Abstract
Definitive diagnosis of malignancy is often challenging due to limited availability of human cell or tissue samples and morphological similarity with certain benign conditions. Our recently developed novel technology—spatial-domain low-coherence quantitative phase microscopy (SL-QPM)—overcomes the technical difficulties and enables us to obtain quantitative information about cell nuclear architectural characteristics with nanoscale sensitivity. We explore its ability to improve the identification of malignancy, especially in cytopathologically non-cancerous-appearing cells. We perform proof-of-concept experiments with an animal model of colorectal carcinogenesis-APCMin mouse model and human cytology specimens of colorectal cancer. We show the ability of in situ nanoscale nuclear architectural characteristics in identifying cancerous cells, especially in those labeled as “indeterminate or normal” by expert cytopathologists. Our approach is based on the quantitative analysis of the cell nucleus on the original cytology slides without additional processing, which can be readily applied in a conventional clinical setting. Our simple and practical optical microscopy technique may lead to the development of novel methods for early detection of cancer.
Keywords: quantitative phase microscopy, cancer diagnosis, pathology
Introduction
Cancer is typically diagnosed based on the microscopic examination of morphological changes in the cell and tissue stained with reagents such as hematoxylin and eosin with a conventional bright-field microscope, whose image contrast is obtained through the differences in the absorption cross section of various stains. Due to its diffraction-limited resolution (∼500 nm), the observed characteristic cytologic alterations in malignant cells for cancer diagnosis are often limited to overall nuclear appearance, such as enlarged nuclear size, irregularity of nuclear contour, and increased nuclear density.1, 2, 3, 4 However, these well-established cytologic characteristics in cancer cells may not be present or significant, especially when only a small amount of human cell or tissue samples are available for examination or in the early course of the tumor development, which may delay definitive diagnosis or lead to repeat procedures to obtain additional cell and tissue samples.
Thanks to the significant advancement in understanding the molecular changes of cancer cells, it is well recognized that tumorigenesis is the result of cumulative effect of multistep genetic and epigenetic alterations. Numerous proteins, RNAs, and genetic markers that are involved in the carcinogenesis of malignant neoplasm have been identified.5 Those cells from cancer patients, even though classified as “indeterminate” or “normal” by pathologists, may still undergo a series of malignancy-associated genetic or molecular alterations. As a result, subtle structural abnormalities may occur, especially in the cell nucleus. For those structural changes at the scale of less than the resolution of conventional optical microscopy (< ∼500 nm), they may not be easily detectable by conventional pathology. If a simple microscopy technique can detect subtle pathologically undetectable cellular alterations in situ, it could potentially improve the ability to accurately diagnose cancer and may lead to the development of novel methods for early detection of cancer.
Phase contrast microscopy and differential interference contrast (DIC) microscopy are capable of detecting subtle subcellular structural alterations. They have been widely used to visualize transparent cells in biological research, in which a minute alteration in the phase or optical path length of internal cell structure, even just a few protein molecules, can be detected through the intensity differences in the image. Despite their ability to visualize transparent cells, the lack of quantitative phase information has become a limiting factor in many biological applications. Due to the significant technical advancement, quantitative phase microscopy has recently emerged as a superior phase microscopy technique, as it provides quantitative phase measurement of a biological cell with ultrasensitivity in detecting subtle dynamic changes in the subcellular structure.6, 7, 8, 9, 10, 11, 12, 13 Despite these significant advances, its utility in clinical diagnosis of cancer is still limited, largely due to the speckle noise, special requirement on sample preparations, and the lack of known diagnostic parameters for cancer.
