Abstract
Restriction enzymes share little or no sequence homology with the exception of isoschizomers, or enzymes that recognize and cleave the same DNA sequence. We present here the structure of a BamHI isoschizomer, OkrAI, bound to the same DNA sequence (TATGGATCCATA) as that cocrystallized with BamHI. We show that OkrAI is a more minimal version of BamHI, lacking not only the N- and C-terminal helices but also an internal 310 helix and containing β-strands that are shorter than those in BamHI. Despite these structural differences, OkrAI recognizes the DNA in a remarkably similar manner to BamHI, including asymmetric contacts via C-terminal ‘arms’ that appear to ‘compete’ for the minor groove. However, the arms are shorter than in BamHI. We observe similar DNA-binding affinities between OkrAI and BamHI but OkrAI has higher star activity (at 37°C) compared to BamHI. Together, the OkrAI and BamHI structures offer a rare opportunity to compare two restriction enzymes that work on exactly the same DNA substrate.
INTRODUCTION
Protein–DNA selectivity is a central event in many biological processes, ranging from transcription and replication to restriction and modification. Type II restriction endonucleases (REases) are ideal systems for studying selectivity because of their high specificity and great variety. More than 3800 Type II restriction enzymes representing 304 unique specificities have now been identified (1). They generally recognize DNA sequences that vary between 4 and 8 bp and require only Mg2+ as a cofactor to catalyze the hydrolysis of DNA (2). Their sequence specificity is remarkable. A single base pair change within the recognition sequence can lead to well over a million-fold reduction in activity (2). BamHI (from Bacillus amyloliquefaciens), is one of the best-studied REase, with structures of the free enzyme and complexes with cognate and non-cognate sites available (3–5). In addition, the co-crystal structures with divalent metals provide snapshots of the pre- and post-reactive states of BamHI (6). Together, these structures grant important insight into DNA recognition, selectivity and the mechanism of cleavage by this endonuclease.
REases share little or no sequence homology with the exception of isoschizomers, or enzymes that recognize and cleave exactly the same DNA sequence (7,8). We have succeeded in expressing and crystallizing a BamHI isoschizomer, OkrAI (from Oceanospirillum kriegii), which recognizes and cleaves the exact same DNA sequence as BamHI (5′ – G↓GATCC – 3′). OkrAI is, however, smaller than BamHI (194 versus 213 amino acids), and from sequence comparisons appears to lack the equivalent of N- and C-terminal helices of BamHI. The absence of a C-terminal helix is particularly intriguing as this helix is crucial for BamHI function. We report here a structure of OkrAI bound to the same DNA fragment (TATGGATCCATA) as that cocrystallized with BamHI. Surprisingly, even though OkrAI lacks some of the secondary structural elements, its mode of DNA recognition is remarkably similar to that of BamHI, including asymmetric interactions via the C-terminal arms. The OkrAI and BamHI structures offer a rare opportunity to compare two REases that work on exactly the same DNA substrate.
MATERIALS AND METHODS
Isolation, expression and purification of OkrAI
An experimental search for BamHI isoschizomers was initiated by screening cultured microorganisms. OkrAI was discovered in Oceanospirillum kriegii (Polisson C., unpublished data, NEB strain collection). The methylase selection method (9) was used to clone the OkrAI methylase gene (okrAIM) and part of the OkrAI endonuclease gene (okrAIR). Inverse polymerase chain reaction (PCR) walk was carried out to obtain the entire okrAIR gene. A PCR fragment carrying the okrAIR gene (flanked by NdeI and SalI sites) was ligated to a T7 expression vector pSYX22 and transformed into M.BamHI premodified Escherichia coli expression host ER2566 [pACYC-bamHIM]. The expression level was estimated at 2 × 106 units of OkrAI per gram of wet cells in clarified cell extracts.
The enzyme was purified by chromatography through phosphocellulose (Whatman P11), hydroxylapatite (Bio-Rad), DEAE Sepharose (Pharmacia), heparin Sepharose (Pharmacia), Q Sepharose (Pharmacia) and Affi-gel Blue (Bio-Rad) columns. The purified OkrAI protein was stored in the following buffer: 20 mM Tris–HCl (pH 7.5), 200 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 50% (v/v) glycerol. The specific activity of OkrAI is determined to be approximately 2 × 106 units/mg protein on λ DNA substrate.
