Abstract
AbrB is a global transcriptional regulator of Bacillus subtilis that represses the expression of many genes during exponential growth. Here, we demonstrate that AbrB and its homolog Abh bind to hundreds of sites throughout the entire B. subtilis genome during exponential growth. Comparison of regional binding of AbrB and Abh in wild-type, ΔabrB and Δabh backgrounds revealed that they bind as homomer and/or heteromer forms with different specificities and affinities. We found four AbrB and Abh binding patterns were major. Three of these contain pairs of TGGNA motifs connected by A/T-rich sequences, differing in arrangement and spacing. We also assessed the direct involvement of these complexes in the control of gene expression. Our data indicate that AbrB usually acts as a repressor, and that the ability of Abh to act as a transcriptional regulator was limited. We found that changes to AbrB/Abh levels affect their binding at several promoters and consequently transcriptional regulation. Surprisingly, most AbrB/Abh binding events had no impact on transcription, suggesting an interesting possibility that AbrB/Abh binding is analogous to nucleoid-associated protein binding in Escherichia coli.
INTRODUCTION
The transition from the exponential to the stationary phase of bacterial growth is induced under suboptimal environmental conditions. This process requires re-direction of the expression of various genes involved in the adaptive response and survival. In the spore-forming Gram-positive bacterium Bacillus subtilis, one of the key regulators controlling gene expression during the transitional phase is the AbrB protein (1,2). This protein primarily functions to prevent inappropriate gene expression in actively growing cells and, during the transition phase, reorganizes the expression of more than 100 post-exponential-phase genes with different biological functions including biofilm formation, antibiotic production, motility, development of competence for DNA uptake, synthesis of extracellular enzymes and sporulation. Although AbrB functions mainly as a repressor of gene expression, AbrB also acts as an activator of some genes (3–5). However, no direct interaction of AbrB with RNA polymerase has yet been demonstrated. Expression of AbrB is growth-phase-dependent; the protein is synthesized at high levels from the lag to the exponential phase but expression levels decrease when the cell enters stationary phase. Reduction of AbrB expression at entry into the stationary phase is mediated by the master regulator for entry to the sporulation process, Spo0A, which directly represses abrB transcription (6,7). In addition, Spo0A activates expression of AbbA, which forms a complex with AbrB and prevents the latter protein from binding to DNA (8).
AbrB is a small protein (10.4 kDa) composed of two domains. The N-terminal domains of two AbrB molecules form a single DNA-binding domain, termed a swapped-hairpin barrel (9,10). The C-terminal domain also has an ability to dimerize, and AbrB thus forms a tetramer via both N-terminal and C-terminal interactions, yielding a protein with a stable DNA-binding site (11–15). AbrB orthologs and paralogs have been found in genomic sequences of all Bacillus, Clostridium and Listeria species, and AbrB has been shown to be involved in induction of virulence factors in Bacillus cereus and Bacillus anthracis (16,17). In addition, AbrB sequences are found sporadically in various bacterial species including archaea (MBGD, http://mbgd.genome.ad.jp/), although the functions of the encoded proteins have not yet been determined. Furthermore, AbrB-like proteins, with DNA-binding motifs similar to those of AbrB have been widely identified in cyanobacteria (10,18), and regulate several physiological and metabolic processes, including carbon and hydrogen metabolism, nitrogen fixation and toxin production (18–22).
Although B. subtilis AbrB plays an essential role in the reorganization of gene expression during the transitional phase, and although the protein has been extensively studied both genetically and biochemically, it is still unclear how AbrB, and related proteins, select binding sites appropriate for achievement of the desired functions. In vitro selection of optimal AbrB-binding sites identified a relevant TGGNA motif (23); however, this motif did not resemble known AbrB binding sites in the B. subtilis genome, for which the consensus sequence WAWWTTTWCAAAAAAW had been suggested (24). To date, examination of more than 40 chromosomal AbrB binding sites has failed to identify a consensus sequence that adequately explains AbrB site selection and recognition, and it has been hypothesized that AbrB binding instead requires a specific three-dimensional conformation of the DNA helix (1,12,25). Recently, nuclear magnetic resonance (NMR) studies of the DNA-binding domains of AbrB and paralogs thereof, together with re-evaluation of previous experimental results, allowed a structural model of the complex between the N-terminal domain of AbrB and the target DNA sequence to be constructed. The model indicates that structural flexibility in the loop regions LP1, which connect β1 and β2, and LP2, which connect α1 and β3, of the DNA-binding domain of the AbrB monomer, allows AbrB to bind to various DNA targets (26).
Several proteins participating in organization of genomic DNA into nucleoids, nucleoid-associated proteins, also act as transcriptional regulators in Escherichia coli; the proteins include HU, H-NS, StpA, Fis and IHF (27). However, homologs of these proteins are absent from B. subtilis, with the exception of HU. AbrB was once considered to be a nucleoid-associated protein because, although the proteins are not related in amino acid sequence, the growth cycle-dependent expression of abrB is similar to that of fis in E. coli and Salmonella typhimurium (6). AbrB possesses the general characteristics of nucleoid proteins identified in E. coli. Such proteins are small in size and abundant, with a high proportion of positively charged amino acids. The proteins participate in transcriptional regulation of various genes with diverse functions. Furthermore, the proteins bind to degenerate consensus sequences biased toward a preponderance of A and T residues, and such a binding event generates a bend in DNA (28). However, the role of AbrB in organization of the B. subtilis nucleoid structure has not been determined.
B. subtilis expresses two other proteins, Abh and SpoVT, with N-terminal DNA-binding domains highly homologous to that of AbrB. The three-dimensional structures of the DNA-binding domains of these proteins are similar, suggesting a common structural basis for DNA binding. However, subtle structural differences between the proteins have also been identified and these may play important roles in the specificity of DNA targeting (25,26). SpoVT is a regulator of forespore-specific genes that are expressed at later stages of sporulation depending on σG activity, and SpoVT is not a transition-state regulator (29). However, Abh has been recently shown to regulate some of AbrB regulons in vivo and to bind to some promoter regions recognized by AbrB in vitro (30–32). Neither the regulatory role of Abh nor the molecular mechanism of Abh action has been systematically studied.
In the present work, we report, for the first time, the in vivo distribution over the entire B. subtilis genome of AbrB-binding sites during exponential growth phase when AbrB principally functions. To this end, we used a modified chromatin immunoprecipitation (ChIP)–chip method, termed the ChAP–chip (Chromatin Affinity Precipitation coupled with tiling chip) approach, developed in our previous studies (33). In parallel, we studied binding of the AbrB paralog Abh, which has not received much research attention. Furthermore, we analyzed changes in the transcriptome profiles of abrB- and abh-deleted cells, to investigate the detailed involvement of AbrB/Abh in transcriptional regulation. Our results reveal novel important properties of AbrB and Abh, and facilitate a deeper understanding of the cellular roles played by these proteins as transcriptional regulators and, possibly, as nucleoid architectural proteins.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The B. subtilis strains used in the present study, the wild-type 168 strain and derivatives thereof, are listed in Supplementary Table S1, and the methods used to construct mutant strains are described in Supplementary Methods. Primers used in the present study are listed in Supplementary Table S2. Luria-Bertani (LB) medium, supplemented with appropriate antibiotics [10 µg/ml kanamycin (Km), 150 µg/ml spectinomycin (Spec) and/or 5 µg/ml chloramphenicol, (Cm)], as appropriate, was used for cell growth. Cells growing exponentially at 37°C in LB medium supplemented with appropriate antibiotic(s) were inoculated into larger volumes of culture medium, without any antibiotic, at a commencement OD600 value of 0.01.
