Abstract
Purpose
In the development of therapeutic vaccines against cancer, it is important to design strategies for antigen cross-presentation to stimulate cell-mediated immune responses against tumor antigens.
Methods
We developed a polyethyleneimine (PEI)-based protein antigen delivery system to promote cross-presentation through the major histocompatibility complex (MHC) I pathway using ovalbumin (OVA) as a model antigen. PEIs formed nanoparticles with OVA by electrostatic interactions, as demonstrated by electrophoresis analysis, scanning electron microscopy, and photon correlation spectroscopy analysis.
Results
The nanoparticles were used to stimulate mouse bone marrow-derived dendritic cells in vitro and resulted in significantly more OVA257–264/MHC I complex presentation on dendritic cell surfaces. The activated dendritic cells interacted specifically with RF33.70 to stimulate interleukin-2 secretion. The cross-presentation promoting effect was more prominent in dendritic cells that had been cultured for longer periods of time (13 days). Further studies comparing the antigen presentation efficacies by other polyanionic agents, such as PLL or lysosomotropic agents, suggested that the unique “proton sponge effect” of PEI facilitated antigen escape from the endosome toward the MHC I pathway.
Conclusion
Such a PEI-based nanoparticle system may have the potential to be developed into an effective therapeutic vaccine delivery system.
Keywords: cross-presentation, polyethyleneimine, dendritic cells, vaccine
Introduction
Cancer vaccines are viewed as promising tools to fight tumors.1,2 The aim of a cancer vaccine is to activate the patient’s own immune system against tumor cells. There have been many cancer vaccine strategies proposed and tested, including tumor cell lysates, tumor antigen-pulsed dendritic cells, recombinant tumor antigens, and various DNA vaccine constructs.3,4 However, the outcomes are highly variable and there have been safety and quality control problems. Based on these concerns, vaccine constructs containing recombinant tumor-associated antigens or epitopes are considered more desirable because of the advantages of easy production, convenient administration, and proven safety.5
Protein-based cancer vaccines in general have limited therapeutic efficacy because they induce predominantly antibody responses instead of antigen-specific cytotoxic T lymphocyte responses, which are critical for immunity against tumors.6 Controlling the antigen-processing process inside antigen-presenting cells and presenting them through the major histocompatibility complex (MHC) I pathway is the key to stimulating cytotoxic T lymphocytes.7,8 Nanoparticle-based delivery systems have shown some efficacy in protecting antigens from degradation and improving uptake by antigen-presenting cells.9 In order to help the antigens to escape from endosomes inside antigen-presenting cells and to access the MHC I pathway, some endosome-disruptive agents were applied. Based on the listeriolysin O protein, Stier et al designed a listeriolysin O-liposome system that could create pores within the endosomal/lysosomal membrane.10 Bungener et al used reconstituted virosomes from the influenza virus and reported membrane fusion between the virosome membrane and the endosome membrane.11 Kwon et al and Murthy et al prepared a responsive polymer microsphere that degraded in the acidic lysosome to release the antigen.12,13 Antigen cross-presentation was considered essential for the efficacy of therapeutic vaccines.
In this study we developed a new antigen delivery system based on the cationic polymer polyethyleneimine (PEI) and demonstrated that its endosome-disruptive effect was sufficient and effective for cross-presentation. Ovalbumin (OVA) was used as a model antigen, and various aspects of immune recognition and stimulation were demonstrated. Mouse bone marrow-derived dendritic cells were used to evaluate cross-presentation efficiency, because dendritic cells were viewed as the most powerful antigen-presenting cells.14,15 Our study indicates that the PEI-based nanoparticles show promise as antigen delivery systems for therapeutic cancer vaccines.
Materials and methods
Materials
OVA257–264 peptide was synthesized by GL Biochem Ltd, Shanghai, China. OVA and PEI (branched, 10 kDa) were purchased from Sigma-Aldrich, St Louis, MO, USA. Goat antimouse IgG–FITC was purchased from the KangChen Bio-tech Company, Shanghai, China. Mouse granulocyte–macrophage colony-stimulating factor and mouse interleukin (IL)-4 were purchased from R & D Systems, Minneapolis, MN, USA. Cell culture media were obtained from Gibco Invitrogen, Carlsbad, CA, USA.
