U74 and U78 were identified. The total sizes of full length snoRNAs are indicated. Reporter gene assays with complementary target sites for the sno-miRNAs indicate a correlation of snoRNA processing to gene silencing activity as displayed in (A). A randomized non-cognate target sequence served as negative control, the ACA45 sno-miRNA as a positive control. The empty psiCHECK-2 vector was used for normalization. Data represent the average outcome of four (HeLa, Jurkat) or three (RPMI8866, MCF7) individual experiments.
miRNA-activity assays for 22 snoRNA derived small RNAs
We tested 21 putative sno-miRNAs, including 18 box C/D and three box H/ACA snoRNA-derived, in dual luciferase assays in three human cell types, i.e. HeLa (cervix carcinoma) line, Jurkat (T cells) and RPMI8866 (B cell). Eleven of the box C/D sno-miRNA candidates tested showed miRNA activity, which was directly comparable to that of the ACA45 sno-miRNA in at least one of the cell lines. All three box H/ACA sno-miRNAs were efficient in gene silencing. A summary for all sno-miRs, including positions within the parental snoRNAs and gene silencing efficiencies is given in Table 1. A more detailed view on the full sequences of box C/D sno-miRNAs and the relevant box C and D elements is provided in Table 2.
Interestingly we identified the functional box C element within the guide sequences of many sno-miRNAs that were capable of gene silencing. The corresponding passenger strands were in most cases stopped prior to or after the first two nucleotides of the box D element. Some passenger strands were in addition flanked by a box D′ element on the 5′-end. Guide and passenger strand positions are shown in Table 2, guides are shown in bold and underlined, passengers only in bold. Box C elements are highlighted in blue, box C and C′ elements in red colour. To confirm that this feature is a general property, we scanned all 402 snoRNAs in snoRNABase version 3 (37) and found 107 candidates where (i) the box C element (UGUGUA) occurs inside an expressed subsequence and/or (ii) the box D element (CUGA) is flanking (and often partly contained in) the small RNA on the 3′-end (Table 3). Appropriate folding of the intron-borne box C/D snoRNAs seems to be crucial for processing of sno-miRNAs. Expression of full length snoRNAs from a H1 polymerase III promotor did not further increase the effects of the endogenous sno-miRNA activity. Five box C/D snoRNAs were tested in overexpression assays combined with the corresponding psiCHECK-2-vectors and resulted in a mild cytotoxicity in HeLa cells without increasing silencing of the Renilla luciferase gene (Supplementary Figure S2).
Table 3.
snoRNAs with box C sequence (completely) inside and/or box D sequence (right) flanking sequences from the small RNA libraries
| snoRNA | C box inside sRNA | D box Flanking sRNA | snoRNA | C box inside sRNA | D box flanking sRNA |
|---|---|---|---|---|---|
| HBI-43 | x | x | U31 | x | x |
| HBII-108B | x | U35A | x | ||
| HBII-13 | x | x | U36B | x | |
| HBII-135 | x | x | U36C | x | |
| HBII-142 | x | x | U37 | x | |