In vivo interaction of AbrB and Abh
Thirty-five milliliter amounts of exponentially growing cells cultured to an OD600 of 0.4 were harvested by centrifugation. Cell pellets were washed with 10 ml of 50 mM HEPES pH 8.0 and resuspended in 1.5 ml of the same buffer. Dithiobis (succinimidyl propionate) (DSP) (Pierce) was added to cell suspensions to a final concentration of 2 mM, and cells were incubated on ice for 5 min, 15 min or 25 min, followed by quenching of the cross-linking reaction by addition of 1 M Tris–HCl (pH 7.5) to a final concentration of 20 mM, followed by incubation on ice for 15 min. Next, cells were washed once with 1.5 ml TBS buffer [50 mM Tris–HCl (pH 7.5) and 150 mM NaCl] and stored frozen at −80°C. The frozen cells were dissolved in 1.4 ml buffer containing 100 mM Tris–HCl, 250 mM NaCl, 20% (v/v) glycerol, 50 mM imidazole with 1 mM PMSF, and 1 × protease inhibitor cocktail (Roche) and disrupted by sonication on ice using an Astrason Ultrasonic Processor XL (Misonix) over 10 min (4 s ‘on’ and 10 s ‘off’, at output level 4.5). Genome DNA and RNA in cell lysates were digested by addition of DNase I to 10 µg/ml and RNase A to 200 µg/ml, respectively, for 30 min at 37°C in the presence of 10 mM MgCl2. Cell debris was removed by centrifugation and each supernatant (80 µl) was mixed with 20 µl of 5× sodium dodecyl sulfate (SDS) sample buffer [250 mM Tris (pH 6.8), 10% (w/v) SDS and 50% (v/v) glycerol] in the absence of the (commonly included) reducing agent β-mercaptoethanol, followed by boiling for 10 min. After separation of 12.5 µl amounts of cell lysates on 10–20% (w/v) SDS-PAGE, proteins on the gels were electrotransferred to Immobilon-PSQ membranes (Millipore) using 100 V for 2 h. Next, the membranes were probed with primary antibodies, either an anti-AbrB or an anti-Abh antibody (see Supplementary Methods for production and purification of the specific antibodies), followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad), and the SNAP ID Protein Detection System (Millipore) was employed to develop protein bands. Finally, membranes were incubated with an ECL substrate (Amersham) and exposed to X-ray film.
ChAP–chip analysis
ChAP–chip analysis of AbrB-2HC and Abh-2HC was performed as described previously (33), with some modifications described in Supplementary Methods.
Data normalization
All probes on the Affymetrix tiling array were mapped to the recently revised B. subtilis genome sequence (GenBank no. NC_000964) using the In Silico Molecular Cloning program, Array Edition (In Silico Biolog), and 242 413 probes that had been uniquely mapped on the genome were selected for study. The signal intensities of perfectly matched probes (only) were used in the following calculations. Signals on ribosomal RNA genes were removed from analysis because of the high copy number of such genes in the genome. Raw data from eight experiments (a duplicate analysis of four strains) were pre-processed using the following steps. First, the signal intensities from DNA in the affinity-purified fraction (ChAP DNA) were divided by those from the DNA of the entire cell extract prior to affinity purification (control DNA), thus identifying enrichment factors in the ChAP fraction. Second, signals considered to be outliers of each dataset were identified and removed using t-testing (P-values ≤0.001), under the assumption that the mean of the signal intensities of six probes around any particular probe should be equal to that of the probe in question. Third, median signal intensity levels in each experiment were normalized to the means of the median values obtained from eight experiments, using the following equation (Supplementary Figure S1A):
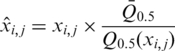 |
where 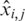 is a normalized value,
is a normalized value, 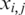 is the relative intensity of the ith probe in experiment j,
is the relative intensity of the ith probe in experiment j,  is the median intensity of all probes used in experiment j and
is the median intensity of all probes used in experiment j and 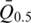 is the mean of
is the mean of 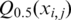 from j=1 to j=k.
from j=1 to j=k.
We observed low contiguous signals, without peaks, in many regions, probably because of contamination of the ChAP fraction with free DNA, but normalization of the median signal intensity values yielded similar background levels for the eight experiments (Supplementary Figure S1B). Finally, the binding signal intensities of AbrB-2HC in wild-type (OC001) and Δabh (OC006) backgrounds, and those of Abh-2HC in the wild-type (OC002) and ΔabrB (OC007) backgrounds, were obtained by averaging of data from two replicate experiments.
Detection and quantitative analysis of AbrB/Abh binding sites
First, we extracted possible protein binding sites in each strain, by searching for regions wherein at least four probes separated by intervals of <100 bp showed binding intensities above the chosen threshold value, which was 0.88 (the 95th percentile of all probe data; Supplementary Figures S1B and S2A). Next, overlapping protein-binding regions extracted for each strain were merged and defined as PBRs. Some peaks were divided by visual examination if several clearly defined peaks were evident. Finally, AbrB and Abh binding intensities to the 928 PBRs of the four strains were calculated as the sums of probe signal intensities within each PBR, after subtraction of background signals, which were defined as those ≤0.4 (the 40th percentile value; see Supplementary Figure S1B).
High-resolution transcriptome analysis
RNA extraction, synthesis of complementary DNA (cDNA), terminal labeling and hybridization with the oligonucleotide tiling chip used for ChAP–chip analysis were all performed according to the Affymetrix instruction manual, as previously reported (34). Processing of hybridization signal data, background correction, data normalization and calculation of expression levels of individual genes were all performed as previously described (35). Finally, the transcriptional signal intensities were obtained by averaging of data from two replicate experiments.
Array design and array data
The array design and the array data in this study have been submitted to ArrayExpress database (http://www.ebi.ac.uk/microarray-as/ae/) under accession code A-AFFY-161(array design), E-MEXP-2774 (ChAP-chip) and E-MEXP-2776 (transcriptome).
RESULTS
AbrB and Abh form homodimers and heterodimers in vivo
Previously, it has been shown that AbrB and Abh share overlapping binding sites in vitro (32), suggesting that these proteins may perhaps bind as heteromers to DNA. To investigate this possibility, we treated exponentially growing B. subtilis cells with DSP [dithiobis (succinimidyl propionate)], a membrane-permeable chemical cross-linker, and analyzed cross-linked proteins in the cell lysate by western blotting using specific antibodies against AbrB and Abh. Because the molecular weights of AbrB and Abh (10.4 kDa and 10.1 kDa, respectively) are similar, the proteins are not adequately separated on SDS-PAGE. Thus, we constructed strains expressing C-terminal 2HC (12 histidines plus a chitin-binding domain)-tagged AbrB (AbrB-2HC) or Abh (Abh-2HC) [designated as strains OC001 and OC002, respectively (Supplementary Table S1)]. Fusion of 2HC to AbrB generates a protein easily distinguishable from Abh on SDS-PAGE, and vice versa. We confirmed that tagging with 2HC did not affect growth rate or gene expression profile during exponential growth (Supplementary Figure S3). We also successfully constructed a further two strains that expressed AbrB-2HC in a Δabh background (strain OC006) and Abh-2HC in a ΔabrB background (strain OC007).
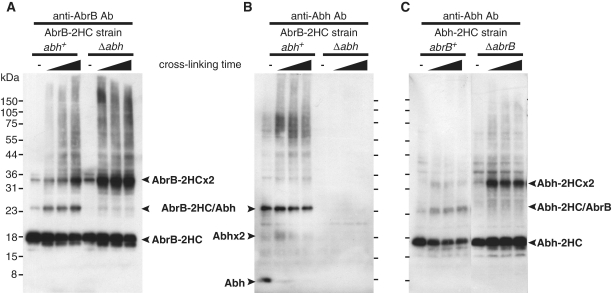
Using an antibody specifically reacting with AbrB (Supplementary Figure S4), western blotting of cell lysates from DSP-treated OC001 cells expressing AbrB-2HC revealed three clear bands (Figure 1A). The molecular weights of two of the bands corresponded to those of the AbrB-2HC monomer and dimer, and that of the remaining band was matched to an AbrB-2HC/Abh heterodimer. Indeed, the third band was not detected in a Δabh background (Figure 1A). In addition, only this band was detected using a specific anti-Abh antibody (Supplementary Figure S4), confirming that the band contained the AbrB-2HC/Abh heterodimer (Figure 1B). These results indicate that a certain proportion of AbrB and Abh exist as a heterodimer complex in vivo. In addition, when the cross-linking time was extended, unresolved high-molecular weight bands were detected. The proportions of these bands increased in parallel with the disappearance of monomer bands when cells were treated with greater concentrations of DSP (data not shown), suggesting that complexes larger than dimers were also formed in vivo. Previous in vitro studies have demonstrated that, although full-length AbrB forms a tetramer through interactions of the C-terminal domains of individual proteins, an AbrBN53 mutant protein lacking the C-terminal domain can form a stable dimer with DNA-binding activity (11). Presently, the predominant molecular forms of AbrB/Abh in vivo are not known, and it is not clear whether the dimer forms detected in the present work were formed via interaction of N-terminal or C-terminal domains.