Cocrystallization and structure determination
OkrAI was cocrystallized with the same palindromic 12-mer (TATGGATCCATA) used to grow the BamHI/DNA cocrystals. The OkrAI/DNA cocrystals grow under similar conditions as the BamHI/DNA cocrystals (from 15–17% PEG 8000, 0.2 M calcium acetate and 0.1 M sodium cacodylate at pH 6.7). All X-ray data were measured at cryogenic temperatures. The OkrAI/DNA cocrystals belong to the same space group (P212121) as BamHI but with unit cell dimensions of a = 119.7 Å, b = 83.1 Å, c = 81.0 Å (as compared to a = 108.8 Å, b = 81.9 Å, c = 68.8 Å for BamHI). The crystals diffract to at least 2.3-Å resolution with synchrotron radiation and an 120o dataset was collected with a 1o oscillation at the Advanced Photon Source (APS). The structure was solved by molecular replacement (MR) method using the L subunit of the BamHI/DNA complex as a search model in program CNS (10) and was refined using CNS (10) and REFMAC (11,12). The C-terminal residues (194–213) were removed to better match the OkrAI subunit structure. The MR solution provided a clear peak distinguishable from all other solutions. Unlike in the BamHI cognate complex structure, there are two OkrAI/DNA complexes in the crystallographic asymmetric unit (subunits A and B bound DNA strands C and D; subunits E and F bound to strands G and H). The final OkrAI structure contains residues 1–194 for monomers A, E and F, residues 1–192 for monomer B, all 12 nt in each of the four DNA strands (C,D,G,H), 710 water molecules, and is refined to a crystallographic R-factor of 16% and Rfree of 23%. The model has an excellent stereochemistry, with over 94% of the residues in the most favored regions of the Ramachandran plot. Data collection and refinement statistics are summarized in Table 1.
Table 1.
Crystallographic data collection and refinement
| A. Data collection and phasing statistics | |
| Total no. of reflections | 131 820 |
| Resolution range (Å) | 20–2.3 |
| No. of unique reflections | 32 438 |
| Rsym (last shell) | 0.042 (0.108) |
| Completeness (%) (last shell) | 89 (84) |
| I/σ(I) | 43.2 (20.8) |
| Redundancy (last shell) | 4.1 (4.0) |
| B. Refinement statistics | |
| Resolution range (Å) | 20–2.3 |
| No. of reflections used in refinement | 30 786 |
| Rcryst/Rfree | 0.16/0.23 |
| No atoms | |
| Protein | 6007 |
| DNA | 972 |
| Water | 710 |
| Average B factors (Å2) | 14.2 |
| R.m.s. deviations | |
| Bonds (Å) | 0.017 |
| Angles (deg.) | 1.9 |
aRsym = ∑|In – < In>/∑In over all h, where In is the intensity of the reflection h.
bRcryst/Rfree = ∑||Fo| – |Fc|| / ∑|Fo|.
Rfree was calculated with 5% of data excluded from refinement.
Structural analysis
Analysis of the stereochemical quality of the protein model and assignment of secondary structure were conducted with PROCHECK (13). DNA analysis was performed with 3DNA (14). Solvent-accessible surface areas were calculated in CNS with the algorithm of Lee and Richards employing a 1.4-Å probe (15). Figures were prepared using PyMOL (www.pymol.org).
Fluorescence anisotropy measurements
5′-Fluorescein (6-carboxyfluorescein)-labeled DNA (5′-TAT GAG CGG ATC CTT ACG AG-3′) was purchased from IDT DNA Technologies. Oligos were re-suspended in buffer with 10 mM Tris–HCl, pH 7.5 at 25°C, 100 mM KCl, 10 mM CaCl2, 1 mM EDTA and 5 mM DTT. The fluorescein-labeled oligo was mixed with the unlabelled complementary strand and annealed to a final concentration of 50 μM. Experiments were performed using Panvera Beacon 2000 fluorescence polarization system (at 25°C). Fluorescence intensity data were collected by setting excitation filter at 490 nm and emission filter at 520 nm. Each reaction sample (volume 200 μl) consisted of 5 nM of 5′-Fluorescein labeled DNA and increasing concentrations of BamHI or OkrAI (0.1–400 nM) in the above-mentioned buffer. Individual reaction samples were equilibrated for more than 30 min before measuring the anisotropy values. The anisotropy values obtained were subtracted from the anisotropy value of the blank (buffer with no protein) and then divided with the maximum value of anisotropy value obtained for the experiment. This normalized anisotropy value yields the fraction of the DNA bound to the protein (y). The fraction of the DNA bound form (y) was plotted against the concentration of BamHI or Okra (x in nM) and fitted to the following equation (using non-linear regression in Origin 7, OriginLab):
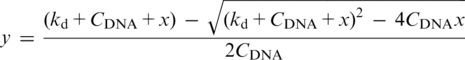 |
where, y is the fraction of the DNA bound to the protein, CDNA is the concentration of DNA used (5 nM), Kd is the dissociation constant and x is the concentration of BamHI or OkrAI.