RF33.70 cells (Kb restricted T cell hybridoma and specific for OVA257–264 ) were kindly provided by Professor Xuetao Cao from the Second Military Medical University, Shanghai, China. These cells were maintained in RPMI-1640 culture medium with 10% heat-inactivated fetal bovine serum and antibiotics.
The hybridoma cells that produced the monoclonal antibody 25-D1.16, specific for the OVA257–264/MHC I complex, were kindly provided by Professor Kyung-Dall Lee from the University of Michigan, Ann Arbor, MI, USA. The cell line was grown in RPMI-1640 with 10% fetal bovine serum.
Five- to six-week-old male C57BL/6 mice (H-2b) were purchased from the Shanghai Laboratory Animal Center, Shanghai, China, and were housed in a specific pathogen-free environment at Shanghai Jiao Tong University School of Pharmacy. All experiments were approved by the Animal Care and Use Committee of Shanghai Jiao Tong University School of Pharmacy, Shanghai, China.
The bone marrow-derived dendritic cells were prepared based on the typical procedure described in the literature.16 Briefly, bone marrow from C57BL/6 mice was collected and cultured in RPMI-1640 complete medium (10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, 100 U/mL streptomycin) supplemented with 10 ng/mL mouse granulocyte–macrophage colony-stimulating factor and 1 ng/mL mouse IL-4. Nonadherent cells were removed after three days, and the adherent cells were replanted in fresh RPMI-1640 complete medium supplemented with mouse granulocyte–macrophage colony-stimulating factor and IL-4. The nonadherent and loosely adherent cells were harvested as dendritic cells after another three days.
Preparation of PEI–OVA and PLL–OVA nanoparticles
The nanoparticles were prepared following the protocols that were used to prepare polymer–DNA complex with modification.17,18 OVA was dissolved in 1 mM HEPES (pH 7.4) solution to get an OVA concentration of 5 mg/mL. Branched PEI10000 (Sigma) or PLL26000 (Sigma) was dissolved in distilled water to yield different concentration. Polymer–OVA particles were prepared by adding OVA solution to polymer solution at equal volume and vortexing for 30 seconds at room temperature.
Nanoparticle characterization
The polymer–OVA nanoparticles were analyzed using both native sodium dodecyl sulfate polyacrylamide gel electrophoresis (PAGE, 8%–16% Tris-Glycine gel) and denatured sodium dodecyl sulfate PAGE (4%–12% Bis-Tris gel from Invitrogen). Gels were stained with Coomassie Blue (Invitrogen).
The surface morphology of nanoparticles was observed by scanning electron microscopy (SEM, FEI SIRION 200 system, FEI Company). After the samples were deposited on silica wafers and dried, they were observed at 5 kV. The size distribution and zeta potential of the nanoparticles were determined using photon correlation spectroscopy (Zetasizer Nano ZS90, Malvern Corp, Malvern, UK) at a scattering angle of 90°C and a temperature of 25°C.
OVA257–264/MHC I complex characterization on dendritic cells
Polymer–OVA nanoparticles were added to dendritic cells at different concentrations. Dendritic cells were incubated overnight at 37°C and then harvested and washed with phosphate-buffered saline. The expression of OVA257–264/MHC I on the surface of the dendritic cells was determined by staining the cells with 25-D1.16 monoclonal antibodies at 4°C. Thirty minutes later, the dendritic cells were washed and incubated with secondary antibody for a further 30 minutes. The dendritic cells were then washed again and analyzed by flow cytometry.
IL-2 secretion by OVA257–264/MHC I complex-specific T cells
After addition of polymer–OVA nanoparticles, dendritic cells were harvested after 12 hours and diluted by RPMI-1640 complete medium. 2 × 104 dendritic cells and 2 × 105 RF33.70 cells per well were mixed and incubated in 96-well plates. After 24 hours of incubation, IL-2 concentration in the culture supernatant was determined by mouse IL-2 enzyme-linked immunosorbent assay (ELISA) kit (Bender MedSystems, Austria).