| HBII-180B | x | U38A | x | x | |
| HBII-180C | x | U38B | x | x | |
| HBII-202 | x | U42B | x | ||
| HBII-210 | x | x | U43 | x | |
| HBII-251 | x | x | U44 | x | x |
| HBII-276 | x | U45B | x | ||
| HBII-289 | x | U45C | x | x | |
| HBII-295 | x | U48 | x | x | |
| HBII-296B | x | U49A | x | ||
| HBII-336 | x | U50 | x | x | |
| HBII-419 | x | U50B | x | x | |
| HBII-429 | x | x | U51 | x | |
| HBII-436 | x | x | U52 | x | |
| HBII-85-12 | x | U55 | x | ||
| HBII-85-16 | x | U58A | x | x | |
| HBII-85-17 | x | U58C | x | ||
| HBII-85-18 | x | U59B | x | x | |
| HBII-85-19 | x | U60 | x | x | |
| HBII-85-21 | x | U62A | x | ||
| HBII-85-22 | x | U62B | x | ||
| HBII-95 | x | U63 | x | ||
| HBII-99B | x | x | U65 | x | |
| SNORD119 | x | x | U74 | x | x |
| SNORD121B | x | U76 | x | ||
| U101 | x | x | U77 | x | |
| U102 | x | U78 | x | ||
| U104 | x | x | U79 | x | x |
| U105 | x | x | U8 | x | x |
| U105B | x | x | U80 | x | x |
| U106 | x | x | U81 | x | x |
| U15A | x | x | U82 | x | x |
| U15B | x | U83A | x | x | |
| U16 | x | U83B | x | ||
| U18A | x | U84 | x | ||
| U18B | x | U86 | x | ||
| U20 | x | x | U87 | x | x |
| U21 | x | x | U88 | x | x |
| U22 | x | U96a | x | ||
| U24 | x | Z17B | x | ||
| U25 | x | x | mgU2-19 | x | x |
| U26 | x | mgU6-47 | x | ||
| U27 | x | x | mgU6-53B | x | |
| U28 | x | mgh18S-121 | x | ||
| U29 | x | snR38A | x | ||
| U3 | x | x | snR38B | x | x |
| U3-2 | x | x | snR38C | x | |
| U3-2B | x | x | snR39B | x | |
| U3-3 | x | x | |||
| U3-4 | x | x | |||
| U30 | x | x |
The expressed subsequences may act as guide or passenger strands of putative sno-miRNAs.
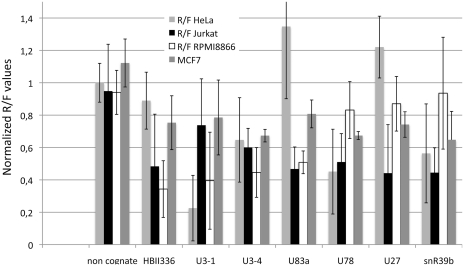
Figure 4 displays the relative silencing activity of the C/D box snoRNA-derived sRNAs in comparison to controls in four cell lines. Apparently some of the sno-miRs were active only in some, hardly ever in all cell types. Examples are snR39b, which is silencing in HeLa and Jurkat, but not in RPMI8866, or U83a and U27, which were silencing well in Jurkat but did not show significant silencing effects in HeLa and MCF7 cells. However, some of the candidates that appeared like promising Dicer substrates (e.g. U21) were incapable of miRNA activity and did not interfere with their complementary targets (Table 1 and Figure 3).
Figure 4.
Functional analysis of snoRNA generated sdRNAs in different human cell lines. Reporter gene assays were performed on human HeLa (light grey), Jurkat T (black), RPMI8866 B (white) and mammary carcinoma cell line MCF7 (dark grey). The snoRNA ACA45 served as positive control, non-cognate target and empty dual luciferase as negative control and for normalization. General gene silencing can be seen for ACA45, U3 and U3-4. HBII-336 and U83a did not induce any reduction of the Rluc gene in HeLa cells, but were very efficiently silencing the same target in both Jurkat and RPMI8866 cells. Note the differences of some sno-miRNA target sensors on the different cell types (U27, HBII-336, U83a, snR39b). Data for HeLa, Jurkat and RPMI8866 cells are from three individual experiments.