OkrAI cleavage activity assays
Purified OkrAI and BamHI were serially diluted in 2-fold series in 20 mM Tris–HCl, pH 7.9, 50 mM NaCl before each assay. Five microliters of the diluted enzymes were incubated with 1 µg of λ DNA in 50 µl reactions containing 20 mM Tris–HCl, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, pH 7.9. The reactions were carried out at the indicated temperature for 1 h. The reactions were stopped by the addition of a loading dye that gave a final concentration of 0.17% for sodium dodecyl sulfate (SDS) and 8.3 mM for EDTA. The cleavage products were analyzed by electrophoresis through 0.8% agarose gels. One unit of endonuclease activity is defined as the minimum amount of enzyme required for the complete cleavage of the substrate. Fidelity index (FI) is defined as the ratio of the maximum enzyme amount showing no star activity to the minimum amount needed to achieve 1 U of activity.
RESULTS
Overall architecture
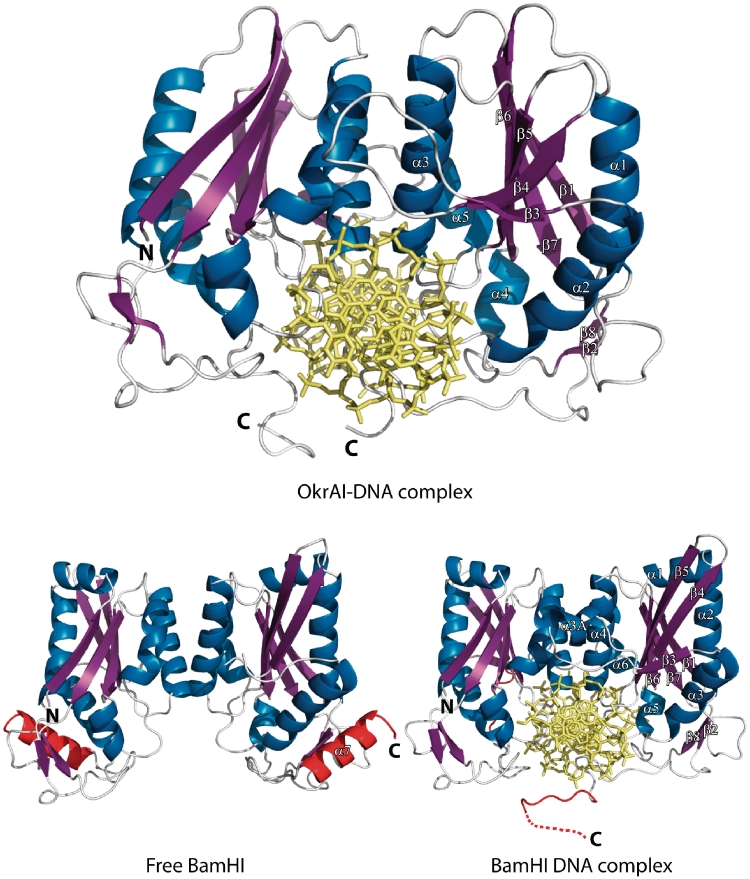
Similar to BamHI, an OkrAI dimer approaches DNA from the major groove side (Figure 1). The subunit consists of a large six-stranded mixed β-sheet, which is sandwiched on both sides by α helices (3). Strands β3, β4 and β5 are antiparallel and form a β meander; strands β5, β6 and β7 are parallel and resemble a Rossman fold, with α3 and α5 acting as the crossover helices. The dimer interface is formed primarily by helices α3 and α5, which pair with the corresponding helices from the symmetry related subunit to form a parallel four-helix bundle (Figure 1). Compared to BamHI, some of the structural elements are either missing or are shorter in OkrAI (Figure 2). In particular, the first helix, α1, of BamHI is entirely missing in OkrAI and β1 is connected to α1 (the equivalent of α2 in BamHI) by a short loop. The loops between β2 and α2 and α2 and β3 are shortened by one residue. The short 310 helix α3A of BamHI between strands β3 and β4 is also missing in OkrAI and is connected by only a loop. Strands β4 and β5 are both shorter by three residues but connected by a slightly longer loop in OkrAI (four versus two residues). There is an insertion of five residues between α5 and β7, the equivalent of α6 and β7 in BamHI. Moreover, the C-terminal extension (containing the ‘arm’) is nine residues shorter in OkrAI than in BamHI. Despite these differences in structure, the way in which OkrAI and BamHI recognize cognate DNA is remarkably similar.
Figure 1.
Schematic representation of the OkrAI–DNA complex (top), the apo form of BamHI (bottom left) and the BamHI–DNA complex (bottom). The secondary structural elements of the R subunit in each complex are labeled and are colored as follows: α helices are blue, β strands are magenta and loops are white. The BamHI C terminus that is α helical in the apo form and unstructured in the DNA bound form is highlighted in red.