Results
Preparation and characterization of PEI–OVA particles
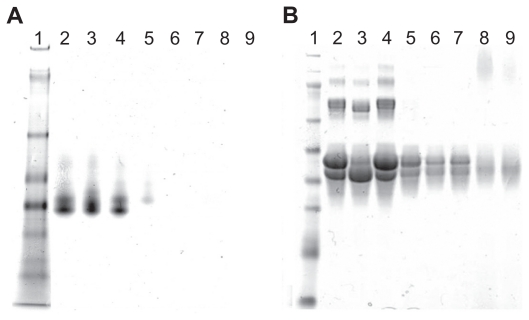
We prepared PEI–OVA nanoparticles based on electrostatic interactions between the positive amino groups on PEI and the negative surface charges on OVA above its pKa. The two agents were mixed in different weight ratios, and the resulting complexes were examined by gel electrophoresis. Figure 1A shows the native gel electrophoresis analysis of the complexes, which indicates that OVA was completely combined with PEI when the PEI/OVA weight ratio was over 0.04. However, the sodium dodecyl sulfate PAGE analysis showed that free OVA still existed at the same weight ratio (Figure 1B). This phenomenon demonstrated that sodium dodecyl sulfate could compete with OVA and disturb the combination of PEI and OVA.
Figure 1.
Combination of PEI and OVA. A) Native gel electrophoresis of PEI–OVA particles in different ratios (PEI/OVA w/w). Lane 1, protein marker; Lane 2, OVA; Lane 3, 0.01; Lane 4, 0.02; Lane 5, 0.04; Lane 6, 0.06; Lane 7, 0.08; Lane 8, 0.12; Lane 9, 0.16. B) SDS–PAGE of PEI–OVA particles in different weight ratios. Lane 1, protein marker; Lane 2, OVA; Lane 3, 0.01; Lane 4, 0.02; Lane 5, 0.04; Lane 6, 0.06; Lane 7, 0.08; Lane 8, 0.12; Lane 9, 0.16.
Abbreviations: PEI, polyethyleneimine; OVA, ovalbumin; SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
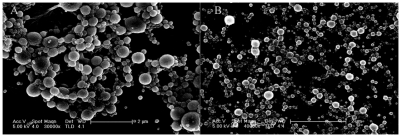
The microscopic morphology of the resulting particles was examined by SEM. The PEI–OVA nanoparticles were, in general, spherical and polydispersed. The SEM image of PEI–OVA 0.04 nanoparticles is shown in Figure 2A.
Figure 2.
SEM image of A) PEI–OVA and B) PLL–OVA particles on silicon wafer.
Abbreviations: PLL, polylysine; PEI, polyethyleneimine; OVA, ovalbumin; SEM, scanning electromicroscopy.
The particles were further evaluated by measuring their size distributions and surface zeta potential. As shown in Table 1, the zeta potentials of the particles increased with increasing ratios of PEI to OVA. The particles were positive when the PEI/OVA ratios were over 0.12. Table 1 also shows the average sizes of these particles, which were around several hundred nanometers between PEI/OVA ratios of 0.01 to 0.16.
Table 1.
Average size and zeta potential of PEI–OVA particles were varied when mixed in different ratios
| PEI/OVA weight ratio | 0.01 | 0.02 | 0.04 | 0.06 | 0.08 | 0.12 | 0.16 |
| Average size (nm) | 69.4 | 104.4 | 169.0 | 342.7 | 359.7 | 262.0 | 297.5 |
| Zeta potential (mV) | −23.1 | −20.3 | −12.4 | −4.6 | −4.4 | 12.4 | 20.0 |
Abbreviations: PEI, polyethyleneimine; OVA, ovalbumin.
For comparison, we also prepared different polycation–antigen nanoparticles. PLL is another kind of widely used cationic polymer in gene transfection.19–21 PLL could form nanoparticles with OVA by electronic interaction. The zeta potential and size distribution were also determined by photon correlation spectroscopy. The results were similar to those we obtained from PEI–OVA nanoparticles (Table 2). SEM results also showed that the surface morphology looks similar (Figure 2B).
Table 2.
Average size and zeta potential of PLL–OVA particles were varied when mixed in different ratios
| PLL/OVA weight ratio | 0.01 | 0.02 | 0.04 | 0.06 | 0.08 | 0.12 | 0.16 |
| Average size (nm) | 51.8 | 109.7 | 172.8 | 124.4 | 92.4 | 82.4 | 80.8 |
| Zeta potential (mV) | −18.4 | −17.6 | −5.8 | 32.9 | 35.6 | 43.5 | 47.7 |
Abbreviations: PLL, polylysine; OVA, ovalbumin.
