Abstract
The complex of the yeast Lsm1p-7p proteins with Pat1p is an important mRNA decay factor that is involved in translational shutdown of deadenylated mRNAs and thus prepares these mRNAs for degradation. While the Lsm proteins are highly conserved, there is no unique mammalian homolog of Pat1p. To identify proteins that interact with human LSm1, we developed a novel immunoprecipitation technique that yields virtually pure immunocomplexes. Mass-spec analysis therefore identifies mostly true positives, avoiding tedious functional screening. The method unambiguously identified the Pat1p homolog in HeLa cells, Pat1b. When targeted to a reporter mRNA, Pat1b represses gene expression by inducing deadenylation of the mRNAs. This demonstrates that Pat1b, unlike yPat1p, acts as an mRNA-specific deadenylation factor, highlighting the emerging importance of deadenylation in the mRNA regulation of higher eukaryotes.
INTRODUCTION
mRNA degradation is a crucial part of the regulation of gene expression, as it limits the amount of protein that can be produced from a given mRNA. The process has been elucidated to considerable detail in yeast. In short, mRNA degradation starts with deadenylation which is either followed by exonucleolytic digestion from the 3′-end of the mRNA or, more important for gene regulation, causes decapping of the transcript with subsequent exonucleolytic digestion from the 5′-end. Decapping is catalyzed by Dcp1p/Dcp2p, which requires a series of auxiliary factors for full activity, such as Dhh1p, Pat1p and the LSm proteins Lsm1p to LSm7p. These factors bind to the mRNA prior to decapping. Pat1p and the LSm proteins form a complex (1,2) that also co-purifies with the 5′-exonuclease Xrn1p (1), even though this enzyme becomes active only after decapping. Both Pat1p and the LSm proteins bind to the target mRNA. The LSm proteins are characterized by the presence of an oligo(U)-specific RNA-binding domain (3), and Pat1p, even though it does not have a recognizable RNA-binding domain (nor any other identifiable domain), also binds to RNA homopolymers with a preference for poly(U) (4). In vitro, the complex preferably binds to mRNAs with a short poly(A) tail (5) suggesting that the complex enters mRNPs after deadenylation. Indeed, the LSm1p to 7p proteins associate preferentially with deadenylated mRNAs in vivo (6). Importantly, Pat1p acts as a translational repressor (7). mRNA translation and degradation via the decapping pathway generally compete with each other [(7), but see also (8)]. Since the Lsm/Pat1p complex does not interact with the decapping enzyme, it is assumed that the Lsm proteins and Pat1p prepare the deadenylated mRNAs for degradation by shifting the equilibrium towards translation arrest and thus facilitating the decapping reaction.
Less is known about mRNA degradation in higher eukaryotes, but the key players are well conserved (9), and like in yeast, metazoan decay pathways start with deadenylation and proceed with decapping and 5′→3′ digestion (10–14). mRNA degradation in higher eukaryotes thus follows the same basic mechanism outlined above.
All components of the decapping-degradation pathway—auxiliary factors, decapping enzyme and 5′-exonuclease—co-localize in cytoplasmic foci called P bodies (PBs), where mRNAs can actually be degraded in yeast (15) and mammalian cells (16,17). In addition, PBs contain the machineries for other mRNA silencing pathways such as non-sense-mediated decay (18,19) and miRNA-mediated silencing (20–22). PBs can also serve as a storage point for silenced mRNAs, which can resume translation after leaving the bodies (23). The generally accepted function of the PBs is to enforce translational silencing by sequestering the mRNPs from the pool of soluble ribosomes/translation factors; the forces that keep the PBs together are less well understood. The current model predicts that translationally silent mRNPs have the propensity to coalesce into the P bodies because they are decorated with protein domains that aggregate via homomeric interactions (24). LSm4p and the enhancer of decapping Edc3p serve this role (25–27), and the responsible domain in Lsm4p is the Gln/Asn-rich carboxy terminus (26,27). A similar domain is found in various other PB factors including Pat1p (27).
While many of the mRNA-decay factors—in particular, the Lsm proteins—are well conserved throughout evolution, the functional homolog of Pat1p in mammals has not been characterized. Two proteins have been proposed based on a weak homology in the C-terminal half (four regions with a total of 13 invariant amino acids) (7,28). Using a novel immunoprecipitation technique, we now unambiguously identify one of the two as the functional Pat1p homolog in HeLa cells. The protein strongly interacts with LSm1, and localizes to PB-like structures even in the absence of functional PBs, presumably due to a Gln/Asn-rich domain. But here, the similarities with yeast Pat1p end: instead of being a general mRNA decay factor, it affects specific mRNAs, and instead of functioning on deadenylated mRNAs, it induces deadenylation. Human Pat1b thus is a novel factor that regulates the expression of specific mRNAs by modulating the length of their poly(A) tails.
MATERIAL AND METHODS
Antibodies
Anti-LSm1 (29), anti-SF3a120 and anti-SF3b155 (30) were polyclonal antibodies raised against peptides derived from the respective protein sequences. They were affinity-purified on a Sulfolink column (Pierce) containing the respective immobilized peptide. The Y12 monoclonal antibody (31) was used directly as a hybridoma supernatant. Polyclonal antibodies directed against DDX6 were from Bethyl Laboratories (TX, USA). For the description of the Pat1b antibodies, see Supplementary data.
The adaptor protein and the double purification procedure
A fragment encoding a calmodulin-binding peptide (CaM-BP) and a double copy of the IgG-binding domain (IGB) was amplified from plasmid pBS1479 (32) and fused with the Strep tag (DWSHPQFEK) at its N-terminus. The resulting fragment was cloned into the NcoI/HindIII sites of the pET-M11 vector (EMBL Heidelberg), further adding a His6 tag. The plasmid sequence was verified by DNA sequencing. The plasmid was introduced into BL21/pLysS cells, and synthesis of the adaptor protein was induced by addition of 0.4 mM IPTG and incubation for 4 h at 37°C. The cells were lysed and the protein was purified by nickel chromatography according to standard procedures.