Functional characterization of box H/ACA sno-miRNAs
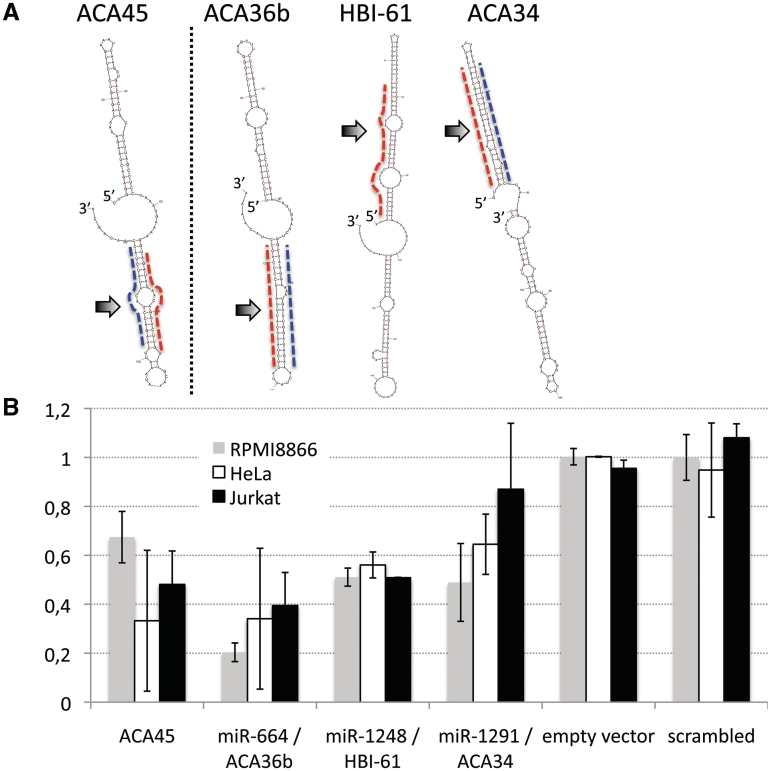
Three miRNA entries in the Sanger miRBase were discovered to originate from H/ACA box snoRNAs with very similar features when compared to ACA45 (Figure 5A). Arrows indicate the location of mature miRNA guide strands. In contrast to previous findings (35) the sno-miRNA guide strands locate either to 3′- or 5′-terminal regions of the snoRNAs. We addressed luciferase targets to the described mature miRNA sequences of miRs 664, 1248 and 1291 and found all of them to be expressed and active in RPMI8866 and HeLa cells (Figure 5B). Sno-miRNA ACA34 performed well only in RPMI8866 cells.
Figure 5.
Box H/ACA originating miRNAs in functional analysis. (A) The predicted secondary structures of three miRbase listed snoRNAs (miR-664/ACA36b, miR-1291/ACA34 and miR-1248/HBI-61, top right) share similar features with ACA45, shown top left. Two rather stable hairpins are connected via a flexible hinge loop, one of the hairpins is giving rise to a sno-miRNA (arrows). Sno-miRNA guide strand positions are marked with dashed red lines and locate to the 3′-stem of ACA45 and ACA36b, but to the 5′-stems of ACA34 and HBI-64. (B) Target sites for the sno-miRNA guide strands were checked in RPMI8866 (grey), Jurkat (black) and Hela (white) cells and compared to silencing activity of ACA45-sdRNA. The most efficient silencing on all cell types was observed for miR-664/ACA36b-sdRNA, miR-1291/ACA34 and miR-1248/HBI-61 displayed only mild silencing, in particular in Jurkat cells. Values represent normalized data from two individual experiments.
Which known miRNAs are actually sno-miRNA?
We also tested whether some known miRNAs are actually derived from snoRNAs. Therefore, the mature human miRNAs in miRBase were matched against the filtered snoRNA fragments (‘Materials and Methods’ section). Among these, we found three miRNAs located in H/ACA box snoRNAs [previously reported in ref. (35) and two in C/D box snoRNAs (Table 1). In all cases, the corresponding precursor sequences in miRBase fully matched the snoRNA sequences, providing additional evidence that these miRNA are misclassified. The predicted secondary structures of the two C/D box snoRNAs represent rather atypical miRNA-precursors or unstable hairpins among the snoRNAs tested here, when comparing global binding patterns (compare Supplementary Figure S1A).