Improved OVA257–264/MHC I complex presentation on dendritic cells
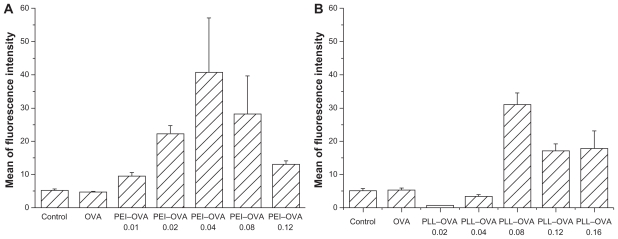
OVA antigen processing and epitope loading by dendritic cells after interacting with the PEI–OVA nanoparticles were examined by determining the expression of OVA257–264/MHC I complexes on dendritic cell surfaces using the specific antibody 25-D1.16 monoclonal antibodies.22 No surface OVA257–264/MHC I complexes were detected by the 25-D1.16 monoclonal antibodies on naïve dendritic cells. Pulsing the dendritic cells with OVA antigen alone would generally result in antigen processing inside the endosome/lysosome and presentation through the MHC I pathway. Therefore, the OVA257–264/MHC I complex signal was very low. Pulsing with PEI–OVA nanoparticles resulted in antigen release from endosomes and cross-presentation through the MHC I pathway. Figure 3A shows the dendritic cell surface OVA 257–264/MHC I signals after pulsing with PEI–OVA nanoparticles at different ratios. The PEI–OVA nanoparticle group that was prepared in a weight ratio of 0.04 had the highest mean signal intensity, approximately eight times higher than that of the OVA group.
Figure 3.
Quantification of dendritic cell surface OVA257–264/MHC I complexes. A) OVA257–264/MHC I complexes on the surface of dendritic cells after pulsing with PEI–OVA nanoparticles which were made using different ratios of PEI and OVA. Data are presented as mean ± standard deviation, n = 3. B) OVA257–264/MHC I complexes on the surface of dendritic cells after pulsing with PLL–OVA nanoparticles which were made using different ratios of PLL and OVA. Data are presented as mean ± standard deviation, n = 3, *P < 0.05 versus OVA solution group, **P < 0.01 versus control group.
Abbreviations: PEI, polyethyleneimine; OVA, ovalbumin; PLL, polylysine; MHC, major histocompatibility complex.
The dendritic cell surface OVA257–264/MHC I signals after pulsing with PLL–OVA nanoparticles at different ratios showed a similar trend. The highest signal intensity was obtained at a weight ratio of 0.08 (Figure 3B).
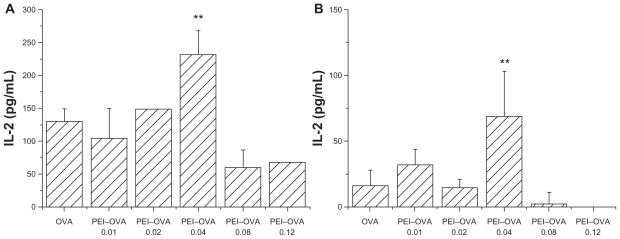
IL-2 secretion by early and late dendritic cells after PEI–OVA pulsing
PEI–OVA pulsed dendritic cells were then incubated with a murine T cell hybridoma specific for OVA257–264 (RF33.70), and the resulting IL-2 concentration in the supernatant was measured as an indicator of the T cell stimulation effect of dendritic cells. Only those dendritic cells that contained antigens cross-presented by MHC I complexes interacted specifically with these T cells and upregulated IL-2 secretion. Two types of pulsed dendritic cells were examined. Bone marrow-derived dendritic cells that were harvested at day 6 were designated as early dendritic cells, whereas the cells harvested at day 13 were designated as late dendritic cells.11 In both cases, dendritic cells pulsed by the PEI–OVA 0.04 nanoparticles showed a significantly improved T cell stimulation effect compared with dendritic cells pulsed by OVA solution at the same concentration (Figure 4). We can see that the increasing extent was more obvious in late dendritic cells, although the value was larger in early dendritic cells.
Figure 4.