Of calmodulin-coated beads, 0.2 ml were incubated for 30 min at 4°C with 20 µg of adaptor protein in 2 ml of CaM-binding buffer (10 mM Tris–HCl pH 8.0; 150 mM NaCl, 1 mM MgCl2, 1 mM imidazole, 2 mM CaCl2, 0.1% Triton, 10 mM β-mercaptoethanol). The beads were washed three times with 1 ml of CaM-binding buffer, and then antibodies were added in 2 ml of CaM-binding buffer. The beads were washed as before. For each precipitation, 1 ml of nuclear extract (∼5 mg of total protein) or 4 ml of cytoplasmic extract (∼14 mg of total protein) was diluted to 10 ml with CaM-binding buffer and cleared by centrifugation at 10 000g for 15 min. The extract was added to the beads and incubated for 1 h at 4°C. The beads were washed three times with 5 ml CaM-binding buffer and then eluted with CaM-elution buffer (10 Tris–HCl pH 8.0; 150 mM NaCl, 1 mM MgCl2, 1 mM Imidazole, 20 mM EGTA, 0.1% Triton, 10 mM β-mercaptoethanol) in five 0.2-ml fractions. Fractions 2–4, containing most of the eluted protein, were pooled, diluted to 2 ml with Strep-binding buffer (20 mM Tris–HCl pH 8.0; 150 mM NaCl, 1.5 mM MgCl2, 0.1% Triton) and applied to 0.2 ml of streptactin-coated beads (Qiagen). The suspensions were incubated for 1 h at 4°C, and then the beads were washed three times with Strep-binding buffer. The purified complexes were eluted with Strep-elution buffer (20 mM Tris–HCl pH 8.0; 150 mM NaCl, 1.5 mM MgCl2, 0.1% Triton, 2 mM desthiobiotin) in five 0.2-ml fractions. Proteins were recovered by acetone precipitation, fractionated by SDS–PAGE and visualized by SYPRO Ruby staining.
Mass spectrometry
Visible bands from silver stained gels were cut out and proteins were digested in-gel with trypsin as described (33). Extracted peptides were sequenced by LC-coupled tandem MS on a Q-ToF (Ultima, Waters) and product ion spectra were searched for protein identification against NCBInr database by using MASCOT as search engine (described in detail at ref. 34).
Immunoprecipitation
Nearly confluent 100-mm dishes of HeLa SS6 cells were rinsed with PBS, scraped into 1 ml of lysis buffer (20 mM Tris–Cl pH 8.0, 300 mM NaCl, 0.2% Triton X-100, 0.5 mM DTT, 0.2 mM PMSF), and centrifuged for 15 min at 13000g at 4°C. Of supernatants, 300 µl were added to proteinA beads containing the respective antibodies and blocked in 1% BSA in PBS. The beads were washed with IP buffer (20 mM Tris–Cl pH 8.0, 300 mM NaCl, 0.1% Triton X-100), and bound proteins were eluted by boiling with SDS-PAGE loading buffer.
GST pull-downs
LSm1 or the mutant lacking the last 33 amino acids were cloned into the NcoI and KpnI sites of the pET M30 vector (EMBL protein expression group). Pat1b fragments from amino acid 381 or 561, respectively, to the C-terminus were cloned into the NcoI and KpnI sites of the pET M11 vector. All peptides were expressed in BL21 by induction with 0.4 mM IPTG at 37°C. The Pat1b peptides were purified by Ni affinity chromatography. LSm1 proteins were immobilized from the crude bacterial lysate on 10 µl glutathione beads (GE healthcare). The beads were washed with precipitation buffer (20 mM Tris–HCl pH 8; 50 mM NaCl; 0.05% NP-40), incubated with Pat1b peptides for 30 min at 4°C, washed again with the same buffer, and eluted by boiling with SDS buffer. Eluted proteins were fractionated by SDS–PAGE and visualized by Coomassie Blue staining.
Cell transfection and immunofluorescence
The entire Pat1b ORF (according to NCBI genebank entry AL831992) was cloned into the XhoI and SacII sites of pEGFP-C1 (Clonetech). COS-7 cells were transfected with the plasmid diluted 1:4 with an empty pBluescript vector, fixed 16 h after transfection, and immunostained as indicated, using Cy3-conjugated anti-rabbit IgG secondary antibodies (GE Healthcare). Pictures were taken with a 60×/1.40 oil immersion objective on a Nikon confocal microscope. For statistical analysis, wide-field micrographs were taken with a 60×/1.35 objective (Olympus), and P bodies were automatically counted using the ‘analyze particles’ algorithm of the ImageJ package (National Institutes of Health; particles with an area of 0.1–1.0 µm2 were considered).
For Pat1b knock-downs, 5 × 106 HeLa SS6 cells in 250 µl Mirus Ingenio® solution were electroporated at 260 V/960 µF with 200 pmol siRNA against Pat1b or with the same amount of control siRNAs directed against the GL2 luciferase. The Pat1b siRNAs were a mixture of three ‘DsiRNA’ (IDT) targeted against positions 2284–2308, 2781–2804 and 2875–2899, respectively (numeration according to National Institutes of Health genebank entry NM_152716). Cells were distributed on three 10-cm dishes, and cultured for 72 h. Cells were harvested with a cell scraper; From a part, the RNA was isolated (see below), from another aliquot, the proteins were extracted by lysis in (20 mM Tris–Cl pH 8, 500 mM NaCl, 0.5% NP-40, 0.2 mM PMSF) and analyzed by western blotting.
MS2 tethering and luciferase assays
The MS2 RNA-binding module was inserted between the GFP and the Pat1b part of the plasmid described earlier. pcLuc MS2x9 was constructed by sub-cloning a fragment of pT7-Luc MS2x9, which contains the luciferase ORF and downstream nine MS2-binding sites (35) into the BglII and EcoRI sites of pcDNA 3 (Invitrogen). The DNA constructs have all been sequenced before usage. COS-7 cells were transfected with a mixture of pcLuc MS2x9, a renilla luciferase control (pRL-tk, Promega), and the GFP-Pat1b plasmid with or without the MS2 module. Cells were harvested 24 h after transfection, and luciferase activity was determined using the appropriate kit (Promega) as detailed by the manufacturer.
Isolation and analysis of RNA
Total RNA was isolated from the equivalent of a 60-mm dish of HeLa or COS-7 cells using the RNeasy kit (Qiagen). If the cells were transfected with luciferase plasmids, the RNA was digested with RQ1 DNase (Promega) and repurified by LiCl precipitation. Plasmid contaminations thus contributed <5% to the total signal in qPCR, as gauged by the control omitting reverse transcriptase.
qPCRs were performed after reverse transcription with M-MLV reverse transcriptase (Invitrogen) using the SYBR Green kit and the LC480 LightCycler (both Roche). The amplification efficiency (AE) of each amplicon was deduced from serial dilution experiments. The RNA quantities relative to the respective control were calculated from the crossing points Cp and the predetermined AE: [RNA] ∼ AE−ΔCp, with ΔCp = Cpunknown − Cpcontrol.