Figure 1.
AbrB and Abh interaction in vivo. Western blot analysis of exponentially growing B. subtilis cells, either untreated or treated with 2 mM DSP for 5, 15 or 25 minutes. Lysates of cells expressing AbrB-2HC in the abh+ or Δabh background were probed using anti-AbrB antibody (A) or anti-Abh antibody (B). Cells expressing Abh-2HC in the abrB+ or ΔabrB background were probed using anti-Abh antibody (C). Electrophoretic mobilities of AbrB/Abh monomers, dimers, and heteromers, as well as those of proteins in a pre-stained broad-range marker mix (Bio-Rad), are indicated.
Notably, in the Δabh background, the level of the AbrB dimer was significantly increased compared with the amount seen in an abh+ background (Figure 1A). When the same experiments were performed using the Abh-2HC-expressing strains OC002 and OC007, Abh homodimer levels were low in the abrB+ background but a marked increase in Abh dimer formation was observed in ΔabrB cells (Figure 1C). The number of Abh molecules per cell under our experimental conditions was 32 700 ± 6500, whereas the figure for AbrB was 65 800 ± 14 200 (Supplementary Figure S5). Although relative quantities of various multimeric forms of AbrB/Abh in the cell cannot be estimated exactly in this method, these results may suggest that many of Abh molecules would be present in a complex with AbrB (an AbrB/Abh heteromer) in wild-type cells. Furthermore, increase in Abh dimer in ΔabrB cells may indicate that Abh would be released from the complexes with AbrB to form Abh homomers when the cellular levels of AbrB decreased.
Profiling of genome-wide AbrB and Abh binding sites
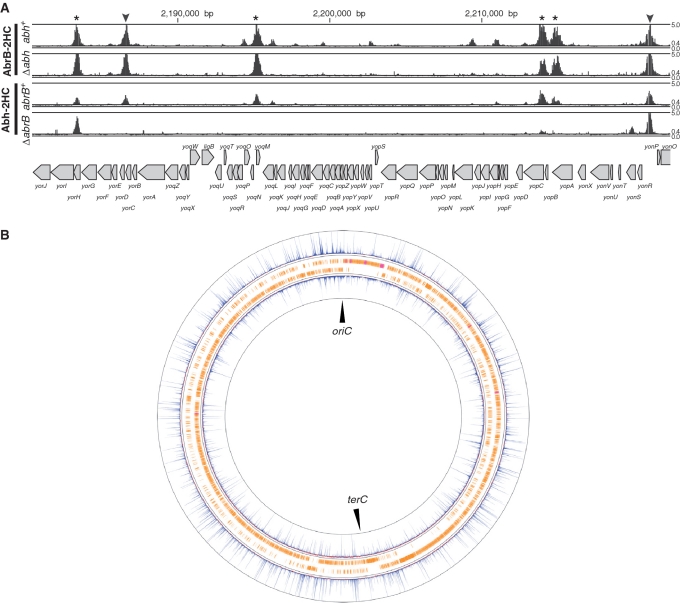
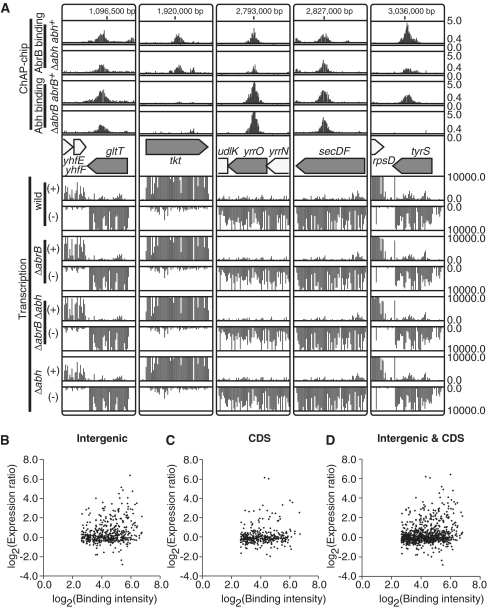
We identified all binding sites for AbrB and Abh in the B. subtilis genome using the ChAP–chip method in the four strains described above: OC001 (abrB-2HC abh+), OC002 (abh-2HC abrB+), OC006 (abrB-2HC Δabh) and OC007 (abh-2HC ΔabrB). The contributions of the AbrB and Abh complexes to the DNA binding were also assessed. Purification of protein–DNA complexes, mapping of co-purified DNA fragments using a custom Affymetrix tiling chip, and comparative quantitative analysis of protein-binding signals were performed as described in ‘Materials and methods’ section. We identified hundreds of AbrB(-2HC) and Abh(-2HC) binding sites distributed throughout the B. subtilis genome (Supplementary Figure S6). Typical examples of AbrB- and Abh-binding signals along the genome in wild-type cells are presented in Figure 2A, and an overview of AbrB and Abh binding signal sites is shown in Figure 2B. In addition, we found that deletion of abrB significantly affected the Abh-binding profile, whereas abh deletion had little impact on AbrB binding (Figure 2A).
Figure 2.
AbrB and Abh binding signals on the B. subtilis genome. (A) Distribution of AbrB binding signals in the abh+ and Δabh backgrounds, and Abh binding signals in abrB+ and ΔabrB backgrounds, in the 2 180 000 bp to 2 222 000 bp region of the genome, are shown. Protein-binding signals for each probe in the region are indicated alongside the genomic coordinates. The top and bottom lines in each column indicate signal intensities of 5 and 0, respectively. The middle line shows the threshold values used to define AbrB and Abh binding peaks (0.4). The gene organization is schematically shown at the bottom. Protein-binding sites located in intergenic regions and coding regions are indicated using triangles and asterisks, respectively. (B) Distributions of AbrB (outer) and Abh (inner) binding signals over the entire genome. Clockwise and counter-clockwise ORFs are schematically indicated.
To quantitatively compare AbrB and Abh binding in the four strains, we normalized the distribution of protein binding signal intensities, as described in ‘Materials and methods’ section and Supplementary Figure S1. We next computationally extracted the AbrB/Abh-binding sites of the various strains in instances where at least four probes at intervals of <100 bp showed signal intensities above a particular threshold (Supplementary Figure S2A), and overlapping binding DNA stretches in different strains were merged, thus defining 928 possible binding regions (PBRs) ranging from 75 to 2167 bp in size (Supplementary Table S3). Additionally, we detected eight broad regions with contiguous binding signals (Supplementary Figure S6, Supplementary Table S3). These stretches overlapped with the wide binding regions of Spo0J and/or Noc around oriC, which are involved in the organization of higher-order nucleoid structure (33,36), suggesting that co-existence of AbrB/Abh and Noc and/or Spo0J at the relevant regions might cross-link these proteins and co-purify DNA of such wide binding regions. In addition, some highly transcribed regions tended to show increased levels of background signal. Such DNA stretches were removed prior to the following analysis.
To quantitatively compare AbrB/Abh binding in the four strains, the sum of AbrB- or Abh-binding signals derived from probes in each PBR was calculated for each strain (a total of 3712 signal intensity values were used in the assessment), and these sums (in arbitrary units) were ranked and grouped into four classes: very low (<2.0, class VL), low (2.0–6.2, class L), middle (6.2–17.8, class M) and high (>17.8, class H), which corresponded to the 25th, 50th, 75th and 100th percentiles, respectively (Supplementary Figure S2B). We defined PBRs with binding intensities greater than 6.2 (classes M and H) as having AbrB/Abh-binding sites, and 753 PBRs satisfied this criterion for either or both AbrB and Abh. This cutoff level detected about 75% of AbrB- and Abh-binding sites previously determined by in vitro experiments (Supplementary Table S4), including overlapping binding sites for AbrB and Abh at sunA, sboA, sdpA, skfA and sigW promoters (32). As for promoter regions where we failed to detect AbrB/Abh binding in Supplementary Table S4, AbrB/Abh-binding affinities to these promoters may be too low to be detectable under our in vivo conditions.