Improved cross-presentation efficiency on early dendritic cells and late dendritic cells. Interleukin-2 concentration in supernatant was measured by ELISA when RF33.70 cells were coincubated for 24 hours with dendritic cells harvested at six days A) or dendritic cells harvested at 13 days, B) pulsed with PEI–OVA nanoparticles. Data are presented as mean ± SD, n = 4, **P < 0.01 versus OVA solution group.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; PEI, polyethyleneimine; OVA, ovalbumin.
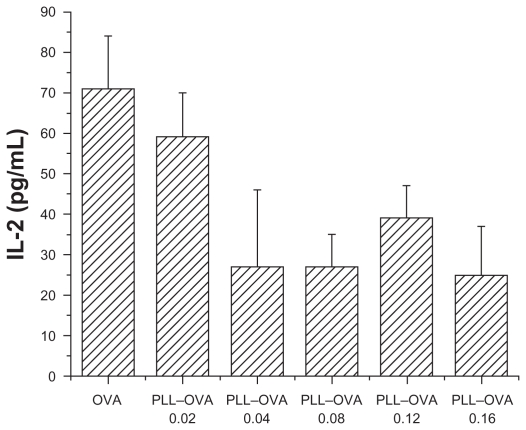
IL-2 secretion by dendritic cells after PLL–OVA pulsing
In contrast, as shown in Figure 5, there was no detectable increase observed in IL-2 secretion when RF33.70 cells were coincubated with dendritic cells pulsed by PLL–OVA nanoparticles prepared at all weight ratios. This indicated that PLL–OVA nanoparticles could not improve antigen cross-presentation or stimulate antigen-specific T cells.
Figure 5.
PLL–OVA nanoparticles did not improve antigen cross presentation effect. Interleukin-2 concentration in supernatant was measured by ELISA when RF33.70 cells were coincubated for 24 hours with dendritic cells harvested at six days. Data are presented as mean ± standard deviation, n = 4, *P < 0.05 versus OVA solution group, **P < 0.01 versus OVA solution group.
Abbreviations: PEI, polyethyleneimine; OVA, ovalbumin; ELISA, enzyme-linked immunosorbent assay.
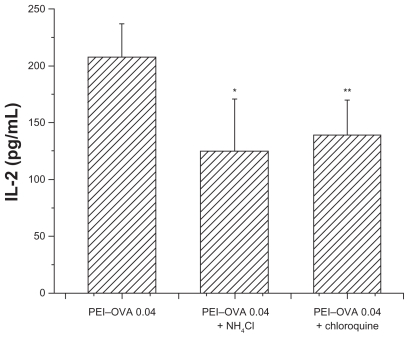
Effect of chloroquine and NH4Cl on OVA processing and cross-presentation
Because compounds such as chloroquine and NH4Cl had been reported to be able to buffer and delay acidification of endosomes and lysosomes, the intracellular fate of exogenous antigens may be changed by addition of these compounds. To study the mechanism of antigen processing, we tested their effects on the outcome of antigen processing and presentation after PEI–OVA pulsing. As shown in Figure 6, these two compounds hindered the cross-presentation efficiency of dendritic cells, which resulted in decreased IL-2 expression following incubation with RF33.70 cells, as compared with the effect when the endosomes/lysosomes had a normal acidification process.
Figure 6.
Interleukin-2 concentration was slightly decreased when RF33.70 cells were coincubated for 24 hours with dendritic cells pulsed with PEI-OVA nanoparticles in the presence of chloroquine or NH4Cl. Data are presented as mean ± standard deviation, n = 4, *P < 0.05, **P < 0.01 versus PEI-OVA nanoparticles group.
Abbreviations: PEI, polyethyleneimine; OVA, ovalbumin.
Discussion
The intracellular fate of antigens is pivotal to the immune effect. Exogenous antigens are usually degraded by enzymes in acidic endosomes, which causes failure to induce the cytotoxic T lymphocyte response. This is one of the main difficulties in the development of therapeutic cancer vaccines. As discussed earlier, methods have been developed to assist antigen escape and showed improved cross-presentation. The aim of our research was to provide a new strategy for antigen delivery.
We prepared PEI–OVA nanoparticles by mixing PEI solution and OVA solution together. The nanoparticles interacted due to electrostatic interactions between the cationic polymer–PEI and the negatively charged protein–OVA. The electrostatic complexation process was confirmed by electrophoresis, SEM, zeta potential, and particle size analysis. The nanoparticles formed were quite stable and their electrophoretic profiles, zeta potential, and size distribution data remained the same for several days. Stability is important in the potential application of these nanoparticles as therapeutic vaccine carriers.