The poly(A)-length test (PAT) was performed as described (36). Briefly, a tag was added to the mRNAs (1 µg of total RNA) by annealing them to the DNA oligo (GCTTCAGATCAAGGTGACCTTTTTTTTTTTT, blocked at its 3′-end by aminohexyl) and extending their 3′-ends using Klenow enyme. The tagged RNAs were retrotranscribed at 48°C using superscript III reverse transcriptase (Invitrogen) and GCTTCAGATCAAGGTGACCTTTT as a primer. The 3′-end was then amplified using a gene-specific primer and the reverse transcription primer. To generate deadenylated controls, 10 µg total RNA were digested with 1.5 µg oligo(dT)15 and 1.25 U RNaseH (Promega). The RNA was re-isolated and quantified prior to the PAT assay. For the graphical representation of the poly(A) length distribution, each lane profile was digitalized. The DNA length corresponding to each pixel was estimated by an exponential function, which was calculated from the DNA markers on the same gel using the Imagequant TL software (GE healthcare). The crude intensities were divided by the DNA length to normalize for the effect that ethidium-bromide staining is relative not to the number of DNA molecules, but to the mass of the DNA. The highest value in each lane was then arbitrarily set to 100, and plotted against the poly(A) length, as deduced from the estimated DNA length and the length of the minimal PCR product.
RESULTS
A novel immunoaffinity purification strategy identifies the human homolog of Pat1p
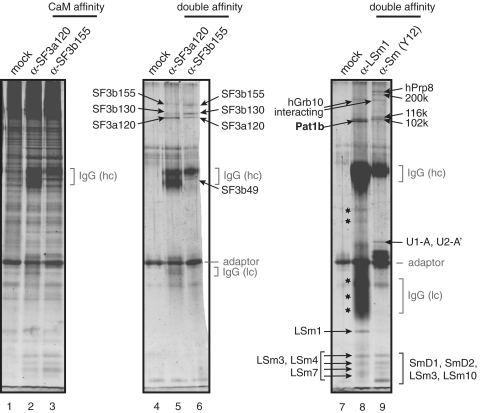
Immunoprecipitations are routinely plagued by significant background precipitation that leads to high numbers of false-positive identifications in mass spectrometry (MS) (see, for example, the gel in Supplementary Figure S1). To avoid the problem, we devised a double-affinity strategy to purify the immunocomplexes (Figure 1). In brief, the antibodies are immobilized on calmodulin beads via an artificial adaptor. The antibodies and bound antigen complexes can thus be recovered under native conditions, and are re-purified using the strep tag that is also present in the adaptor. The strategy was validated by purification of previously characterized RNP complexes from HeLa nuclear extracts. First, we purified the 17S U2 snRNP, which is part of the spliceosome, targeting the SF3a120 and SF3b155 subunits of the complex (30). The intermediate step after EGTA elution from the calmodulin beads does not sufficiently enrich the SF3 complexes. In fact, many proteins are also observed in the mock purification that was performed in the presence of the adaptor, but without antibodies, and only the IgG heavy chains are visibly enriched (Figure 2, compare lanes 2 and 3 with lane 1). The second round of purification, in contrast, yielded a very clear pattern with only few visible bands in the mock control (lane 4). Bands not detected in the control were thus attributed to specific binding; these were cut out, and the proteins present in the bands were identified by MS. The main protein component (as judged by the number of peptides identified, see Supplementary Table S1) of each band is indicated in Figure 2. Each band corresponds to a known component of the 17S U2 snRNP. Few additional proteins were identified in the bands, and they are listed in Supplementary Table S1. Most of them are also part of the SF3 complexes, and only the SF3a120 band in lane 6 contains two proteins that were not previously described as components of the 17S U2 snRNP and that therefore may represent contaminations. As judged by the number of identified peptides, these presumptive contaminations can be considered to be minor. The method thus is well suited to unambiguously identify proteins that are co-purified with the α-SF3b120 and α-SF3b155 antibodies and to distinguish these from residual contamination.
Figure 1.
Outline of the tandem-affinity-immunopurification strategy. An artificial adaptor protein combines an IGB with a calmodulin-binding peptide (C) and a strep tag (S). The adaptor is used to immobilize antibodies on calmodulin beads, which are then incubated with extracts containing the antigen. Immunocomplexes are recovered by native elution with EGTA and are re-purified on Streptactin beads.
Figure 2.
Results of the tandem-affinity immunopurification. The procedure was carried out using α-SF3a120 (lane 2), α-SF3b155 (lane 3) or no antibodies (lane 1). Proteins were recovered by acetone precipitation from 10% of the calmodulin eluates, fractionated by SDS–PAGE and stained with SYPRO Ruby. The remainder was further purified on streptactin beads, eluted, and stained in a similar way (lanes 4–6). Bands that were excised for MS are indicated by arrows showing the major protein component found in the respective band. Double purification of the LSm1 and Sm complexes from HeLa cytoplasmic extracts is shown in lanes 7–9. As before, the protein composition of the final eluate is shown for the precipitation with the α-LSm1 polyclonal antibody (lane 8), the Y12 α-Sm monoclonal antibody (lane 9) or no antibodies (lane 7). Regions in lane 8 marked by asterisks were found to contain only IgGs or fragments thereof.
Having validated the method, it was next used to characterize the cytoplasmic Sm and like-Sm (LSm) complexes. The seven Sm proteins form the core of the spliceosomal snRNPs; they assemble on the snRNAs in the cytoplasm and are then imported into the nucleus. We used a monoclonal antibody, Y12, that preferentially recognizes native assembled Sm proteins (37) to purify the Sm complexes from HeLa cytoplasmic extracts, with the aim to gain further insights on the cytoplasmic phase of the Sm complex. The result of the purification is shown in Figure 2, lane 9: several proteins specifically co-precipitated with the Sm proteins, but not the control (lane 7). All proteins belong to the known nuclear snRNP complexes U1 (U1-A), U2 (U2-A′) and U5 (220, 200, 116 and 102K). Again, the preparation was very clean, as only a few additional proteins were identified underneath the main components of the bands (Supplementary Table S1). Most can be explained by the presence of nuclear Sm snRNPs U1 (U1-70K), U2 (SF3b14) and U4/U6 (LSm3 and hPrp24). LSm10 is part of the Sm complex found on U7 snRNA, which, although divergent from the canonical Sm core snRNP, nevertheless binds to anti-Sm type antibodies (38). In summary, the major proteins and most of the minor proteins identified in each band are known components of the nuclear snRNPs, and we conclude that we have indeed purified nuclear snRNPs from the cytoplasmic extract, probably due to the presence of mitotic cells that lack a nuclear compartment, and/or owed to leaking of the nuclei during the extraction procedure. Nevertheless, the detection of a nuclear contamination in the cytoplasm demonstrates the good yield that can be obtained with this technique. The high purity is again underlined by the fact that the major protein in each band is a known snRNP protein; the only exception is the Grb10-interacting protein. Since this protein is also co-purified with LSm1, it may be a contaminant that generally co-purifies due to the particular steps of this protocol (‘Discussion’ section).