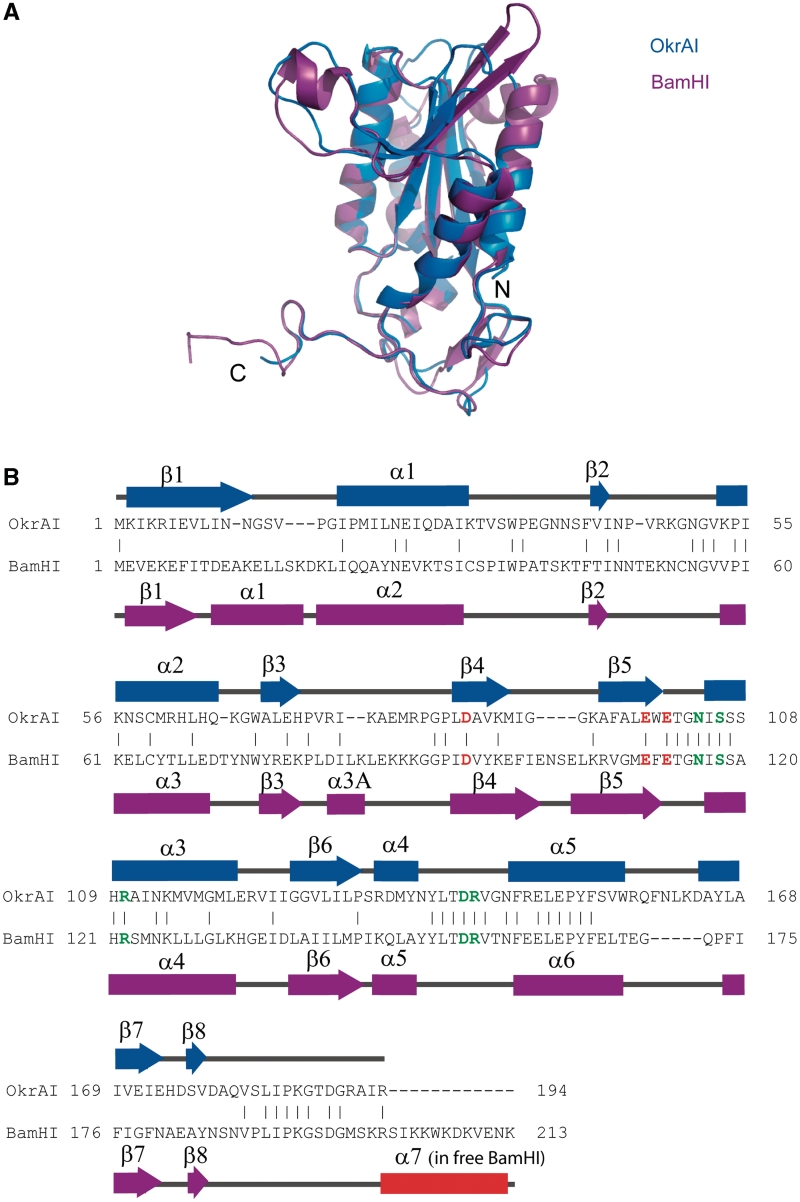
Figure 2.
Comparisons between OkrAI and BamHI subunits. (A) The monomer of OkrAI (blue) superimposed on the monomer of BamHI (magenta). (B) Sequence alignment of OkrAI and BamHI based on the structural alignment performed by DALI (36). The catalytic residues of both enzymes are shown in red and the DNA-binding residues in green. The secondary structural elements are labeled and shown above (OkrAI) and below (BamHI) the alignment, respectively. The BamHI C terminus that is α helical in the apo form and unstructured in the DNA-bound form is highlighted in red. This segment is missing in OkrAI.
DNA conformation
The OkrAI DNA retains a B-DNA conformation over the central 10 bp with average helical twist and rise of 32.26o and 3.33 Å, respectively, compared to a helical twist and rise of 33.44o and 3.43 Å for BamHI. There are no kinks in the OkrAI DNA axis of the type seen in EcoRV and EcoRI DNAs, or any major unwinding at the central base-pair step as seen in EcoRV (16), EcoRI (17) and BglII (18) DNAs. The direction of curvature is the same as in most restriction enzyme complexes, namely away from the α/β core. In contrast, dimeric transcription factors such as phage 434 repressor (19) or E. coli Trp repressor (20) bend DNA toward the body of the protein. As a consequence, the minor groove at the center of the DNA is significantly narrower (∼3 Å) in the phage 434 DNA complex than in the OkrAI–DNA complex (∼7 Å).