DISCUSSION
We identified numerous so far undiscovered box C/D sno-miRNAs that exhibit processing, and in particular, mRNA silencing features similar to those of miRNA molecules. These sno-miRNAs originate from relatively short snoRNAs, such as U27 or HBII-336 as well as from structurally more complex snoRNAs with >150 nt in length, e.g. U3. The latter is particularly interesting since U3 is a very well characterized snoRNA in terms of its functions in pre-rRNA processing, including pre-rRNA capping (40), the shuttling of U3–snRNP complex between cytoplasm and nucleoli (41) and also the post-transcriptional regulation of the snoRNA U3 itself (42). Structure predictions and experimental determination of U3 snoRNP (43,44) characterize the U3 snoRNA as complex and highly structured RNA molecule (compare also Supplementary Figure S1B), assembling into the U3 snoRNP complex. However, we find sequences of the U3 3′-hairpin structure that are, at least in HeLa and RPMI8866 cells, capable of targeting complementary sequences for efficient RNA silencing.
In functional analysis we demonstrate that in total 11 candidate sequences from C/D box snoRNAs snR39b, U3 (2 sno-miRNAs), U78, HBII-336, HBII-429, HBII-142, U27, U83a, U74 and U15a, are capable of entering the silencing machinery and perform efficient gene silencing on reporter-gene mRNAs (Table 1). Our results are indicating that snoRNA-derived small RNAs can function like miRNAs and may therefore be termed sno-miRNAs. We tested a broad variety of human cell lines, including T- and B-lymphocytes, HeLa cells and the mammary carcinoma cell line MCF7 and observed general gene-silencing activity for 14 sno-miRNAs including box H/ACA sno-miRNAs. In some cases we observed a positive regulation of the target gene. This effect may be due to an increased stability of the transcribed mRNA after insertion of the target site. In absence of a loaded RISC attacking this mRNA R/F values >1 may occur (compare Figure 4). However, a number of the candidate sno-miRNAs tested were only active in some cell types such as sno-miRNA U3-1 only in HeLa and RPMI8866, sno-miRNA U83a in Jurkat and RPMI8866 cells or sno-miR U27 only in Jurkat (Figure 3).
The silencing activity differs among cell types, indicating discrete processing of snoRNAs that resembles the post-transcriptional regulation of pre-miRNAs described for rodent miRNAs (45). Differential snoRNA expression among different cell types and tissues was shown for 5S snoRNA genes (46) and regulation can be linked to host genes in which the snoRNA are located. One example for this effect is the increased level of U14 snoRNA that can be observed upon co-expression of its host gene HSC70 during stress response in Chinese hamster ovary cells (47). In general snoRNAs appear to have an extremely high degree of expression flexibility considering the modes of transcription (individual and intronic), genomic organization and processivity (48). A profiling of the expression levels of box C/D snoRNAs in 11 different human tissues was recently published and displayed strong variations of copy numbers for some snoRNAs/sno-miRNAs from our study (including snR39b and U3) (49). By testing total RNA samples from human cells on northern blots with radioactively labelled probes for different sno-miRNA guide strands we see indications of different expression levels of the snoRNAs between human HeLa and Jurkat cells. Also, we succeeded in correlating existence of the sno-miRNA to its function in RNA silencing. Due to the low abundance of snoRNAs when compared to e.g. miRNAs the signals are rather faint and only slightly above the detection threshold. However, determinants for generating sno-miRNAs from snoRNAs remain elusive and will be subjected to future studies. Minimum free energy based secondary structure predictions for all snoRNAs that were tested for miRNA activity are provided in the Supplementary data. These structures are indicating possible alternatives to the functional snoRNAs, which may be structurally different in their native nucleolar environment. Increasing the expression levels of the ‘naked’ snoRNAs by plasmid driven polymerase III transcription did not lead to additional processing of sno-miRNAs and an increase of the gene silencing activity. This finding indicates that native processing of the naturally intron-borne box C/D snoRNAs is a crucial pre-requisite for sno-miRNA production.