Finally, the Sm-like protein LSm1 was purified from HeLa cytoplasmic extracts. So far, LSm1 is known only to bind to the LSm proteins LSm 2–7, and its yeast counterpart binds to the degradation factor Pat1p, the functional mammalian homolog of which remains to be identified. In this case, a lot of antibody was used, and IgGs or fragments thereof overshadow parts of the gel (‘Discussion’ section). Several gel slices (indicated by asterisks in Figure 2, lane 8) therefore yielded only IgG sequences. Several bands in the lower part of the gel were identified as the antigen LSm1 and other members of the LSm family, confirming that they form stable complexes with LSm1. Two bands are specifically enriched in the upper part of the gel: one corresponds to the Grb10-interacting protein (see above), while the other identified one of the two Pat1p homologs present in the human database. In the following, we call this protein Pat1b—in Xenopus, the other homolog is expressed first during development and therefore gets the letter ‘a’ (39).
Characterization of Pat1b
To verify the LSm1-Pat1b interaction, anti-Pat1b antibodies were generated (described in Supplementary Figure S2) to co-precipitate LSm1 from COS-7 cell lysates. As shown in Figure 3A, both the anti-LSm1 and the new anti-Pat1b antibodies efficiently precipitate their own antigen (lane 3, top panel, and lane 4, bottom). In addition, LSm1 efficiently co-precipitates with the anti-Pat1b antibodies, but not with mock IgGs (lanes 3 and 2, respectively). Likewise, Pat1b co-precipitates with anti-LSm1 antibodies (lanes 4 and 2). In order to determine if the interaction depends on the formation of mRNPs or P bodies, the immunoprecipitations were repeated in the presence of RNase (lanes 7 and 8), or from extracts of cells that were treated with cyclohexamide, which strongly reduces the number of P bodies (see Figure 4, and refs 16 and 17). Neither of the two treatments affects the efficiency of LSm1–Pat1b co-precipitation (lanes 7, 8 and 11, 12). To verify if the interaction occurs via direct binding of the two proteins, we reconstituted the complex from recombinant proteins expressed in Escherichia coli. Full-length Pat1b proved impossible to express in a soluble form. We therefore expressed two fragments that contain the central, highly conserved domain and the C terminus, but has the putative aggregation domain in the N terminus deleted (see below), or only the C terminus. Binding of these fragments was tested to LSm1 full length immobilized to glutathione beads, or to the naked LSm1 Sm domain (deletion of 33 amino acids from the C terminus). Pat1b ΔN binds to both LSm1 peptides, but not an unrelated protein (compare Figure 3B, lanes 3 and 4 with 2; the weaker precipitation with full-length LSm1 should not be overvalued, since there was less GST fusion protein immobilized in this assay). This demonstrates that Pat1b directly binds to LSm1, forming contacts predominantly with the Sm domain. The C terminus of Pat1b is also sufficient for this interaction, albeit to a lesser extent when compared to the input (Figure 3B, lanes 5, 7 and 8). We conclude that Pat1b and LSm1 bind to each other via protein–protein interactions that are independent of P-body formation.
Figure 3.
LSm1 co-precipitates with Pat1b independently of RNA or PBs. (A) COS-7 cell lysates were immunoprecipitated with mock IgGs (lanes 2, 6, 10), with anti-Pat1b antibodies (lanes 3, 7, 11) or with anti-LSm1 antibodies (lanes 4, 8, 12). Co-precipitating proteins were fractionated by SDS–PAGE and blotted. The upper part of the blot was immunostained for Pat1b, the lower part for LSm1. Of the respective input, 10% was loaded in lanes 1, 5 and 9. Lanes 1–4 show the experiment performed with untreated lysates. In lanes 5–8, 20 µg/ml RNase A and 50 U/ml RNase T1 was added to the cell lysate before immunoprecipitation, and in lanes 9–12, cells were treated with 25 µg/ml cycloheximide for 30 min at 37°C prior to lysis. The migration of molecular weight markers is indicated on the left, and the position of the proteins of interest on the right. (B) GST-DCoH (an unrelated protein encoded by the original pET M30 plasmid; lanes 2 and 6), GST-LSm1 ΔC (lanes 3 and 7) as well as GST-LSm1 (lanes 4 and 8) was immobilized to glutathione beads, and incubated with Pat1b ΔN (2–4), or with the Pat1b C-terminal domain (6–8). Proteins bound to the beads were eluted and fractionated by SDS–PAGE. Shown is the Coomassie-stained gel; lanes 1 and 5 show the input Pat1b fragments.
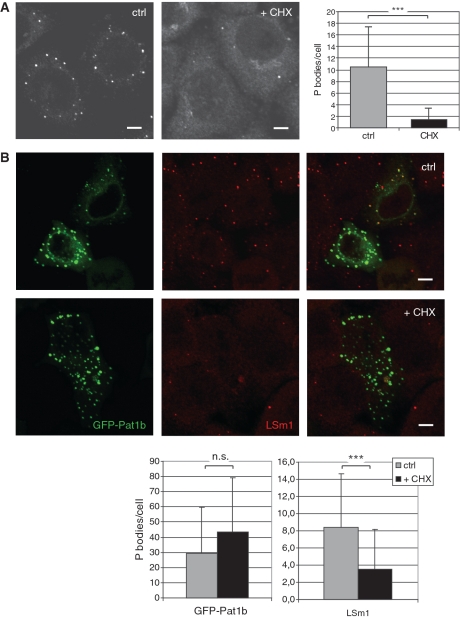
Figure 4.