DNA recognition
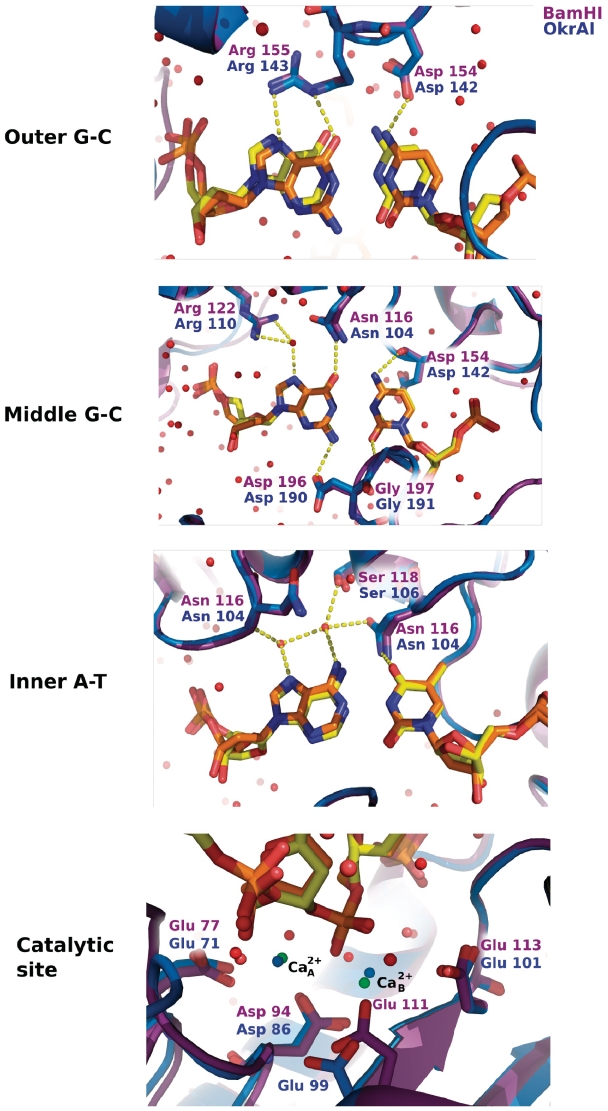
The DNA is bound in the cleft formed by the two OkrAI monomers (Figure 1). There are extensive protein–DNA interactions, with side chain and main chain atoms, and tightly bound water molecules all contributing toward recognition of the GGATCC sequence. Interactions with DNA occur both in the major and minor grooves. In the major groove, every hydrogen bond donor and acceptor group takes part in direct or water-mediated hydrogen bonds with the protein (Figure 3). These interactions are remarkably similar to those observed in the BamHI cognate DNA complex In particular, the major groove contacts are made primarily by regions at the N-terminal ends of helices α3 and α5, the equivalent of helices α4 and α6 in the BamHI complex. The outer G•C base pair (GGA) is contacted by Arg143 and Asp142 from the 140–146 loop that precedes helix α5, the equivalent of residues Arg155 and Asp154 in BamHI. The middle G•C base pair (GGA) is contacted by Asp142, Arg110 and Asn104, the equivalent of residues Asp154, Arg122 and Asn116 of BamHI. The inner A•T base pair (ATC) is contacted primarily through water mediated hydrogen bonds as well as by Asn104. Consistent with the structural data, mutating this amino acid residue in BamHI (N116H) resulted in a mutant enzyme that prefers to cleave a methylated site (GGN6mATCC) (21). In the minor groove, contacts to bases are made by the C-terminal arm of the R subunit. There is insufficient room to accommodate arms from both subunits if they were to lay symmetrically in the groove. The R arm makes specific interactions with bases in both DNA half-sites via residues Asp189, Gly190 and Arg191, the equivalent of residues Asp196, Gly197 and Met198 of BamHI. Thus, in a similar manner to BamHI, OkrAI makes contacts in the minor groove in an asymmetric fashion, with the arm from one subunit (R) entering the groove, and the arm from the other subunit (L) following and making contacts with the DNA sugar-phosphate backbone. Altogether, there are extensive interactions between OkrAI and the sugar-phosphate backbone, including contacts from residues at the NH2-terminal ends of helices α2, α3 and α5, the ordered region before strand β4 (residues 83–85), and the arm of the L subunit (residues 190 and 191).
Figure 3.
Base-specific DNA interactions and active sites in OkrAI and BamHI. The proteins are shown in ribbon representation (OkrAI in blue and BamHI in magenta). The nucleotides (OkrAI DNA in orange and BamHI DNA in yellow) and the base specific residues are shown in ‘stick’ representation. In each picture, only the base-pairs in question are shown, other nucleotides are omitted for clarity. In the lower panel, the active site residues of OkrAI (Glu71, Asp86, Glu99 and Glu101) and BamHI (Glu77, Asp94, Glu111 and Glu113) are shown in ‘stick’ representation. The Ca2+ ions of OkrAI are shown in green and that of BamHI in blue. The two metal sites are separated by 3.8 Å in OkrAI and 4.3 Å in BamHI.