Most Abh binding sites overlap with those of AbrB in wild-type cells
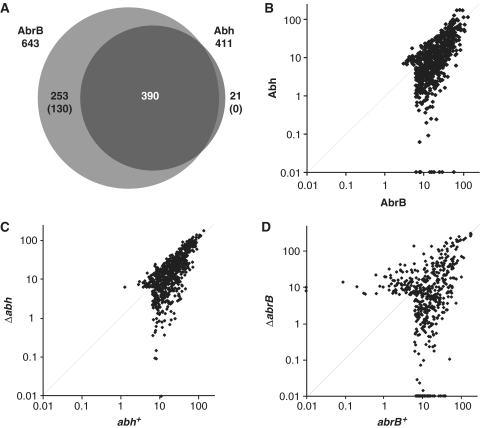
Using the criteria outlined above, we detected 643 AbrB- and 411 Abh-binding sites in exponentially growing wild-type cells (Figure 3A). We found that most Abh-binding sites (390) overlapped with those for AbrB. This result is consistent with the presence of the AbrB/Abh heteromer in such cells, although simultaneous bindings of both AbrB- and Abh-homomers are also possible. We detected 21 Abh-specific binding sites with middle-level signal intensities (class M), but low-level signal intensities (class L) of AbrB binding were also observed at these positions. This was also seen for AbrB-specific binding sites; 123 of 253 such sites were associated with low-level Abh-binding signals. Thus, it seems that 130 AbrB-binding sites were AbrB-specific, whereas Abh-specific binding sites were not detected in wild-type cells. The presence of AbrB-specific binding sites and the absence of Abh-specific binding sites in wild-type cells were also evident when the scatter plots of AbrB and Abh binding intensities were viewed (Figure 3B).
Figure 3.
Comparison of AbrB and Abh-binding signals in various strains. (A) A Venn diagram of overlapping binding sites for AbrB and Abh in a wild-type background. The numbers in parentheses indicate the levels of confidence associated with the identification of particular AbrB- and Abh-specific binding sites (please see main text). (B) A log-scale scatter plot of the binding intensities of AbrB (x-axis) and Abh (y-axis) in 664 PBRs in a wild-type background. (C) A log-scale scatter plot of AbrB binding intensities in abh+ (x-axis) and Δabh (y-axis) cells. (D) A log-scale scatter plot of Abh binding intensities in abrB+ (x-axis) and ΔabrB (y-axis) cells.
We next evaluated the positions of AbrB- and Abh-binding sites (near the centers of PBRs). We found that 58% of binding sites were located in protein-encoding regions, whereas 42% were in intergenic regions (Supplementary Table S3, Supplementary Figure S6). Although AbrB/Abh-binding sites were concentrated in intergenic regions (forming 13% of genomic DNA), a significant number of sites were in coding regions, unlike observations in general transcriptional regulators, which usually bind to the promoter regions of target genes.
AbrB markedly influences Abh-binding profile, whereas Abh has a minor effect on AbrB binding
To examine the contribution of AbrB/Abh interactions to DNA binding, we compared AbrB binding in abh+ and Δabh cells and Abh binding in abrB+ and ΔabrB cells. The scatter plots of AbrB binding intensities in the presence and absence of Abh indicated that, although some AbrB binding intensities decreased in the absence of Abh, most such intensities were unaffected (Figure 3C). In contrast, AbrB deletion resulted in a re-organization of Abh binding intensities (Figure 3D). These results indicate that AbrB has the ability to bind to most target loci independent of Abh, whereas many Abh-binding events are dependent on the presence of AbrB.
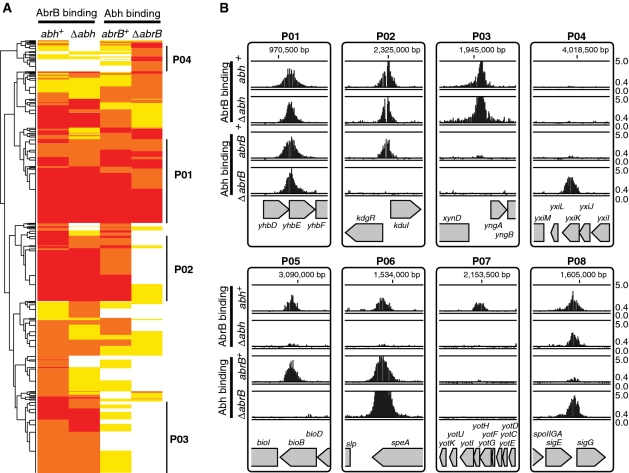
To gain further insight into the effects of AbrB/Abh interactions on DNA binding, we classified PBRs based on AbrB and Abh binding intensities in the four strains. We first performed a hierarchical clustering analysis of all 753 PBRs based on AbrB/Abh binding classes in the four strains using the R statistical environment software (37). The PBRs formed several clusters (Figure 4A). However, it was apparent that the cluster positioning of many sites was ambiguous, probably because adjacent binding sites with different AbrB/Abh-binding properties were merged into a single PBR.
Figure 4.
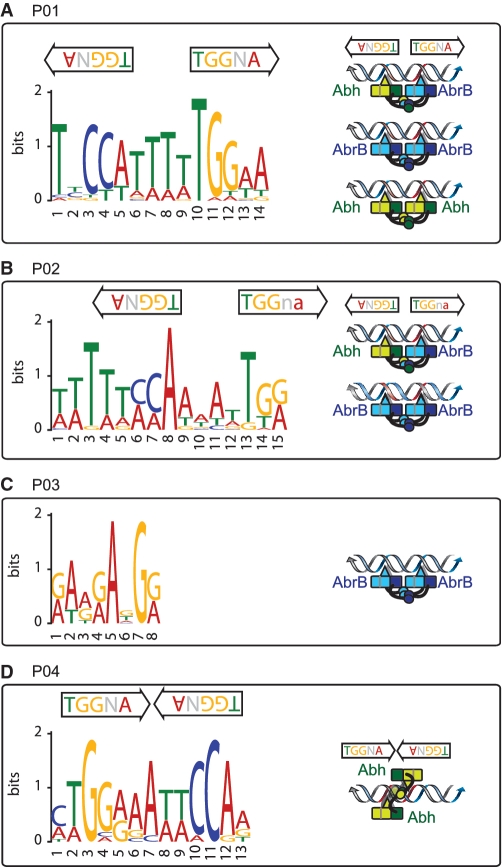
Classification of PBRs according to AbrB and Abh binding profiles. (A) A cluster diagram of 753 PBRs based on AbrB-binding intensities (in various classes) in abh+ and Δabh backgrounds, and Abh-binding classes in abrB+ and ΔabrB backgrounds. The VL, L, M and H classes of AbrB/Abh binding are indicated in white, yellow, orange and red, respectively. Clusters containing PBRs belonging to the four major binding profiles of the selected unambiguous PBRs summarized in Table 1 are marked by bold lines at the right of the diagram. (B) Typical examples of AbrB and Abh binding signals in PBRs belonging to classes P01–P08. The binding signals are shown as in Figure 2A. Gene organizations around selected PBRs are indicated at the bottom of each panel.