Pat1b localization in PBs. (A) Endogenous hPat1 was revealed by immunostaining COS-7 with anti-hPat1 antibodies. The left micrograph shows a representative confocal slice of control cells, the right one of cells that were treated with 25 µg/ml cycloheximide (CHX) for 30 min prior to fixation. The decrease in observable cytoplasmic foci (from 10.5 to 1.5, counted on 85 and 66 cells, respectively) is highly significant (***P < 0.0001, Student’s t-test). Scale bars, 5 µm. (B) COS-7 cells were transfected with GFP-Pat1b (green in the panels on the left) and immunostained for LSm1 (red in the middle panels). The upper row shows a confocal image of control cells, the lower row of cells treated with CHX. Scale bars, 5 µm. The statistical analysis is shown at the bottom: the count of GFP-Pat1b foci does not significantly change (average of 29.5 in 44 control cells versus 43.6 in 25 CHX-treated cells; P = 0.085 in Student’s t-test), while the count of LSm1 foci decreases from 8.4 to 3.5 (in 84 control and 65 treated cells; ***P < 0.0001). Note that the count of LSm1-positive foci also decreases in the GFP-Pat1b-transfected cells (P < 0.0002; data not shown).
As an LSm1-interacting protein, Pat1b is expected to localize to the P bodies (PBs, 29). In fact, COS-7 cells all exhibited bright cytoplasmic foci in an anti-Pat1b immunofluorescence (Figure 4A). As observed for other P body markers (16,17), arrest of protein translation by cycloheximide greatly reduces the number of Pat1b-positive P bodies (from an average of 10.5 per cell to 1.5, Figure 4A), indicating that these foci are bona fide P bodies. To visualize LSm1-Pat1b co-localization, a GFP-Pat1b fusion protein was expressed in COS-7 cells and counter stained with anti-LSm1 antibodies. Interestingly, GFP-Pat1b forms many more foci than endogenous Pat1b or LSm1 (an average of 29.5 versus 10.5 and 8.4, respectively: Figure 4B, upper row). Endogenous LSm1 is found in GFP-Pat1b-positive PBs (which thus appear yellow in the merged picture, see in particular the cell on the upper right), but the excess of GFP-Pat1b PBs does not contain detectable levels of endogenous LSm1 and thus appears green in the overlay image, indicating that most GFP-Pat1b foci do not represent bona fide PBs. This notion is supported by the finding that cycloheximide treatment disperses the LSm1, but not the GFP-Pat1b foci (see Figure 4B, lower row, and the statistical evaluation at the bottom). Very much the same picture is observed when counterstaining GFP-Pat1b-transfected cells with the P-body marker Ddx6/RCK (Supplementary Figure S3). We therefore conclude that Pat1b can form P body-like foci in the cytoplasm that are independent of the presence of natural P bodies, as they do not dissolve with cycloheximide, and do not contain endogenous P body markers. These aggregates were observed despite the fact that we limited expression of GFP-Pat1b by diluting the plasmid (‘Materials and Methods’ section). It is probably no coincidence that Pat1b can form PB-like structures, as the protein contains a Q/N-rich putative aggregation domain in its N-terminal half (data not shown; see also ‘Discussion’ section).
Pat1b represses translation by inducing deadenylation and degradation of the target mRNA
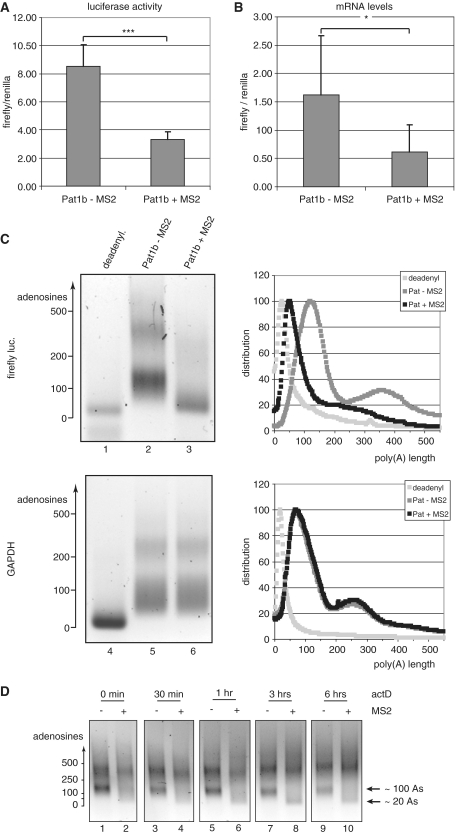
To elucidate the function of Pat1b, we used the MS2 protein to tether Pat1b to a firefly luciferase reporter that contains nine MS2-binding sites in its 3′-UTR. COS-7 cells were transfected with a mixture of the reporter plasmid, a Renilla luciferase cDNA that lacks MS2-binding sites, and the Pat1b constructs with or without the MS2 fusion. After 24 h, the cells were lysed and the activity of the firefly luciferase relative to the Renilla enzyme was determined. As shown in Figure 5A, targeting of Pat1b drastically lowered expression of the firefly luciferase with respect to the Pat1b construct lacking the MS2 domain (down by 61%: P < 0.001; n = 14). To determine whether this is due to a destabilization of the mRNA or to translational arrest, we measured the mRNA levels by quantitative PCR (qPCR). The reduced expression can be entirely explained by a reduced abundance of the firefly luciferase mRNA relative to its Renilla counterpart (Figure 5B—the amount of mRNA is reduced by 62%: P < 0.05; n = 6). This strongly suggests that the stability of the targeted mRNA is compromized. Usually, the first step in mRNA decay is the shortening of the poly(A) tail. The lengths of the poly(A) tails were measured by the tag-addition method (36). Figure 5C shows the poly(A) length distribution of the firefly luciferase reporter and, as a control, of the endogenous GAPDH mRNA. The trace of the intensities in each lane is plotted on the right of the gel images (for their calculation, see ‘Materials and Methods’ section). Both the reporter and the control mRNA show two broad peaks at ∼100 and 300–400 adenosines (as judged from the migration of DNA markers; lanes 2 and 5), and both populations reflect differentially polyadenylated mRNAs, as artificial deadenylation condenses both peaks into one single, sharp band migrating at the expected height. When cells are transfected with the targeted MS2-Pat1b construct, the firefly luciferase mRNA, but not the GAPDH control, is severely deadenylated: the peak at 350 adenosines becomes less intense and shifts to shorter lengths, and the main peak is shifted to <50 adenosines, suggesting that Pat1b binding primarily induces deadenylation.
Figure 5.