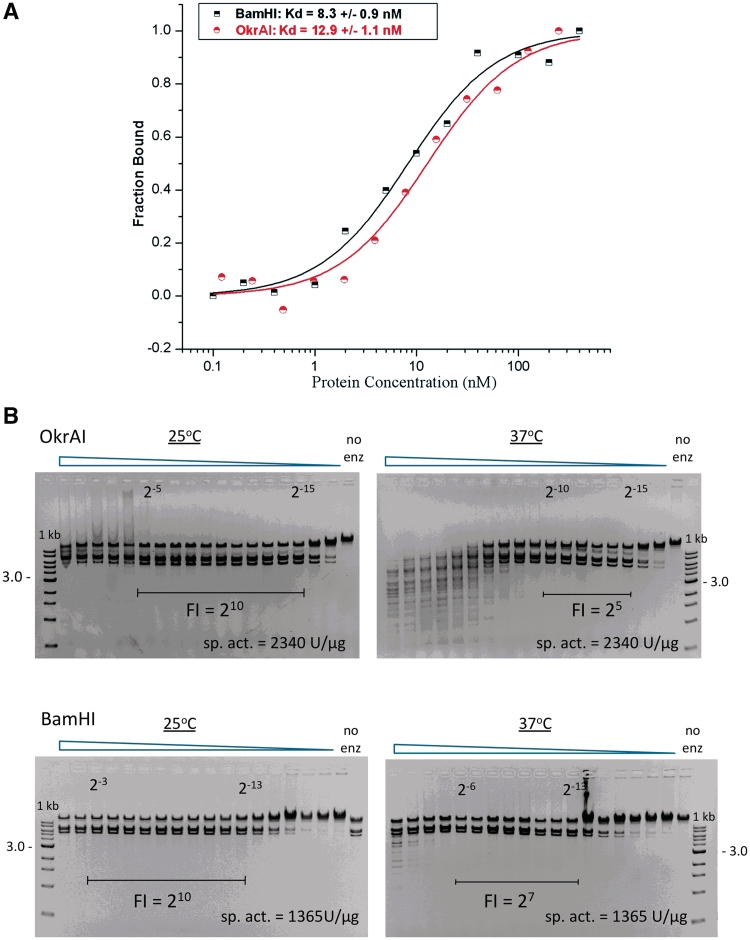
The structural similarity between OkrAI and BamHI in recognizing the cognate GGATCC sequence is reflected in their similar binding constants (with a Kd of 12.9 and 8.3 nM for OkrAI and BamHI, respectively), as measured by fluorescence anisotropy (Figure 4A). However, when we compare the DNA cleavage properties of the two enzymes there are differences in how they cleave non-cognate or ‘star’ sites at different temperatures. We find that both enzymes have reduced star activity at 25°C as compared to 37°C (Figure 4B), but OkrAI has a significantly higher star activity (4-fold higher) at 37°C compared to BamHI.
Figure 4.
Binding and cleavage data. (A) Fluorescence anisotropy results for OkrAI and BamHI. The fraction of the bound DNA is plotted against the concentration of the protein. The data points for BamHI are shown as black half squares and for OkrAI as red half circles, and the fitted curves as solid lines. (B) Cleavage activity data for OkrAI (top) and BamHI (bottom) at 25°C and 37°C using λ DNA as the substrate. The fidelity index (FI) is defined as the ratio of the maximum enzyme amount showing no star activity to the minimum amount needed to achieve 1 U of activity.
Active site
Most members of the PD-(D/E)XK family of restriction endonucleases, to which BamHI and OkrAI belong, utilize Mg2+ for cleavage. The active site geometry of the R subunit of the OkrAI/DNA complex is almost identical to that of the R subunit of the BamHI cognate complex (Figure 3), with residues of Glu71, Asp86, Glu99 and Glu101 aligning with Glu77, Asp94, Glu111 and Glu113 of BamHI. Other than BamHI and its isoscizomers (OkrAI, Bsp98I and DdsI) only one other REase, BtsCI is (22) known to contain a glutamate at the last catalytic residue position (Glu113 in BamHI and Glu101 in OkrAI), whereas the majority of restriction enzymes contain a lysine at this position. Interestingly, there is a severe loss of cleavage activity when Glu113 in BamHI is substituted by a lysine residue (23) or conversely, when Lys92 in EcoRV (24) or Lys113 in EcoRI (25) is substituted by a glutamate residue, even though the geometry of the active sites of these restriction enzymes is generally similar. Based on the structures of the pre- and post-reactive complexes of BamHI (6), a two-metal mechanism has been proposed for BamHI, in which metal A activates the attacking water molecule, while metal B stabilizes the buildup of negative charge on the leaving O3′ atom. At the same time, both metals (acting as Lewis acids) are proposed to help stabilize the pentacovalent transition state. In the OkrAI complex, we observe two calcium ions at positions almost identical to the metal A and metal B sites in BamHI, suggesting that OkrAI also utilizes a two-metal mechanism for cleavage. Curiously, in the pre- and post-reactive complexes of BamHI (6), only the R subunit contains metals, the L subunit does not. Interestingly, in OkrAI the L subunit contains a single metal site roughly halfway between metal A and metal B. Thus, in BamHI and OkrAI, the binding of two metals is correlated to the active site of the subunit that has its C-terminal arm in the minor groove. This asymmetry in DNA binding and the binding of metals have been suggested to correlate with an observed sequential DNA cleavage mechanism in BamHI (26).