It is still very likely, that both snoRNAs and miRNAs share a common ancestor, or even more plausible that miRNAs evolved from the early ‘housekeeping’-snoRNAs, as was claimed by Scott et al. (35). An overview on ancestral RNA-silencing mechanisms is provided in (50) and is indicating common origins for various classes of regulatory non-protein coding small RNAs. The miRNA function of a sno-miRNA (miRNA2) originating from the box C/D snoRNA GlsR17 in the ancient Giardia lamblia is supporting the idea that biogenesis and processing of both snoRNAs and miRNAs is performed by the same machinery (32).
However, mammalian miRNA biogenesis is rather restricted when considering secondary structural features of pri-miRNAs and pre-miRNAs. MiRNA function through accurate RISC loading is dependent on successful processing in the nucleus by Drosha/Pasha as already described in 2004 (23), subsequent export of the pre-miRNA to the cytoplasm followed by Dicer processing prior to RISC loading (51). The processing of the H/ACA box sno-miRNA ACA45 is Drosha-independent (16), indicating alternative routes for the biogenesis of small RNAs with miRNA-like functions. Box C/D sno-miRNAs display a variety of predicted structures, including rather unstable hairpins (Supplementary Figure S1). A proper Dicer processing remains questionable since the preferred structure for the ribonuclease III is a stable RNA stem (52,53) and structural requirements for accurate miRNA processing are well defined in mammals (54). Recently a novel processing scheme for miRNA biogenesis was introduced and could probably explain processing and RISC loading of snoRNAs. Processing of the human miR-451 resulted in overlapping and loop-spanning mature miRNAs in a presumably ago2 mediated and Dicer independent mechanism (55,56). MiRNA biogenesis in plants harbours some principle differences and the secondary structure of appropriately processed miRNA precursors is much more diverse when compared to animal miRNAs (57,58). A link from snoRNA structure to a miRNA-like function in gene silencing cannot be concluded from our data. Nevertheless, we provide an alternative, which is based on the location of box D elements (CUGA) right flanking one mature sno-miRNA strand in most of the snoRNAs tested here (Table 2). Box C elements with the consensus sequence UGAUGA are often located within the other mature sno-miRNA sequence. The same pattern could be identified in 107 from 402 snoRNA sequences which are currently contained in snoRNABase (Table 3). This may indicate a controlled RNA processing based on recognition of the functional elements of box C/D snoRNAs rather than on structural elements as it is the case for miRNA biogenesis. We find some sno-miRs originating from RNA hairpin regions; some do not at all display this feature. However, for both types of secondary structures in the parental snoRNAs we identified candidates with and without miRNA like functions.
Carefully observing the recent literature the reader will find numerous publications on the rapidly growing number of novel non-coding RNA species with regulatory or so far unknown function (59). New members of this family are tiRNAs, which apparently are involved in promoter transcriptional gene regulation via TSS interaction (14) or, very recently, tRNA transcript-derived RNA fragments, or tRFs, which are not yet functionally characterized, but aberrantly induce proliferative defects when lost (60). Our study is now adding an additional class of dual-action non-coding RNAs by introducing the gene regulatory effects of human box C/D snoRNAs.
The spectra of novel ncRNA, the bandwidth of their functions, their abundance through the kingdoms of life and the ever increasing range of new discoveries is implying a Kuhnian revolution in understanding the molecular processes in life sciences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charges: German Federal Ministry of Education and Research joint research project ‘RNomics in Infectious Diseases’ in the frame of the National Genome Research Net (NGFN-Plus).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Timofey Rhozdestvensky and Prof. Jürgen Brosius for fruitful discussion. Christina Albrecht and Meike Hermes contributed valuable comments on the article and hands-on help. Ellen Eckermann-Felkl contributed expert technical assistance, Martina Reitz, Alexandra Juckert and Michaela Micke contributed to experimental work.