Pat1b down-regulates the expression of target genes by inducing deadenylation and destabilization of the mRNAs. COS-7 cells were co-transfected with a firefly luciferase construct containing MS2-binding sites, Renilla luciferase control and either GFP-Pat1b or GFP-MS2-Pat1b. (A) The ratio of firefly to Renilla luciferases in arbitrary units, with the error bar indicating the standard deviation. ***P < 0.001 in Student’s t-test. (B) The abundance of the firefly mRNA relative to the Renilla reporter, as determined by qPCR, with the error bar indicating the standard deviation. *P < 0.05 in Student’s t-test. The left panels of (C) show the poly(A) tests of the firefly luciferase (lanes 1–3) and of the endogenous GAPDH mRNA (lanes 4–6). The sample analyzed in lanes 2 and 5 stemmed from cells transfected with GFP-Pat1b, in lanes 3 and 6 from cells expressing GFP-MS2-Pat1b. In lanes 1 and 4, the RNA was artificially deadenylated by RNaseH digestion in the presence of oligo(dT). The length of the poly(A) tail, as estimated from the migration of DNA markers, is indicated on the left panels. The profiles of lanes 1, 4 are shown in light gray, lanes 2, 5 in dark gray, and 3, 6 in black. The ordinate axes report arbitrary units of intensity, which was normalized for fragment length in order to reflect molar distribution. (D) Time course of the poly(A) decay. Transfected cells were treated with actinomycin D (5 µg/ml) for the indicated times, total mRNA was isolated, and the poly(A) length of the firefly luciferase mRNAs was determined as in 5C. The mRNA populations containing tails of ∼20 and 100 adenosines, respectively, are indicated on the right.
To verify this hypothesis, we performed a deadenylation time course (Figure 5D). Synthesis of new mRNAs was shut down by actinomycin D, and the poly(A) length distribution was monitored over 6 h (Figure 5D). For the non-targeted reporter construct (odd lanes), the distribution does not change much: only the population of mRNAs with poly(A) tails of ∼100 nt decreases relative to the population with longer poly(A) tails, indicating that the mRNAs with long poly(A) tails decay slower. In contrast, the population with ∼100 A residues is much less prominent for the reporter mRNA targeted by MS2 already at the steady state level (Figure 5D, lane 2; see also Figure 5C). During the time course, this population further decreases and another population with critically short poly(A) tails (around 20 nt) builds up. This shows that upon Pat1b tethering, a step downstream of deadenylation becomes rate limiting and therefore demonstrates that Pat1b primarily induces a shortening of the poly(A) tail, which then leads to the destabilization of the mRNA. No obvious changes are observed in the mRNA population with the long poly(A) tails, suggesting that Pat1b targets primarily mRNAs with tails of ∼100 As and/or that mRNAs with longer tails are protected, e.g. by active translation.
Knock-down of Pat1b increases the poly(A) tail of specific mRNAs
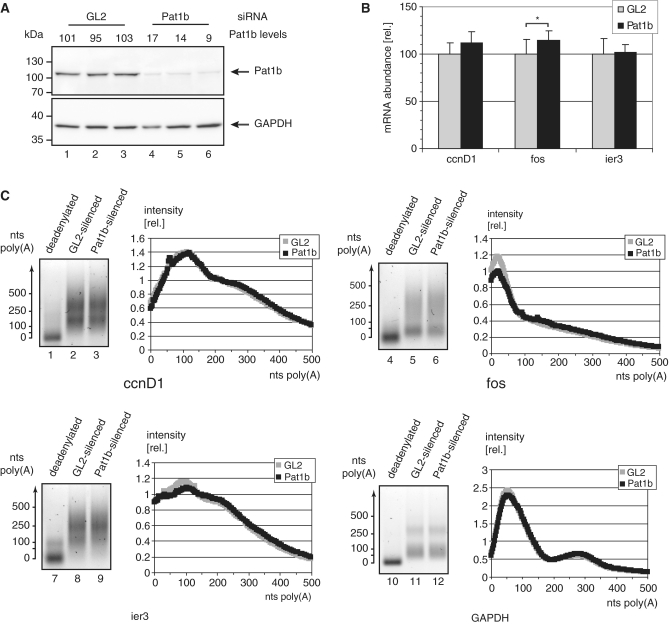
To see if Pat1b has a similar function under physiological conditions, the protein was depleted by siRNA transfection to <20% of the mock-treated levels (Figure 6A). Three unstable mRNAs were selected, namely cyclin D1 (ccnD1), c-fos (fos) and the immediate-early response mRNA ier3. All three contain characterized AU-rich elements (AREs) in their 3′-UTRs. The levels of these mRNAs were determined by qPCR. Comparing mock-treated and Pat1b-depleted cells, the ccnD1 mRNA tends to be more stable and therefore more abundant, but the trend is not statistically significant. fos mRNA levels increase by 15%, and this is significant (P < 0.05; n = 9). The amount of the immediate-early response mRNA ier3, instead, stays perfectly the same (Figure 6B). Pat1b depletion thus affects some but not all unstable mRNAs. If Pat1b indeed induces deadenylation, then depletion of the protein should have parallel effects on deadenylation and mRNA abundance. In fact, the poly(A) length distributions for the mRNAs that do not change in quantity nicely superimpose between the mock-treated and the Pat1b-depleted samples (Figure 6C, see the GAPDH—used for normalization in 6B—and the ier3 and ccnD1 mRNA panels). Only the fos mRNA behaves differently in that a population of this mRNA has critically short poly(A) tails already in the mock-treated sample (Peak at 25 adenosines, upper-right panel in Figure 6C). This population decreases by ∼20% when Pat1b is depleted. In summary, Pat1b depletion affects the poly(A) tail length of only one of the instable mRNAs tested here. That we did not find a more striking effect may be due to the residual Pat1b left after siRNA depletion (Figure 6A). Alternatively, Pat1b could regulate only a subset of mRNAs under specific physiological or developmental conditions (‘Discussion’ section).
Figure 6.
Pat1b stabilizes the poly(A) tail of select mRNAs. HeLa cells were electroporated in the presence of siRNAs directed against the firefly luciferase (GL2) or against Pat1b. After 72 h, both proteins and RNA were extracted. (A) Total proteins were separated by SDS–PAGE, transferred onto a membrane and stained with antibodies against Pat1b (top) or against GAPDH (bottom). The band intensities were quantified, and Pat1b was normalized first against GAPDH, and then against the average of Pat1b in the GL2-treated extracts. The resulting Pat1b levels are indicated above each lane. The migration of marker proteins is indicated to the right. (B) The total RNAs were reverse-transcribed and the levels of the ccnD1, fos, and ier3 mRNAs were quantified by qPCR. Shown are the relative quantities after normalization for GAPDH mRNA and the average of the respective mRNA levels in the GL2-treated samples. *P < 0.05 in Student’s t-test (n = 9). (C) Poly(A) lengths of the mRNAs encoding ccnD1 (lanes 1 to 3), fos (4–6), ier3 (7–9) and GAPDH (10–12) were determined. Lanes 2, 5, 8 and 11 show the PAT reaction for the GL2-treated sample, lanes 3, 6, 9, 12 for the Pat1b-depleted sample. Lanes 1, 4, 7 and 10 also show the Pat1b-depleted mRNA, but the RNAs were deadenylated prior to the PAT reaction. To the right of each block, the intensity of the GL2 and Pat1b-transfected sample is displayed. The values represent arbitrary units, normalized for the total signal of each lane.