DISCUSSION
The OkrAI and BamHI structures offer a rare opportunity to compare two REases that work on exactly the same DNA substrate. To our knowledge, the only other pair of isoschizomers whose structures have been determined are the homing endonucleases I-CreI and I-MsoI (27). However, unlike OkrAI and BamHI, these two enzymes show considerable divergence in residues that partake in DNA recognition and the protein–DNA interface as a whole is strikingly different in the two structures. In contrast, OkrAI emerges here as essentially a more minimal version of BamHI. OkrAI lacks not only the N- and C-terminal helices but also the 310 helix between strands β3 and β4, and containing β-strands (β4 and β5) that are shorter than those in BamHI. Despite these structural differences, OkrAI recognizes the GGATCC sequence in a remarkably similar manner to BamHI. All of the the DNA recognition residues (as well as the catalytic residues) are conserved between the two enzymes and make the same pattern of hydrogen bonds to bases in the major and minor grooves. Not all of the BamHI isoschizomers may be so similar, however; with several isoschizomers having been identified (28–32) that differ in size from 22 kDa to 43 kDa and ranging from monomer (28) to tetramer (31). It will be interesting to compare the structures of these isoschizomers to know which structural elements are conserved and which have diverged over the course of the evolution of these type II restriction enzymes.
One of the most surprising features of the OkrAI structure is asymmetric DNA binding. This was unexpected because OkrAI appeared to lack, from sequence alignments, the equivalent of a C-terminal helix in BamHI, which is present in ‘free’ BamHI and when the enzyme binds to non-cognate DNA, but which unfolds on binding to cognate DNA to form long partially disordered arms, with the arm from one subunit (R) fitting into the minor groove and the arm from the other subunit (L) following the DNA sugar-phosphate backbone. As such, the C-terminal residues in BamHI are believed to fulfill a dual role: first, as helices, to aid in the initial binding and the diffusion of the enzyme on nonspecific DNA; second, by unfolding to increase the lifetime of the specific complex for the subsequent cleavage reaction. The lack of a C-terminal helix in OkrAI lends to C-terminal arms that are much shorter than in BamHI, and also, unlike BamHI, they are likely to be unstructured in free OkrAI or when the enzyme binds to non-cognate DNA. However, despite the truncated arms, OkrAI recognizes cognate DNA in the same asymmetric manner as BamHI, with one arm making base contacts in the minor groove and the other making contacts with the DNA backbone. As in the case of the BamHI–DNA complex, there is insufficient room in the minor groove to accommodate arms from both subunits. Asymmetric DNA binding is not limited to BamHI and OkrAI but extends to other proteins, including the transcription factor HAP1 (33). In HAP1 the two subunits bind in a head-to-tail fashion to a symmetric recognition site resulting in a dramatically asymmetric complex. A different kind of asymmetry is observed when monomeric MspI endonuclease binds its palindromic DNA sequence (34,35), utilizing different parts of the monomer to recognize symmetric half sites of the DNA.
Curiously, OkrAI and BamHI have increased ‘star’ activity at higher temperatures that may be related to the flexibility of their C-terminal arms. That is, at the higher temperature (37°C versus 25°C), we envisage that the BamHI C-terminal helix is more prone to unfolding and thereby in inducing a pseudo-specific complex competent for cleavage at the ‘star’ sequences; in free BamHI, the C-terminal helix is the most mobile secondary structural element (with an average B factor of 33 Å2). Interestingly, at this higher temperature (37°C), OkrAI has even higher star activity than BamHI, which may be due to the lack of a C-terminal α-helix and the presence of shorter arms that are already unfolded and, as such, require lower activation energy to induce a pseudo-specific complex for cleavage at star sequences. Mutational studies are under way to test the role of the C-terminal residues on OkrAI and BamHI cleavage activities. Taken together, OkrAI and BamHI provide for the first time a structural framework for comparing two type II restriction enzymes that work on exactly the same DNA substrate.
ACCESSION NUMBER
Coordinates have been submitted to the RCSB Protein Data Bank with accession code 3ODH.