The OVA257–264 peptide is a class I (Kb)-restricted peptide epitope of OVA that is presented by the MHC I complex on antigen-presenting cells. We measured the amount of OVA257–264/MHC I complexes on dendritic cell surfaces as an indicator of the efficiency of antigen cross-presentation after OVA antigen pulsing. The 25-D1.16 antibody specifically recognizes OVA257–264/MHC I complexes.23 Our data indicated that the expression of OVA257–264/MHC I increased significantly after pulsing with PEI–OVA nanoparticles overnight. The extent of this increment was also found to be related to the PEI–OVA mixing weight ratio. The nanoparticles with best immune effect were prepared at a weight ratio of 0.04. The cross-presentation effect was further confirmed by IL-2 secretion of RF33.70 cells that were OVA257–264/MHC I-specific hybridoma cells. The dendritic cells pulsed by PEI–OVA nanoparticles at a weight ratio of 0.04 stimulated the most significant IL-2 secretion by RF33.70 cells. Other PEI–OVA ratio particles had very little effect. We suspect this may be because PEI had some cytotoxicity, which interfered with the interaction between dendritic cells and RF33.70 cells. If a less toxic cationic polymer can be developed with similar functions, we may see greater improvements in antigen processing and cross-presentation efficiencies.
On the contrary, the cross-presentation effect of the PLL–OVA nanoparticles was somewhat different. We did observe a significant OVA257–264/MHC I signal increase with 25-D1.16 antibody staining, but there was no stimulation of IL-2 secretion by RF33.70 cells. The highest 25-D1.16 antibody staining signal was observed at a PLL to OVA weight ratio of 0.08 and above. Those nanoparticles had quite high positive surface charges, which may attract nonspecific binding of 25-D1.16 antibodies, resulting in false signal increase. However, because the PLL–OVA pulsed dendritic cells had no effect on stimulating IL-2 secretion by RF33.70 cells, we concluded that PLL–OVA nanoparticles were not capable of promoting antigen cross-presentation in dendritic cells.
To understand the mechanism of PEI-mediated antigen cross-presentation, Yang and Hsu, and Yan et al proposed that it may be due to the positive surface charge that promoted uptake by dendritic cells and promoted the production of reactive oxygen species. The main evidence was the increased expression of CD80/CD86 on dendritic cell surfaces.24,25 However, our findings were different. The PEI–OVA 0.04 nanoparticles had a negative surface potential, and there was no significant upregulation of CD80/86 expression on dendritic cells. However, the improvement of specific OVA257–264/ MHC I presentation and functional stimulation of IL-2 secretion was significant. We believe that the PEI–OVA nanoparticles we prepared may be effective as a result of the special endosome-disruptive effect of PEI.
PEI is well known for its proton sponge effect, whereby the unprotonated amines of PEI can buffer influent protons, leading to osmotic swelling and rupture of endosomes.26 Such an endosomal rupturing effect may help antigen release from the endosome and its subsequent cross-presentation by the MHC I pathway. To find support for such a hypothesis, we further examined the effect of endosomal pH on cross-presentation efficiency. We made an interesting observation that the PEI effect on cross-presentation with dendritic cells harvested at 13 days was more prominent than that with dendritic cells harvested at six days. This may be due to the different endosome acidification characteristics in dendritic cells harvested at six days versus dendritic cells harvested at 13 days. Endosomal acidification was believed to control exogenous antigen degradation by influencing the activity of lysosomal proteases.27 Hotta et al reported that late dendritic cells were usually not capable of cross-presenting exogenous antigens because the endocytic compartments would acidify rapidly in late dendritic cells after antigen uptake, whereas in early dendritic cells the pH would be maintained at a mildly acidic level and allow some antigen escape from endosomes for cross-presentation.28 Therefore, our data suggest that PEI can buffer the influent protons in late dendritic cells and prevent rapid acidification, which resulted in greatly improved cross-presentation efficiency in dendritic cells harvested at 13 days.