DISCUSSION
In the present study, we have identified the human ortholog of the yeast auxiliary decapping factor Pat1p by virtue of its interaction with LSm1, utilizing a novel immunoaffinity method. Immunopurification is a straightforward approach to the isolation of macromolecular complexes, as immobilized antibodies recognize their target with high specificity and binding strength. One of the shortcomings of immunoprecipitation is the high level of non-specifically bound proteins. Mass-spec analysis of the precipitate therefore routinely identifies many false positives, and the strategy requires extensive screening of the identified proteins, e.g. with a functional assay. To overcome this problem, quantitative MS has been used to measure which proteins are enriched relative to a control precipitation (SILAC) (40). This approach is cost-intensive and limited to experimental setups that permit isotope labelling. Moreover, without a highly accurate MS instrument, data evaluation is rather time consuming. Therefore, it is desirable to perform mass-spec analysis on highly pure complexes in order to exclude false positives in the first place. To this end, tagging strategies have been developed, such as the FLAG (41), or the ‘tandem affinity purification’ tag (32). The latter is particularly suited, as the complexes are purified by two subsequent steps, eliminating most of the background. Every tagging method, however, has several disadvantages. The tag may interfere with the function and/or interactions of the target protein. Even when the tag does not block the function, the untagged protein is often preferentially incorporated into native complexes, so that the endogenous copy of the gene must be replaced or inactivated (42). Finally, the tagged gene has to be introduced into, and expressed in, cells of the parent organism, with all the attendant practical difficulties. This can be achieved in cell culture by viral transfection (43), but is all but impossible in a mammalian organism yet.
We have combined the advantages of immunopurification and TAP tag affinity purification by moving the tag from the target protein to the antibody (Figure 1). The strategy was validated with antibodies directed against members of the spliceosomal SF3 complex (30). We placed emphasis on the identification of genuine complex components with the highest possible confidence; no effort was undertaken to obtain a complete list of the proteins that bind to our target proteins. Indeed, the purification of the SF3a120 and SF3b155 proteins led almost exclusively to the identification of components of the SF3a and SF3b splice factors. Only one band contained, in addition to SF3a120, minor quantities of proteins not related to SF3 (Supplementary Table S1). Thus, the two antigen proteins were correctly and unambiguously identified as parts of the 17S U2-splicing complex. This particle, however, contains additional protein components that we did not identify here that might have been, for example, hidden under the IgG bands. Also, we did not identify all LSm proteins known to be associated with LSm1. Sm/LSm proteins are mostly hard to detect as they are very small and thus yield only a limited number of useful tryptic peptides; hLSm5 has not one in the 400–2000 Da range that is typically analyzed. Our protocol can in principle be followed for any macromolecular complex. The amount of antibodies must be closely controlled in each experiment: a certain excess is necessary to obtain efficient precipitation. With a large excess, however, the IgG bands overshadow a great part of the gel (Figure 2, lane 8). If a complete analysis were desired, the procedure could be modified to subtract the antibodies, or to use less stringent conditions, resulting in a bigger yield of intact complexes. Also, the two tags employed should be modified according to the particular needs: in brain extracts, for example, recovery rates of the calmodulin adapter are virtually nil (C.Mattioli and T.Achsel, unpublished data), most likely due to the high levels of endogenous calmodulin in the extracts. The basic outline of our method, however, will be very useful in purifying any macromolecular complex for which suitable antibodies are available.
When using the protocol to purify LSm1-associated factors, we identified only two non-LSm proteins. This again underlines the advantage of the double-purification method: conventional immunoprecipitation with the same antibodies showed many bands, and virtually all are also observed in the mock control (Supplementary Figure S1). Of the two proteins, the ‘Grb-10 interacting protein’ was also observed in the Y12 purification of the snRNPs and thus either represents a protein that always co-purifies when our method is applied to cytoplasmic extracts—it would be the only such background protein—or it may generally interact with complexes of the Sm/LSm family. In support of the latter notion, the protein contains a GYF domain. Such a domain is also found in the snRNP protein 52K (44), and the 52K GYF domain can interact with the Sm protein B/B’ (45). The other hit identified one of two potential mammalian orthologs of the yeast factor Pat1p (28). We confirmed this interaction using conventional immunoprecipitation: The protein co-precipitates with anti-LSm1 antibodies, and the reciprocal precipitation also works efficiently (Figure 3). The interaction of the two proteins is independent of RNA and of P-body formation. Together with the high efficiency of co-precipitation this suggests that the two proteins directly interact with each other. The other potential homolog (7,28) was not identified in our screen, indicating that it is absent from or at least underrepresented in the HeLa LSm complex. Either, the protein is not expressed in HeLa cells, or it does not efficiently bind to LSm1. In favor of the first notion, the Xenopus ortholog of the other Pat protein was identified as an oocyte-specific protein. It also argues against the second idea that the central domain that is responsible for the Pat1p-LSm1p interaction in yeast (4) is conserved in both mammalian Pat1 proteins (data not shown). We conclude that the main LSm1-interacting protein in undifferentiated mammalian cells is the factor that we have identified here, Pat1b.
As expected, endogenous Pat1b is highly concentrated in cytoplasmic foci (Figure 4), and the co-localization of GFP-Pat1b with LSm1 and Ddx6 identifies these foci as the P bodies (Figure 4) (28). Interestingly, cycloheximide-induced reduction of the PBs affected only the distribution of endogenous Pat1b; GFP-Pat1b remained concentrated in cytoplasmic foci of similar size and number. Since these foci are void of Lsm1 and Ddx6, we interpret them as aggregates caused by over-expression of the protein—even though we tried to limit the amount of overexpressed protein. These foci tell us that Pat1b is prone to aggregation into structures that resemble PBs. PBs are seen as self-organized assemblies of translationally silent mRNPs induced by self-interacting proteins on these complexes (24). In yeast, the Gln/Asn-rich C terminus of LSm4p has been incriminated (26,27). Several other yeast P body factors contain domains that are strikingly rich in glutamine and asparagine, including the Pat1p amino terminus (27). This domain is not evolutionarily conserved, but the amino-terminal half of Pat1b is also rich in glutamine and asparagine: 17 amide residues in the highest-scoring 80-mer peptide (data not shown), against 23 in the yeast protein, and eight in an average protein (27). It therefore appears that the driving force of P body assembly, aggregation of Gln/Asn-rich domains, has remained the same in yeast and man, even if the individual domains are not conserved.