We recently developed a novel optical microscopy technique—spatial-domain low-coherence quantitative phase microscopy (SL-QPM)—that overcomes the limitation of conventional phase microscopy and provides a speckle-free, nanoscale-sensitive map that quantifies the spatial variation of optical path length or refractive index differences of the subcellular architecture.14 The low spatial coherence length from a thermal light source and common-path configuration suppress the notorious noise artifacts associated with quantitative phase microscopy. Here we explore the value of SL-QPM-derived in situ nanoscale nuclear architectural characteristics for cancer diagnosis. We perform the experiments with two model systems: an animal model of colorectal carcinogenesis and cytological specimens from colorectal cancer. In a proof-of-principle experiment with cytologically normal-appearing intestinal epithelial cells from an animal model from intestinal carcinogenesis—the APCMin mouse model—we show that despite their indistinct cytological features, the changes in in situ nanoscale nuclear architectural heterogeneity can be detected using SL-QPM in small intestinal epithelial cells from the 4 to 5-month-old APCMin mice with tumors, compared with the wild-type mice. We further demonstrate that these changes in the nanoscale-sensitive architectural heterogeneity of a cell nucleus can differentiate benign and malignant cells in human cytological specimens from patients with colorectal cancer, especially its capability to identify malignancy in cells characterized by pathologists as cytologically indeterminate.
Materials and Methods
SL-QPM
SL-QPM utilizes14 a low spatial-coherence thermal light source and common-path interferometer configuration to produce a speckle-free optical path length map of nuclear architecture with subnanometer sensitivity from the original unmodified clinical cytology specimens. The high sensitivity achieved by SL-QPM derives from its reflectance-mode configuration, low-coherence broadband illumination, and spectroscopic detection. The reflectance mode highlights the interference signals from elastic light scattering properties, while suppressing the effect of absorption in most cytology specimens. The spectroscopic detection enables us to analyze the interference signals due to refractive index variations within the object at every pixel of a microscopic image. With the glass substrate and biological cell of the cytology slides as a reference and a sample, respectively, that share a common path, SL-QPM effectively minimizes the slightest external disruptions that may compromise the ultrahigh sensitivity. The low-spatial-coherence illumination serves as a virtual aperture to remove the speckle noise, thus producing a speckle-free phase image.
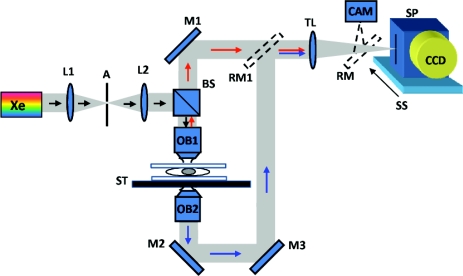
The hardware design of the SL-QPM is similar to the previously reported elastic backscattering spectroscopic microscopy,15 as shown in Fig. 1. The SL-QPM system was described in detail in our previous publication.14 Briefly, a broadband white light from a Xe arc lamp was collimated by a 4f imaging system and focused on the sample by a low-numerical-aperture (NA) objective (NA = 0.4). The resulting backscattering light was collected and projected by a tube lens onto the slit of an imaging spectrograph (Acton Research, Massachusetts) coupled with CCD camera (Andor Technology, Connecticut), which is mounted on a scanning stage. The magnification of the system is about 44. By linearly scanning the slit of the spectrograph with a 10-μm step size, the backscattering image is acquired. In each scanning step, the CCD camera records a matrix with the x axis corresponding to the wavelength and the y axis corresponding to the spatial position, resulting in a 3-D intensity cube I(x, y, k), where k represents the wave number. A transmission-mode microscope was used to record the conventional cytology image.
Figure 1.
Schematics of SL-QPM instrument: Xe, xenon arc lamp; L1, and L2, lenses; A, aperture; BS, beamsplitter; OB1, objective lens; M, mirror; ST, sample stage; TL, tube lens; RM, removal mirror; CAM, camera; SP, spectrograph; SS, scanning stage; and CCD, charged-coupled device. The additional transmission-mode microscopic components (OB2-M2-M3-RM1) are used to record the conventional cytology images for comparison.