We also examined the effects of two other lysosomotropic agents, chloroquine and NH4Cl, on cross-presentation efficiency. Both chloroquine and NH4Cl inhibit acidification of endosomes and have been reported to increase crosspresentation of exogenous antigens.29–31 However, treatment with chloroquine or NH4Cl actually reduced PEI–OVA nanoparticle-mediated cross-presentation (Figure 6). We think this is because the inhibition of endosome acidification by chloroquine and NH4Cl reduced the proton influx and prevented endosome rupture and antigen escape. This is consistent with a previous report that the use of chloroquine slightly decreased the gene transfection activity of PEI.17
Conclusion
In summary, our data demonstrate that PEI–OVA nanoparticles were formed by electronic interactions, and the proton sponge effect of PEI was essential for promoting antigen cross-presentation for therapeutic vaccine application. This system is attractive due to its simplicity, flexibility, stability, and high loading efficiency. The main limitation is the toxicity of PEI.32 Further studies are needed to synthesize new polymers that have the proton sponge effect but lower toxicity. Additionally, a targeting strategy may be necessary for in vivo applications.33
Acknowledgments
This work was supported by grants from the National High Technology Research and Development Program of China (No. 2007AA021104) and the Natural Science Foundation of China (No. 30825045). We thank Professor Xuetao Cao from the Second Military Medical University, China, and Kyung-Dall Lee from the University of Michigan, Ann Arbor, MI, USA, for providing key reagents for this study.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Rescigno M, Avogadri F, Curigliano G. Challenges and prospects of immunotherapy as cancer treatment. Biochim Biophys Acta. 2007;1776(1):108–123. doi: 10.1016/j.bbcan.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Itoh K, Yamada A, Mine T, Noguchi M. Recent advances in cancer vaccines: An overview. Jpn J Clin Oncol. 2009;39(2):73–80. doi: 10.1093/jjco/hyn132. [DOI] [PubMed] [Google Scholar]
- 3.Dermime S, Armstrong A, Hawkins RE, Stern PL. Cancer vaccines and immunotherapy. Br Med Bull. 2002;62:149–162. doi: 10.1093/bmb/62.1.149. [DOI] [PubMed] [Google Scholar]
- 4.Espinoza-Delgado I. Cancer vaccines. Oncologist. 2002;7(Suppl 3):20–33. doi: 10.1634/theoncologist.7-suppl_3-20. [DOI] [PubMed] [Google Scholar]
- 5.Pejawar-Gaddy S, Finn OJ. Cancer vaccines: Accomplishments and challenges. Crit Rev Oncol Hematol. 2008;67(2):93–102. doi: 10.1016/j.critrevonc.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Conejo-Garcia JR, Katsaros D, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 7.Guermonprez P, Amigorena S. Pathways for antigen cross presentation. Springer Semin Immunopathol. 2005;26(3):257–271. doi: 10.1007/s00281-004-0176-0. [DOI] [PubMed] [Google Scholar]
- 8.Pavelic V, Matter MS, Mumprecht S, Breyer I, Ochsenbein AF. CTL induction by cross-priming is restricted to immunodominant epitopes. Eur J Immunol. 2009;39(3):704–716. doi: 10.1002/eji.200838901. [DOI] [PubMed] [Google Scholar]
- 9.Nayak JV, Hokey DA, Larregina A, et al. Phagocytosis induces lysosome remodeling and regulated presentation of particulate antigens by activated dendritic cells. J Immunol. 2006;177(12):8493–8503. doi: 10.4049/jimmunol.177.12.8493. [DOI] [PubMed] [Google Scholar]
- 10.Stier EM, Mandal M, Lee KD. Differential cytosolic delivery and presentation of antigen by listeriolysin O-liposomes to macrophages and dendritic cells. Mol Pharm. 2005;2(1):74–82. doi: 10.1021/mp049896v. [DOI] [PubMed] [Google Scholar]
- 11.Bungener L, Serre K, Bijl L, et al. Virosome-mediated delivery of protein antigens to dendritic cells. Vaccine. 2002;20:17–18. 2287–2295. doi: 10.1016/s0264-410x(02)00103-2. [DOI] [PubMed] [Google Scholar]
- 12.Kwon YJ, Standley SM, Goodwin AP, Gillies ER, Frechet JM. Directed antigen presentation using polymeric microparticulate carriers degradable at lysosomal pH for controlled immune responses. Mol Pharm. 2005;2(1):83–91. doi: 10.1021/mp0498953. [DOI] [PubMed] [Google Scholar]