FUNDING
National Institutes of Health grant GM44006 (to A.K.A.). Funding for open access charge: National Institutes of Health grant GM44006.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the staff at CHESS and at the APS for help with data collection. We thank Elinor Vinecombe for assistance with the structure determination; Jim Samuelson, Jianping Xiao and Becky Kucera for technical assistance; Ira Schildkraut for initiation of the OkrAI cloning project; Don Comb for support and encouragement. OkrAI expression strain request should be directed to xus@neb.com.
REFERENCES
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE – a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts RJ, Halford SE. In: Nucleases. Linn SM, Lloyd RS, Roberts RJ, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 3.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 4.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 5.Viadiu H, Kucera R, Schildkraut I, Aggarwal AK. Crystallization of restriction endonuclease BamHI with nonspecific DNA. J. Struct. Biol. 2000;130:81–85. doi: 10.1006/jsbi.2000.4235. [DOI] [PubMed] [Google Scholar]
- 6.Viadiu H, Aggarwal AK. The role of metals in catalysis by the restriction endonuclease BamHI. Nat. Struct. Biol. 1998;5:910–916. doi: 10.1038/2352. [DOI] [PubMed] [Google Scholar]
- 7.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 8.Roberts RJ, Macelis D. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1991;19(Suppl.):2077–2109. doi: 10.1093/nar/19.suppl.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szomolanyi E, Kiss A, Venetianer P. Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene. 1980;10:219–225. doi: 10.1016/0378-1119(80)90051-7. [DOI] [PubMed] [Google Scholar]
- 10.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 11.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 12.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 13.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 14.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 16.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 18.Lukacs CM, Kucera R, Schildkraut I, Aggarwal AK. Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution. Nat. Struct. Biol. 2000;7:134–140. doi: 10.1038/72405. [DOI] [PubMed] [Google Scholar]
- 19.Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 21.Whitaker RD, Dorner LF, Schildkraut I. A mutant of BamHI restriction endonuclease which requires N6-methyladenine for cleavage. J. Mol. Biol. 1999;285:1525–1536. doi: 10.1006/jmbi.1998.2409. [DOI] [PubMed] [Google Scholar]
- 22.Too PH, Zhu Z, Chan SH, Xu SY. Engineering Nt.BtsCI and Nb.BtsCI nicking enzymes and applications in generating long overhangs. Nucleic Acids Res. 2010;38:1294–1303. doi: 10.1093/nar/gkp1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu SY, Schildkraut I. Isolation of BamHI variants with reduced cleavage activities. J. Biol. Chem. 1991;266:4425–4429. [PubMed] [Google Scholar]
- 24.Selent U, Ruter T, Kohler E, Liedtke M, Thielking V, Alves J, Oelgeschlager T, Wolfes H, Peters F, Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992;31:4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- 25.Grabowski G, Jeltsch A, Wolfes H, Maass G, Alves J. Site-directed mutagenesis in the catalytic center of the restriction endonuclease EcoRI. Gene. 1995;157:113–118. doi: 10.1016/0378-1119(94)00714-4. [DOI] [PubMed] [Google Scholar]
- 26.Hensley P, Nardone G, Chirikjian JG, Wastney ME. The time-resolved kinetics of superhelical DNA cleavage by BamHI restriction endonuclease. J. Biol. Chem. 1990;265:15300–15307. [PubMed] [Google Scholar]
- 27.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 28.Murakami M, Mizuno H, Yamada Y. The restriction endonuclease GceinI of Gluconobacter cerinus IFO 3260, an isoschizomer of BamHI, has a monomeric structure. Agric. Biol. Chem. 1990;54:2747–2749. [PubMed] [Google Scholar]
- 29.Hwang HY, Yim J. SolI, a novel isoschizomer of BamHI isolated from Streptoverticillium olivoverticillatum. Nucleic Acids Res. 1994;22:2197. doi: 10.1093/nar/22.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grones J, Turna J. ApaCI, an isoschizomer of BamHI isolated from Acetobacter pasteurianus. Nucleic Acids Res. 1992;20:3513. doi: 10.1093/nar/20.13.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catterall JF, Welker NE. Isolation and properties of a thermostable restriction endonuclease (ENDO R-Bst1503) J. Bacteriol. 1977;129:1110–1120. doi: 10.1128/jb.129.2.1110-1120.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts RJ. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1985;13(Suppl.):r165–r200. doi: 10.1093/nar/13.suppl.r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King DA, Zhang L, Guarente L, Marmorstein R. Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein. Nat. Struct. Biol. 1999;6:64–71. doi: 10.1038/4940. [DOI] [PubMed] [Google Scholar]
- 34.Xu QS, Kucera RB, Roberts RJ, Guo HC. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Xu QS, Roberts RJ, Guo HC. Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure. Protein Sci. 2005;14:2590–2600. doi: 10.1110/ps.051565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]