Theoretically, AbrB and Abh binding abilities to particular sites can be classified into 15 patterns (P01–P15) as shown in Table 1. To select PBRs with unambiguous binding patterns, we searched for PBRs that could be clearly assigned to 1 of the 15 patterns. To this end, we considered signal intensities of <6.2 (classes VL and L) as protein-binding-negative and those with intensities of more than 17.8 (class H) as positive. PBRs with intermediate signal intensities were not included in further analysis. As a result of this exercise, 160 PBRs with unambiguous binding patterns were extracted, possibly reflecting a fundamental mode of AbrB and Abh binding to target sequences (Table 1). The results of Table 1 indicate that AbrB/Abh binding to DNA falls into four major patterns, P01–P04, and four minor patterns, P05–P08. These results are generally consistent with the results of the hierarchical clustering analysis of all PBRs shown in Figure 4A. Typical examples of AbrB- and Abh-binding signals assigned to each profile are shown in Figure 4B, and the expected molecular species with the ability to bind to DNA within each pattern are listed in Table 1. Among the 160 PBRs, AbrB-binding sites observed in wild-type cells were mostly retained in the absence of Abh (136/148), whereas about 44% of Abh binding sites in wild-type cells disappeared in the absence of AbrB (51/115). AbrB-homomer-specific binding sites in wild-type cells formed one of the major patterns (P03). However, Abh-homomer-specific binding was evident only in the absence of AbrB (P04), probably due to an increased concentration of Abh homomer. The binding profile P07, AbrB homomer binding with assistance from Abh, was unexpected and the molecular mechanism of P07 is not yet clear. These results indicate that AbrB/Abh-binding sites include various sequences that differ in specificities and affinities for homomers and heteromers of AbrB and Abh.
Table 1.
Classification of 160 PBRs with clear binding profiles
| Possible binding profiles |
Molecules expected to be bounda |
Assigned PBRs |
Binding motif | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Profile ID | AbrB in wild | AbrB in Δabh | Abh in wild | Abh in ΔabrB | AbrB homomer | AbrB/Abh heteromer | Abh homomer | Number | % | |
| P01 | • | • | • | • | • | • | • | 60 | 37.3 | TNCCAWWWWTGGNA |
| P02 | • | • | • | – | • | • | – | 46 | 28.6 | WWWWWCCAWWWWTGG |
| P03 | • | • | – | – | • | – | – | 29 | 18 | not clear |
| P04 | – | – | – | • | – | – | • (ΔabrB) | 13 | 8.1 | TGGNAWTNCCA |
| P05 | • | – | • | – | – | • | – | 5 | 3.1 | |
| P06 | • | – | • | • | – | • | • | 4 | 2.5 | |
| P07 | • | – | – | – | • (wild) | – | – | 3 | 1.9 | |
| P08 | • | • | – | • | • | – | • (ΔabrB) | 1 | 0.6 | |
| P09 | • | – | – | • | • (wild) | – | • (ΔabrB) | 0 | 0 | |
| P10 | – | • | • | • | • (Δabh) | – | • | 0 | 0 | |
| P11 | – | • | • | – | • (Δabh) | • (wild) | 0 | 0 | ||
| P12 | – | • | – | • | • (Δabh) | – | • (ΔabrB) | 0 | 0 | |
| P13 | – | • | – | – | • (Δabh) | – | – | 0 | 0 | |
| P14 | – | – | • | • | – | – | • | 0 | 0 | |
| P15 | – | – | • | – | – | – | • (wild) | 0 | 0 | |
aIf AbrB/Abh binding is expected to occur in a particular genetic background, it is indicated in parentheses.
A TGGNA motif identified in vitro acts as a determinant not only for AbrB but also for Abh binding in vivo
No consensus sequence has been identified that adequately explains AbrB site selection and recognition. We also failed to find consensus sequences in the 160 unambiguous PBRs or in any of the major binding patterns, P01–P04, using the MEME program [http://meme.nbcr.net (38)]. We suspected that the selected PBRs still included multiple AbrB/Abh-binding sites of different binding types. Thus, to eliminate such a possibility, we further selected only sites at which binding signal peaks were clearly triangular in shape, reflecting a simple AbrB/Abh-binding profile, by visual inspection (Supplementary Table S5) and manually selected 100-bp regions around the peak positions. This allowed identification of motifs specific for P01, P02 and P04 (Table 1 and Figure 5).
Figure 5.
Consensus sequences for AbrB/Abh binding. Consensus sequences identified in selected PBRs belonging to classes P01 (A), P02 (B), P03 (C) and P04 (D) are presented, together with the position(s) and direction(s) of the TGGNA motif(s) in each sequence. Possible binding modes of AbrB/Abh homotetramers and heterotetramers to each sequence are schematically presented (please see ‘Discussion’ section). Red coloration of double-stranded DNA indicates the TGGNA motif. Light and dark-blue boxes show the N-terminal domains of two AbrB molecules that interact to form a DNA-binding domain, whereas the light- and dark-blue arm-like structures extending from the boxes depict C-terminal domains that form tetramers. Loop regions, which have been predicted to contact DNA and contribute to DNA recognition, are shown using triangles. Similar structures in Abh are shown in green.
The consensus motif for sequences of the P01 pattern was TNCCA-WWWW-TGGNA, which was present in 21 of the 37 selected PBRs. Interestingly, this is an inverted repeat, with a 4-bp interval, of TGGNA, which was identified as the basic motif for AbrB binding using in vitro selection experiments (23). Furthermore, inverted repeats of the TGGNA motif, with inversion of direction compared with the P01 sequence, which were separated by 1 bp, were also found in 100% (12 of 12 selected PBRs) of Abh-homomer-specific binding sequences (class P04). From sequences of the P02 pattern, a variant of the P01-type motif was extracted, which lacked the terminal A or T on one end but had an additional series of W bases at the other end, thus forming WWWWW-CCA-WWWW-TGG. This sequence was present in 14 of 18 selected PBRs. The Abh homomer does not bind to this sequence motif, suggesting that Abh binding may require strict conservation of the TGGNA motif.
In sequences of pattern P03, the AbrB-homomer-specific binding sequences, neither the TGGNA motif nor any other consensus sequence was detected, but A- and G-rich sequences were evident (Figure 5). This may indicate that, in addition to TGGNA motif-dependent binding, AbrB has an ability to bind to other sequences, possibly by recognizing a particular conformation of three-dimensional DNA, as previously proposed.
AbrB plays a major role in control of gene expression whereas Abh has a minor effect
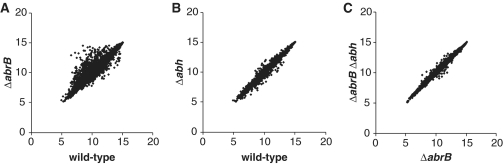
To understand the correlation between AbrB/Abh binding and transcriptional regulation, we analyzed genome-wide transcriptional profiles in wild-type (strain 168), ΔabrB (OC003), Δabh (OC004) and ΔabrB Δabh (OC005) cells during exponential growth, using the Affymetrix tiling chip, and the results are summarized in Supplementary Table S3. Scatter plots of the transcriptional intensities of each gene in deletion mutant cells compared with wild-type cells (Figure 6A and B) indicated that, as expected, deletion of abrB affected the expression of many genes, whereas abh deletion had little impact on the gene expression profile. Furthermore, the effect of the abrB/abh double deletion on the transcriptome was similar to that of the abrB deletion alone (Figure 6C).
Figure 6.
Transcriptome analysis in ΔabrB and Δabh mutants. Log-scale scatter plots of transcriptional intensities of each gene in ΔabrB (A, y-axis) and Δabh (B, y-axis) cells compared to those of wild-type cells (A and B, x-axis), and ΔabrBΔabh cells (C, y-axis), in comparison with that of ΔabrB cells (C, x-axis). The averaged signal intensities from two independent experiments using each strain are plotted.
Next, we searched for genes that were up- or downregulated by more than 2.5-fold in mutant cells compared with wild-type cells and investigated if AbrB/Abh-binding signals overlapped with the transcription start sites (TSSs) identified in our transcriptome analysis. We found that 90 transcriptional units (TUs) downregulated by more than 2.5-fold in ΔabrB mutant cells are associated with AbrB at their TSSs, suggesting that these TUs involving 171 genes with various functions are specifically repressed by AbrB probably via protein binding to promoter regions (Supplementary Table S6A). Notably, and consistent with the observation that abh deletion did not affect (or only marginally affected) expression, Abh-binding signals in the promoter regions of the 90 TUs were generally low (classes VL or L) in an abrB deletion background. However, significant levels of Abh binding (classes M or H) were detected in 20 operons, indicating that Abh binding in these regions did not significantly affect transcription.