In yeast, Pat1p represses translation and induces packaging of the mRNPs into PBs, with mRNA degradation following downstream (7). We therefore expected that tethering of Pat1b to a reporter mRNA would repress its expression primarily by inhibiting translation. To our surprise, we found that the decrease in protein expression is precisely matched by a reduction in mRNA abundance (Figure 5A and B). Furthermore, Pat1b modulates the level of mRNAs with oligo(A) tails (Figures 5C and D, and 6C). If Pat1p enhanced degradation after deadenylation, its presence should decrease the steady-state level of deadenylated mRNAs, and its absence should increase them. The opposite was observed (Figures 5C and 6C). Therefore, our data can only be explained assuming that Pat1b increases the steady-state level of mRNAs with short poly(A) tails, i.e. it has a role in mRNA deadenylation. This notion is strongly supported by the actinomycin D time course that shows that in mRNAs targeted by Pat1b, a deadenylated decay intermediate accumulates (Figure 5D). While this article was in final preparation, a description of the Drosophila Pat1 homolog was published (46). This protein associates with the Ccr4/Not deadenylase complex, which greatly supports our hypothesis.
Although yeast Pat1p binds RNA with a preference for poly(U) (4), the yeast factor does not specifically induce degradation of some mRNAs, nor is it involved in specific regulatory pathways. Assuming a similarly general function, we tested three randomly picked unstable mRNAs. Of these three, Pat1b depletion affected only one, namely the fos mRNA. Both mRNA abundance and poly(A) length distribution of the fos mRNA are affected, even though the effect is minor in both cases: in fact, the evident decrease in the mRNA population with very short mRNAs (Figure 6C) should be matched by a concomitant increase of longer tails, but this is too small and diluted to be discerned. Thus, the small amplitude of the effect suggests that the fos mRNA may even not be the main target for Pat1b regulation. In mammals, the best-described mRNA destabilization elements are the AREs and miRNA-binding sites. AREs do not specify Pat1b-mediated deadenylation, because all three mRNAs checked here contain active AREs, but only fos mRNA was affected by Pat1b depletion. Furthermore, the ARE-binding TTP recruits the Ccr4/CAF1 deadenylase to the mRNA and thereby directly induces shortening of the poly(A) tail without the need for additional factors such as Pat1b (47). miRNAs and the associated RISC complex likewise associate with Ccr4/CAF1, and therefore trigger deadenylation independently of other factors (12,48,49). While we cannot formally rule out that Pat1b has a mere auxiliary role in one of the two mRNA-destabilizing pathways, we think it is much more likely that it is involved in a third, as yet unknown deadenylation pathway.
In conclusion, Pat1b is a deadenylating factor that acts on specific mRNAs. Deadenylation is becoming ever more important for gene regulation. Examples include the miRNA pathway, the regulation of AREs by TTP (see above), or the anti-proliferative pathway regulated by the Tob family of proteins (50,51). Pat1b—in contrast to what is known about its yeast counterpart—therefore has all that it takes to be a specific regulator of gene expression. It will be interesting to see, in future experiments, in which pathways Pat1b is involved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Ministry of Health (‘progetto finalizzato’ grant); ‘Methusalem’ grant to Bart De Strooper (Katholieke Universiteit Leuven and Flemish government). Funding for open access charge: Flemish Institute for Biotechnology (VIB).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Claudia Bagni for critically reading the article. We gratefully acknowledge the laboratory of Prof. Reinhard Lührmann for the gift of HeLa cell extracts, and Gaby Heyne and Cindy L. Will of the same laboratory for affinity-purified antibodies directed against SF3a120 and SF3b155. We thank Fátima Gebauer for the pLuc-MS2x9 plasmid, and Nicki Gray for the permission to use it.
REFERENCES
- 1.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 3.Achsel T, Stark H, Lührmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl Acad. Sci. USA. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 7.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 10.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 12.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bönisch C, Temme C, Moritz B, Wahle E. Degradation of hsp70 and other mRNAs in Drosophila via the 5′ 3′ pathway and its regulation by heat shock. J. Biol. Chem. 2007;282:21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- 15.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 23.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franks TM, Lykke-Andersen J. The control of mRNA decapping and p-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazzoni C, D’Addario I, Falcone C. The C-terminus of the yeast Lsm4p is required for the association to P-bodies. FEBS Lett. 2007;581:4836–4840. doi: 10.1016/j.febslet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, Díez J. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim. Biophys. Acta. 2007;1773:1786–1792. doi: 10.1016/j.bbamcr.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 30.Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Lührmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 34.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 35.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.di Penta A, Mercaldo V, Florenzano F, Munck S, Ciotti MT, Zalfa F, Mercanti D, Molinari M, Bagni C, Achsel T. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J. Cell Biol. 2009;184:423–435. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brahms H, Raker VA, van Venrooij WJ, Lührmann R. A major, novel systemic lupus erythematosus autoantibody class recognizes the E, F, and G Sm snRNP proteins as an E-F-G complex but not in their denatured states. Arthritis Rheum. 1997;40:672–682. doi: 10.1002/art.1780400412. [DOI] [PubMed] [Google Scholar]
- 38.Strub K, Birnstiel ML. Genetic complementation in the Xenopus oocyte: co-expression of sea urchin histone and U7 RNAs restores 3′ processing of H3 pre-mRNA in the oocyte. EMBO J. 1986;5:1675–1682. doi: 10.1002/j.1460-2075.1986.tb04411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marnef A, Maldonado M, Bugaut A, Balasubramanian S, Kress M, Weil D, Standart N. Distinct functions of maternal and somatic Pat1 protein paralogues. RNA. doi: 10.1261/rna.2295410. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selbach M, Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK) Nat. Methods. 2006;3:981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 41.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 42.Forler D, Köcher T, Rode M, Gentzel M, Izaurralde E, Wilm M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- 43.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 44.Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Lührmann R. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA. 2005;11:598–608. doi: 10.1261/rna.2300805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kofler M, Heuer K, Zech T, Freund C. Recognition sequences for the GYF domain reveal a possible spliceosomal function of CD2BP2. J. Biol. Chem. 2004;279:28292–28297. doi: 10.1074/jbc.M402008200. [DOI] [PubMed] [Google Scholar]
- 46.Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J. Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 49.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mauxion F, Faux C, Séraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.