- 13.Murthy N, Xu M, Schuck S, Kunisawa J, Shastri N, Frechet JM. A macromolecular delivery vehicle for protein-based vaccines: Acid- degradable protein-loaded microgels. Proc Natl Acad Sci U S A. 2003;100(9):4995–5000. doi: 10.1073/pnas.0930644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S. Antigen presentation and T cell stimulation by dendritic cells. Annu Rev Immunol. 2002;20:621–667. doi: 10.1146/annurev.immunol.20.100301.064828. [DOI] [PubMed] [Google Scholar]
- 15.Heath WR, Carbone FR. Cross-presentation, dendritic cells, tolerance and immunity. Annu Rev Immunol. 2001;19:47–64. doi: 10.1146/annurev.immunol.19.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Pan J, Zhang M, Wang J, et al. Interferon-gamma is an autocrine mediator for dendritic cell maturation. Immunol Lett. 2004;94:1–2. 141–151. doi: 10.1016/j.imlet.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 18.Mann A, Richa R, Ganguli M. DNA condensation by poly-L-lysine at the single molecule level: Role of DNA concentration and polymer length. J Control Release. 2008;125(3):252–262. doi: 10.1016/j.jconrel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Trentin D, Hubbell J, Hall H. Non-viral gene delivery for local and controlled DNA release. J Control Release. 2005;102(1):263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Mannisto M, Ronkko S, Matto M, et al. The role of cell cycle on polyplex-mediated gene transfer into a retinal pigment epithelial cell line. J Gene Med. 2005;7(4):466–476. doi: 10.1002/jgm.693. [DOI] [PubMed] [Google Scholar]
- 21.Ward CM, Pechar M, Oupicky D, Ulbrich K, Seymour LW. Modification of pLL/DNA complexes with a multivalent hydrophilic polymer permits folate-mediated targeting in vitro and prolonged plasma circulation in vivo. J Gene Med. 2002;4(5):536–547. doi: 10.1002/jgm.296. [DOI] [PubMed] [Google Scholar]
- 22.Shen H, Ackerman AL, Cody V, et al. Enhanced and prolonged cross-presentation following endosomal escape of exogenous antigens encapsulated in biodegradable nanoparticles. Immunology. 2006;117(1):78–88. doi: 10.1111/j.1365-2567.2005.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6(6):715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 24.Yang YW, Hsu PY. The effect of poly(D,L-lactide-co-glycolide) microparticles with polyelectrolyte self-assembled multilayer surfaces on the cross-presentation of exogenous antigens. Biomaterials. 2008;29(6):2516–2526. doi: 10.1016/j.biomaterials.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Yan W, Chen W, Huang L. Reactive oxygen species play a central role in the activity of cationic liposome based cancer vaccine. J Control Release. 2008;130(1):22–28. doi: 10.1016/j.jconrel.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 26.De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm Res. 2000;17(2):113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 27.Savina A, Jancic C, Hugues S, et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126(1):205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 28.Hotta C, Fujimaki H, Yoshinari M, Nakazawa M, Minami M. The delivery of an antigen from the endocytic compartment into the cytosol for cross-presentation is restricted to early immature dendritic cells. Immunology. 2006;117(1):97–107. doi: 10.1111/j.1365-2567.2005.02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Accapezzato D, Visco V, Francavilla V, et al. Chloroquine enhances human CD8+ T cell responses against soluble antigens in vivo. J Exp Med. 2005;202(6):817–828. doi: 10.1084/jem.20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran KK, Shen H. The role of phagosomal pH on the size-dependent efficiency of cross-presentation by dendritic cells. Biomaterials. 2009;30(7):1356–1362. doi: 10.1016/j.biomaterials.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garulli B, Stillitano MG, Barnaba V, Castrucci MR. Primary CD8+ T-cell response to soluble ovalbumin is improved by chloroquine treatment in vivo. Clin Vaccine Immunol. 2008;15(10):1497–1504. doi: 10.1128/CVI.00166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 33.Tacken PJ, de Vries IJ, Torensma R, Figdor CG. Dendritic-cell immunotherapy: From ex vivo loading to in vivo targeting. Nat Rev Immunol. 2007;7(10):790–802. doi: 10.1038/nri2173. [DOI] [PubMed] [Google Scholar]