The AbrB-regulated TUs identified included 11 operons/genes previously reported to be directly repressed by AbrB; these were spoVG, spo0E, sboAX-albABCDEFG, yknWXYZ, yxzE, tasA-sipW-yqxM, sunA-bdbB, skf(ybcO-ybcE), sdp(yvaWXY), yxaAB and eps(yvfF-yvek) (Supplementary Table S4). Derepression of the five TUs shown in Supplementary Table S4, sigH, sinIR, sigW-ybbM, com, and slr, was also seen in the abrB mutant, although the enhancement values (1.4–2.4-fold) were below our inclusion criterion. It is probable that expression levels of such genes during the exponential growth phase is lower than that in stationary phase, which has been examined in previous reports. No data supporting the previously reported direct repression by AbrB of a further six operons were obtained in the present work, although our data indicate that the pbpE-racX, lia and yvlABCD operons were indirectly affected by abrB deletion. The reasons for these inconsistencies are not presently clear.
Conversely, AbrB has previously been reported to act as a transcriptional activator of the ribose uptake (rbs) operon (4), and our data support this conclusion (Supplementary Table S6B). In addition, we found that four other operons involved in carbohydrate utilization, glp, gnt, gmu and amyE, were activated. Expression of the citB and hut operons has also been reported to be activated by AbrB (3,5). However, we found no effect of AbrB on hutB expression and indirect repression of citB expression by AbrB.
We also identified possible promoters directly regulated by Abh, although the number of such promoters was limited, as expected based on the overall effect of abh deletion on the transcriptome profile. In the case of the glp operon described above, strong Abh binding in a wild-type background and a moderate reduction of expression in an abh-deletion background were evident, indicating that the Abh homomer may act as an activator of this operon. At the sunA promoter, Abh bound with the same profile as shown by AbrB but had a different effect; Abh acted as an activator, whereas AbrB was a repressor, as previously reported (30,32) (Supplementary Table S6C). We also found that Abh repressed the expression of six operons/genes, yflA, ylaE, ctaCDEFG, yojL, ycdA and ywoF, in a manner additive to the effect of AbrB (Supplementary Table S6D). Finally, abh deletion resulted in stronger derepression of the srf operon compared with that seen with abrB deletion (Supplementary Table S6E). However, Abh binding to the srf promoter in the abrB-deletion background was weak, whereas AbrB binding was retained in the abh-deletion strain. Thus, the molecular mechanism by which srf operon expression is regulated by AbrB/Abh remains unclear.
These results clearly indicate that AbrB regulates the expression of many operons, acting mainly as a repressor but also as an activator in a limited number of instances. In contrast, Abh binding affords transcriptional regulation of only a small number of operons, at least during exponential growth.
Genome-wide correlation between AbrB/Abh binding and transcription levels
In the present study, we identified 643 AbrB- and 411 Abh-binding sites. However, only 103 AbrB- and 7 Abh-binding sites have been suggested to directly affect transcription. Our analysis may underestimate the number of TUs directly regulated by AbrB/Abh because AbrB/Abh sites involved in regulation of TUs specifically induced at the transition phase would not have been detected in our system. Abh is under the control of the ECF sigma factors σX, σM and σW (30,31); hence, cell wall stress may modulate AbrB/Abh regulation. Even when these possibilities are considered, our results indicate that many AbrB/Abh-binding events have no impact on transcription. In support of this idea, some binding sites are located in actively transcribed regions, such as those of the secDF, gltT, tkt, yrrO and tyrS genes (Figure 7A). Indeed, scatter plots of AbrB-binding intensities to PBRs, and ratios of PBR transcription intensities in abrB-deleted cells compared with wild-type cells, showed that most AbrB-binding events had no impact on transcriptional level, regardless of whether the binding occurred in intergenic or coding regions (Figure 7B–D). Thus, the cellular roles of many AbrB/Abh-binding events await further examination.
Figure 7.
AbrB/Abh binding to actively transcribed regions. (A) Examples of AbrB/Abh binding to coding regions of highly transcribed genes are shown. The AbrB/Abh binding profiles in four strains are depicted as in Figure 2A. The gene organization of selected regions is indicated in the middle. Below the gene map, transcriptional signals (y-axis) of each probe in the selected regions are presented separately for Watson (+) and Crick (–) strands. (B) A scatter plot of log2 binding intensity values of AbrB to intergenic regions (x-axis) and log2 expression ratios of downstream genes in ΔabrB cells compared to those in wild-type cells (y-axis). If AbrB bound to the intergenic region of divergently transcribed genes, we plotted the expression ratios of both genes. (C) Scatter plot of log2 binding intensity values of AbrB to coding regions (x-axis) and log2 expression ratios of AbrB-bound genes in ΔabrB cells compared to those in wild-type cells (y-axis). (D) The results shown in (B) and (C) are merged.
DISCUSSION
We demonstrated that AbrB and Abh bound to hundreds of sites throughout the genome in wild-type B. subtilis cells, and that almost all Abh-binding sites overlapped with sites for AbrB. The results of in vivo cross-linking experiments suggested that many of Abh molecules may exist in complex with AbrB in wild-type cells, and that Abh would be released from complexes with AbrB to form Abh homomers when the cellular levels of AbrB decreased. Consistent with this hypothesis, Abh-specific binding sites were detected only in the absence of AbrB. Thus, our results strongly suggest that the context of oligomeric forms of AbrB/Abh, and binding of these forms to the genome, are intimately related. Although previous in vitro experiments suggested that tetramers were the basic functional units of AbrB/Abh, the predominant molecular species in cells requires further examination. In addition, the molecular basis of AbrB/Abh heteromer formation, and whether C-terminal and/or N-terminal interaction(s) are involved, is an important issue requiring clarification.
Quantitative comparison of AbrB/Abh-binding intensities to relevant binding sites, defined here as PBRs, in wild-type, ΔabrB and Δabh backgrounds revealed that PBRs contain various types of sequences with different specificities and affinities for AbrB/Abh homomers and heteromers. We attempted to identify the basic patterns of AbrB/Abh binding to PBR sequences, and found four major (P01–P04 in Table 1) and four minor (P05–P08) patterns. Binding sites in the classes P01, P02 and P03 were recognizable by AbrB homomers. In addition, P01 sites were recognized by both Abh homomers and AbrB/Abh heteromers, and sites in the P02 class bound AbrB/Abh heteromers. P03 sites were specific for AbrB homomers. Interestingly, P04 sites were specific for Abh homomers, and Abh binding to these sites was detected only in the absence of AbrB. These results demonstrate that, although AbrB and Abh are similar in primary and tertiary structure, subtle structural differences are reflected in variations in target sequences, as might be anticipated.
Furthermore, by carefully selecting PBRs with single protein-binding peaks, we were able to extract consensus binding sequences for the P01, P02 and P04 classes. A previous in vitro SELEX study suggested that the TGGNA motif was a determinant of AbrB binding (23). Interestingly, our consensus sequences for the P01, P02, and P04 sites each contain two TGGNA motifs, differing in arrangement and spacing: TNCCA-WWWW-TGGNA for P01 sequences, WWWWW-CCA-WWWW-TGG for P02 sequences and TGGNAWTTCCA for P04 sequences. These results indicate that, in vivo, a pair of TGGNA sequences acts as a determinant of both AbrB and Abh binding, at least in part, although direct demonstration of AbrB/Abh binding to the sequences identified in the present work requires further investigation. The finding that Abh does not bind as a homomer to P02 sequences may indicate that Abh has a strict requirement for the TGGNA motif, whereas AbrB recognizes more diverse DNA sequences. The rather nonspecific AG-rich consensus sequence of the P03 class, the AbrB-homomer-specific binding site, might also support this idea.