REFERENCES
- 1.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 3.Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol. Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 5.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 6.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DH, Rossi JJ. Transcriptional gene silencing using small RNAs. Methods Mol. Biol. 2009;555:119–125. doi: 10.1007/978-1-60327-295-7_9. [DOI] [PubMed] [Google Scholar]
- 11.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 13.Kawasaki H, Taira K, Morris KV. siRNA induced transcriptional gene silencing in mammalian cells. Cell Cycle. 2005;4:442–448. doi: 10.4161/cc.4.3.1520. [DOI] [PubMed] [Google Scholar]
- 14.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 15.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle. 2009;8:2332–2338. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 16.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell. Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 18.Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 19.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliceiri GL. Small nucleolar RNAs. Cell Mol. Life Sci. 1999;56:22–31. doi: 10.1007/s000180050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 22.Hirose T, Shu MD, Steitz JA. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 23.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 24.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell. Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 28.Triboulet R, Chang HM, Lapierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 30.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolev NG, Ullu E. snoRNAs in Giardia lamblia: a novel role in RNA silencing? Trends Parasitol. 2009;25:348–350. doi: 10.1016/j.pt.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 37.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Culver GM. Sno-capped: 5′ ends of preribosomal RNAs are decorated with a U3 SnoRNP. Chem. Biol. 2002;9:777–779. doi: 10.1016/s1074-5521(02)00171-0. [DOI] [PubMed] [Google Scholar]
- 41.Leary DJ, Terns MP, Huang S. Components of U3 snoRNA-containing complexes shuttle between nuclei and the cytoplasm and differentially localize in nucleoli: implications for assembly and function. Mol. Biol. Cell. 2004;15:281–293. doi: 10.1091/mbc.E03-06-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nabavi S, Nellimarla S, Nazar RN. Post-transcriptional regulation of the U3 small nucleolar RNA. J. Biol. Chem. 2008;283:21404–21410. doi: 10.1074/jbc.M802189200. [DOI] [PubMed] [Google Scholar]
- 43.Antal M, Mougin A, Kis M, Boros E, Steger G, Jakab G, Solymosy F, Branlant C. Molecular characterization at the RNA and gene levels of U3 snoRNA from a unicellular green alga, Chlamydomonas reinhardtii. Nucleic Acids Res. 2000;28:2959–2968. doi: 10.1093/nar/28.15.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clery A, Senty-Segault V, Leclerc F, Raue HA, Branlant C. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol. Cell. Biol. 2007;27:1191–1206. doi: 10.1128/MCB.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang LS, Lin SY, Lieu AS, Wu TL. Differential expression of human 5S snoRNA genes. Biochem. Biophys. Res. Commun. 2002;299:196–200. doi: 10.1016/s0006-291x(02)02623-2. [DOI] [PubMed] [Google Scholar]
- 47.Chen MS, Goswami PC, Laszlo A. Differential accumulation of U14 snoRNA and hsc70 mRNA in Chinese hamster cells after exposure to various stress conditions. Cell Stress Chaperones. 2002;7:65–72. doi: 10.1379/1466-1268(2002)007<0065:daousa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Castle JC, Armour CD, Lower M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS ONE. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soifer HS, Sano M, Sakurai K, Chomchan P, Saetrom P, Sherman MA, Collingwood MA, Behlke MA, Rossi JJ. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res. 2008;36:6511–6522. doi: 10.1093/nar/gkn687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collingwood MA, Rose SD, Huang L, Hillier C, Amarzguioui M, Wiiger MT, Soifer HS, Rossi JJ, Behlke MA. Chemical modification patterns compatible with high potency Dicer-substrate small interfering RNAs. Oligonucleotides. 2008;18:187–200. doi: 10.1089/oli.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saetrom P, Snove O, Nedland M, Grunfeld TB, Lin Y, Bass MB, Canon JR. Conserved microRNA characteristics in mammals. Oligonucleotides. 2006;16:115–144. doi: 10.1089/oli.2006.16.115. [DOI] [PubMed] [Google Scholar]
- 55.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 2009;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 58.Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2009;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 59.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.