To further evaluate the contribution of the TGGNA motif to genome-wide AbrB/Abh binding, we searched for the presence of two TGGNA motifs, in palindromic or tandem orientation, separated by 0 to 6 bp of W (A or T), allowing a single base mismatch, and determined if such sites were enriched in the 100-bp regions around the centers of 753 PBRs (Supplementary Table S7). Although our AbrB/Abh-binding motifs contained inverted repeats of the TGGNA sequence, in vitro experiments have shown that AbrB also binds to tandem repeats (23). Supporting the involvement of the TGGNA motif in AbrB/Abh-binding, palindromic or tandem motif pairs connected by 4–5 W bases were found to be enriched in PBR sequences. However, such sequences were detected in only 166 of 753 PBRs (22%). This may indicate that TGGNA-motif-dependent AbrB/Abh binding is relatively relaxed in specificity and that other constraints are required to restrict the targets to PBR regions because a search for possible binding sites allowing a two base-pair mismatch resulted in loss of site enrichment in PBR regions. In vitro footprinting experiments indicated that an AbrB/Abh complex often covers more than 40 bp of sequence, and it has been recently proposed that such observations indicate simultaneous binding of more than two AbrB tetramers (26,32). Multiple arrangements of sequences containing two TGGNA motifs, such as the M-5bp-M-4bp-M-5bp-M sequence identified in the SELEX study of Xu and Strauch (1997), may control AbrB/Abh binding to the genome. Our results also indicate that the AbrB homomer binds independently of the TGGNA motif, possibly recognizing a specific substructure of the DNA helix.
Although the exact contribution of the TGGNA motif to genome-wide AbrB/Abh binding requires further investigation, our results indicate that an AbrB/Abh complex has the ability to recognize various configurations of the two motifs, as shown schematically in Figure 5. The crystal structure of full-length SpoVT has been recently reported (39), in which two monomers dimerized by N-terminal domain interactions form swapped-hairpin β-barrels and then tetramerize through formation of mixed helix bundles between the C-terminal domains. Dimerized N-terminal DNA-binding domains and C-terminal domains are connected by flexible linker sequences, and such flexibility would also allow the two DNA-binding domains of AbrB/Abh tetramers to adopt various conformations.
We assessed the direct contributions of AbrB and Abh binding to control of gene expression by comparing AbrB/Abh binding in, and the transcriptome profiles of, abrB and/or abh deletion mutant cells. Our data indicate that expression of at least 90 TUs (171 genes) would be specifically repressed by AbrB in exponentially growing cells. Newly identified AbrB-repressed genes include genes involved in cell-wall biosynthesis (pbpH, dacF), membrane biogenesis (cccA), chemotaxis (tlpA), antibiotic production (ppsABCDE), metabolism of amino acids and related molecules (rocA, rocG), detoxification (ykuU, yocD), protein modification (tkmA, ptkA, ptpZ), phage-related functions and genes of unknown function (Supplementary Table S6). In addition, it has been suggested that AbrB is involved in the activation of five operons (22 genes) related to carbohydrate utilization. Although Abh-binding signals in these genetic regions were generally low (in the classes VL or L) in ΔabrB cells, signals in the M and H classes, but without any apparent effect on transcription level, were also observed. However, Abh has been suggested to repress the expression of six TUs (10 genes), in a manner additive to repression by AbrB. In addition, our data support the direct activation by Abh, and repression by AbrB, of the sun operon (30,32). The different effects of Abh binding on transcription may be explained by the levels of Abh bound to promoter regions. However, no clear correlation between Abh binding intensity and derepression of transcription was observed in ΔabrB cells (Supplementary Table S6). It is also possible that the relative location of Abh-binding sites with respect to promoters is important when the detailed effects on transcription are considered, although the limited resolution of our protein binding maps did not allow us to examine this possibility. The AbrB-binding profiles on promoters repressed and activated by the protein were also apparently indistinguishable. Thus, the molecular basis by which AbrB/Abh binding promotes repression or activation of transcription requires further detailed analysis.
We found that most Abh molecules were in a complex with AbrB in cells growing exponentially in LB medium, and Abh-homomer-specific binding sites were detected only in the absence of AbrB. Although we did not observe any changes in transcriptional levels around binding sites in exponentially growing ΔabrB cells, Abh-homomer-specific binding sites may appear in wild-type cells in the stationary phase, after AbrB expression is repressed, to regulate expression of growth phase-dependent gene transcription. Supporting this idea, an alteration in biofilm architecture was observed in abh-deleted cells compared with wild-type cells during biofilm formation (31,40). Conversely, a decrease in Abh level will increase AbrB homomer level and enhance protein binding to AbrB-homomer-specific binding sites. Interestingly, we found that the ydjL and yolA genes might be under this form of control (Supplementary Table S6). The expression of these genes is repressed specifically by the AbrB homomer, and this repression is further enhanced in a Δabh background. This may indicate that reduction in the level of the AbrB homomer is one of the biological functions of Abh during the exponential growth phase under normal conditions. These observations indicate that regulation of the relative levels of AbrB and Abh is one of the strategies used to modulate the global gene expression profile and cope with environmental changes, by reorganizing AbrB/Abh binding along the whole genome.
Finally, and importantly, we found that most AbrB and Abh binding events did not affect transcription, although we accept that our data may underestimate the number of genes directly regulated by AbrB and/or Abh. About half of the binding sites for these proteins are located in ORFs, and some such ORFs are actively transcribed in the presence or absence of AbrB/Abh. The other half of the binding sites are located in intergenic regions. However, binding to many such sites had no effect on transcription, even though the binding intensities were high. Recently, it has been shown that E. coli RutR, regulator for pyrimidine catabolism, binds mainly to coding regions with little or no effect on transcript levels (41), and that NsrR, regulator for adaptive responses to nitric oxide, also binds many sites in coding regions (42). It is possible that AbrB/Abh are transcriptional regulators with similar binding site preferences, although numbers of the binding sites of RutR and NsrR on the genome were 20 and 62, respectively, much smaller than those of AbrB and Abh.
Conversely, AbrB/Abh share some properties with E. coli nucleoid-associated proteins, which are abundant proteins of low molecular weight with a low sequence specificity for DNA binding, and the numerous AbrB/Abh-binding sites along the whole genome found in the present work further extend the similarity. Furthermore, the mutual dependence of AbrB/Abh for DNA binding is reminiscent of a property of the E. coli nucleoid protein H-NS and its paralog StpA; H-NS binding is apparently StpA-independent, whereas many of StpA binding is H-NS-dependent (43). These results imply another interesting possibility that, although sequence similarities are lacking, AbrB and Abh might be functional homologs of H-NS and StpA. E. coli nucleoid proteins participate in diverse DNA-dependent functions, including transcription, replication, recombination and the creation of higher-order structures in genomic DNA. AbrB/Abh may thus have function(s) other than transcriptional regulation. However, as yet there is no experimental evidence to support this hypothesis because abrB abh double mutant cells show no relevant phenotypic characteristics, such as an altered nucleoid morphology (data not shown).
Another possible hypothesis is that binding sites that are not involved in direct transcriptional regulation might act as pools of AbrB and Abh and enable gradual changes of AbrB/Abh bindings to promoter regions when AbrB/Abh levels are changed. Thus, even abrupt change in their levels might be followed by a gradual alteration of transcriptome, providing a time toward an overall response required to adapt to new conditions.
In summary, our ChAP–chip analysis, together with transcriptome profiling, revealed that AbrB and Abh are not simple transcriptional regulators. We disclosed a close relationship between AbrB and Abh with respect to genome-wide binding and transcriptional regulation. Thus, regulation of the relative levels of AbrB and Abh would be one of the strategies used to modulate the global gene expression profile and cope with environmental changes. Furthermore, it is possible that these proteins play roles similar to those of E. coli nucleoid-associated proteins. This insight may further our understanding of the molecular mechanism of nucleoid structure formation in B. subtilis, which is poorly understood at present.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A Grant-in-Aid for Scientific Research (KAKENHI) on Priority Areas, Systems Genomics, from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and a Japanese Government Scholarship for Foreign Research Students (to C.O.). Funding for open access charge: Research fund from Nara Institute of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Phillips ZE, Strauch MA. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 2002;59:392–402. doi: 10.1007/s00018-002-8431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauch MA, Hoch JA. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol. Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 3.Kim HJ, Kim SI, Ratnayake-Lecamwasam M, Tachikawa K, Sonenshein AL, Strauch M. Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 2003;185:1672–1680. doi: 10.1128/JB.185.5.1672-1680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauch MA. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J. Bacteriol. 1995;177:6727–6731. doi: 10.1128/jb.177.23.6727-6731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher SH, Strauch MA, Atkinson MR, Wray LV., Jr Modulation of Bacillus subtilis catabolite repression by transition state regulatory protein AbrB. J. Bacteriol. 1994;176:1903–1912. doi: 10.1128/jb.176.7.1903-1912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Reilly M, Devine KM. Expression of AbrB, a transition state regulator from Bacillus subtilis, is growth phase dependent in a manner resembling that of Fis, the nucleoid binding protein from Escherichia coli. J. Bacteriol. 1997;179:522–529. doi: 10.1128/jb.179.2.522-529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauch M, Webb V, Spiegelman G, Hoch JA. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl Acad. Sci. USA. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banse AV, Chastanet A, Rahn-Lee L, Hobbs EC, Losick R. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc. Natl Acad. Sci. USA. 2008;105:15547–15552. doi: 10.1073/pnas.0805203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobay BG, Andreeva A, Mueller GA, Cavanagh J, Murzin AG. Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins. FEBS Lett. 2005;579:5669–5674. doi: 10.1016/j.febslet.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 10.Coles M, Djuranovic S, Soding J, Frickey T, Koretke K, Truffault V, Martin J, Lupas AN. AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi beta barrels. Structure. 2005;13:919–928. doi: 10.1016/j.str.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Benson LM, Vaughn JL, Strauch MA, Bobay BG, Thompson R, Naylor S, Cavanagh J. Macromolecular assembly of the transition state regulator AbrB in its unbound and complexed states probed by microelectrospray ionization mass spectrometry. Anal. Biochem. 2002;306:222–227. doi: 10.1006/abio.2002.5704. [DOI] [PubMed] [Google Scholar]
- 12.Bobay BG, Benson L, Naylor S, Feeney B, Clark AC, Goshe MB, Strauch MA, Thompson R, Cavanagh J. Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry. 2004;43:16106–16118. doi: 10.1021/bi048399h. [DOI] [PubMed] [Google Scholar]
- 13.Vaughn JL, Feher VA, Bracken C, Cavanagh J. The DNA-binding domain in the Bacillus subtilis transition-state regulator AbrB employs significant motion for promiscuous DNA recognition. J. Mol. Biol. 2001;305:429–439. doi: 10.1006/jmbi.2000.4305. [DOI] [PubMed] [Google Scholar]
- 14.Xu K, Strauch MA. DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein. J. Bacteriol. 2001;183:4094–4098. doi: 10.1128/JB.183.13.4094-4098.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao F, Strauch MA. Independent and interchangeable multimerization domains of the AbrB, Abh, and SpoVT global regulatory proteins. J. Bacteriol. 2005;187:6354–6362. doi: 10.1128/JB.187.18.6354-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucking G, Dommel MK, Scherer S, Fouet A, Ehling-Schulz M. Cereulide synthesis in emetic Bacillus cereus is controlled by the transition state regulator AbrB, but not by the virulence regulator PlcR. Microbiology. 2009;155:922–931. doi: 10.1099/mic.0.024125-0. [DOI] [PubMed] [Google Scholar]
- 17.Saile E, Koehler TM. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 2002;184:370–380. doi: 10.1128/JB.184.2.370-380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii A, Hihara Y. An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803. Plant Physiol. 2008;148:660–670. doi: 10.1104/pp.108.123505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agervald A, Zhang X, Stensjo K, Devine E, Lindblad P. CalA, a cyanobacterial AbrB protein, interacts with the upstream region of hypC and acts as a repressor of its transcription in the cyanobacterium Nostoc sp. strain PCC 7120. Appl. Environ. Microbiol. 2010;76:880–890. doi: 10.1128/AEM.02521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieman-Hurwitz J, Haimovich M, Shalev-Malul G, Ishii A, Hihara Y, Gaathon A, Lebendiker M, Kaplan A. A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 2009;11:927–936. doi: 10.1111/j.1462-2920.2008.01818.x. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira P, Lindblad P. An AbrB-Like protein regulates the expression of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2008;190:1011–1019. doi: 10.1128/JB.01605-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shalev-Malul G, Lieman-Hurwitz J, Viner-Mozzini Y, Sukenik A, Gaathon A, Lebendiker M, Kaplan A. An AbrB-like protein might be involved in the regulation of cylindrospermopsin production by Aphanizomenon ovalisporum. Environ. Microbiol. 2008;10:988–999. doi: 10.1111/j.1462-2920.2007.01519.x. [DOI] [PubMed] [Google Scholar]
- 23.Xu K, Strauch MA. In vitro selection of optimal AbrB-binding sites: comparison to known in vivo sites indicates flexibility in AbrB binding and recognition of three-dimensional DNA structures. Mol. Microbiol. 1996;19:145–158. doi: 10.1046/j.1365-2958.1996.358882.x. [DOI] [PubMed] [Google Scholar]
- 24.Strauch MA. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promote. J. Bacteriol. 1995;177:6999–7002. doi: 10.1128/jb.177.23.6999-7002.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bobay BG, Mueller GA, Thompson RJ, Murzin AG, Venters RA, Strauch MA, Cavanagh J. NMR structure of AbhN and comparison with AbrBN: FIRST insights into the DNA binding promiscuity and specificity of AbrB-like transition state regulator proteins. J. Biol. Chem. 2006;281:21399–21409. doi: 10.1074/jbc.M601963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan DM, Bobay BG, Kojetin DJ, Thompson RJ, Rance M, Strauch MA, Cavanagh J. Insights into the nature of DNA binding of AbrB-like transcription factors. Structure. 2008;16:1702–1713. doi: 10.1016/j.str.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauch MA, Ayazifar M. Bent DNA is found in some, but not all, regions recognized by the Bacillus subtilis AbrB protein. Mol. Gen. Genet. 1995;246:756–760. doi: 10.1007/BF00290723. [DOI] [PubMed] [Google Scholar]
- 29.Bagyan I, Hobot J, Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 1996;178:4500–4507. doi: 10.1128/jb.178.15.4500-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Y, Helmann JD. Extracytoplasmic function σ factors with overlapping promoter specificity regulate sublancin production in Bacillus subtilis. J. Bacteriol. 2009;191:4951–4958. doi: 10.1128/JB.00549-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray EJ, Strauch MA, Stanley-Wall NR. σX is involved in controlling Bacillus subtilis biofilm architecture through the AbrB homologue Abh. J. Bacteriol. 2009;191:6822–6832. doi: 10.1128/JB.00618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauch MA, Bobay BG, Cavanagh J, Yao F, Wilson A, Le Breton Y. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J. Bacteriol. 2007;189:7720–7732. doi: 10.1128/JB.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishikawa S, Ogura Y, Yoshimura M, Okumura H, Cho E, Kawai Y, Kurokawa K, Oshima T, Ogasawara N. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 2007;14:155–168. doi: 10.1093/dnares/dsm017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morimoto T, Kadoya R, Endo K, Tohata M, Sawada K, Liu S, Ozawa T, Kodama T, Kakeshita H, Kageyama Y, et al. Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis. DNA Res. 2008;15:73–81. doi: 10.1093/dnares/dsn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rukmana A, Morimoto T, Takahashi H, Giyanto, Ogasawara N. Assessment of transcriptional responses of Bacillus subtilis cells to the antibiotic enduracidin, which interferes with cell wall synthesis, using a high-density tiling chip. Genes Genet. Syst. 2009;84:253–267. doi: 10.1266/ggs.84.253. [DOI] [PubMed] [Google Scholar]
- 36.Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2004. [Google Scholar]
- 38.Bailey TL, Elkan CP. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, California: AAAI Press; 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- 39.Asen I, Djuranovic S, Lupas AN, Zeth K. Crystal structure of SpoVT, the final modulator of gene expression during spore development in Bacillus subtilis. J. Mol. Biol. 2009;386:962–975. doi: 10.1016/j.jmb.2008.10.061. [DOI] [PubMed] [Google Scholar]
- 40.Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimada T, Ishihama A, Busby SJ, Grainger DC. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008;36:3950–3955. doi: 10.1093/nar/gkn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009;73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- 43.Uyar E, Kurokawa K, Yoshimura M, Ishikawa S, Ogasawara N, Oshima T. Differential binding profiles of StpA in wild-type and h-ns mutant cells: a comparative analysis of cooperative partners by chromatin immunoprecipitation-microarray analysis. J. Bacteriol. 2009;191:2388–2391. doi: 10.1128/JB.01594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.