The detected signal I(x, y, k) comes from interference from the reference (i.e., the glass substrate) and sample signals (i.e., light traveling a round-trip along the axial dimension of the cell) with a shared common path and can be related to the signal and reference field (Es ,Er) as follows:
| (1) |
The backscattering spectrum I(x, y, k) from each pixel was normalized by the spectral profile of the optical system, which accounts for the wavelength-dependent response of the light source and optical components. Each spectrum was numerically resampled to evenly spaced wave numbers and multiplied by a Hanning window before we applied a fast Fourier transform. The Fourier transformed data at the prominent peak corresponding to the optical path length of interest were selected for phase processing. We confirmed that the selected optical path length of interest is not affected by the absorption of the cytology stains, as shown in Fig. 2. After taking the discrete Fourier transform, a complex-valued F is obtained, and the phase can be extracted by taking the argument of
| (2) |
The phase map can be obtained by plotting the phase variation in a certain optical path length plane. The axial optical path length variation can be derived by δ(OPL) = (λ0 /4π)ϕ(x, y) where λ0 is the source central wavelength (550 nm), and φ(x, y) is the phase at each pixel. No phase-unwrapping program was used. The measured phase stability of the SL-QPM was 0.9 nm, as determined by the standard deviation of the histogram of measured phase performed for ∼3 min. The final optical path length for each pixel was calculated by adding the optical depth of interest to its shifted (due to phase variation) path length.
Figure 2.
Fourier transformed spectrum |F(OPL)| of the original backscattering spectrum I(x, y, k) from (a) the Diff-Quik staining solution and (b) a cell nucleus stained with Diff-Quik stains. (a) The prominent peak at the low-spatial-frequency component of |F(OPL)| comes from the absorption profile of the Diff-Quik staining solution. (b) Evidently, the |F(OPL)| from the Diff-Quik-stained cell nucleus has a distinctly different peak, corresponding to the optical path length of the cell nucleus (as indicated by the black arrow). These results confirmed that our selected optical path length of interest is not affected by the absorption profile of the Diff-Quik staining solution.
Optical Path Length (OPL) Map
The SL-QPM essentially records a microscopic image with a reflectance interferometric intensity cube I(x, y, k). The phase value at every pixel can be obtained by Fourier analysis of I(x, y, k) and is converted to the corresponding OPL [OPL(x, y) = ⟨n(x, y)⟩ L(x, y), where ⟨n(x, y)⟩ is the average refractive index along the axial direction (i.e., the z direction) at the specific pixel (x, y), and L(x, y) is the physical thickness]. As a result, we obtain a 2-D, nanoscale spatial distribution of the OPL (OPL map). To quantify the image characteristics of the OPL map in the nucleus, we extracted three statistical parameters: average OPL ⟨OPL⟩ over the 2-D spatial distribution of OPL(x, y); standard deviation σOPL over OPL(x, y), representing a global variation; and entropy EOPL , which is a measure of randomness, representing a more localized heterogeneity derived from texture analysis. Both σOPL and EOPL quantify the architectural heterogeneity.
Texture Analysis of OPL Map of a Cell Nucleus
Based on the spatial distribution of the OPL from a single nucleus, we performed a texture analysis and quantified three statistical parameters. The average OPL ⟨OPL⟩ and standard deviation σOPL were calculated by taking the mean and standard deviation of all OPL values from a single nucleus. The entropy EOPL , a parameter describing the randomness, is defined as , where Ii is a random variable indicating intensity, p(Ii) is the histogram of the intensity levels in a region, and N is the number of possible intensity levels.16 The intranuclear ⟨OPL⟩ describes the average OPL or average density with a single nucleus (assuming the same nuclear thickness). The intranuclear standard deviation σOPL and intranuclear entropy EOPL describe the global and local heterogeneity of the OPL distribution, respectively.
Animal Model of Colorectal Carcinogenesis
All animal studies were performed in accordance with the institutional Animal Care and Use Committee of University of Pittsburgh. All mice were housed in microisolator cages in a room illuminated from 7:00 AM to 7:00 PM (12:12-h light-dark cycle), with access to water and ad libitum. Three C57BL APC wild-type mice and three age-matched C57BL APCMin mice at 4 to 5 months old were sacrificed. The small intestines were removed, longitudinally opened, and washed with phosphate-buffered saline (PBS). Epithelial cells were obtained from visually normal mucosa of the small intestine with a cytology brush. The cytology brush was immersed into the Cytolyt solution (Cytec Corporation, Boxborough, Massachusetts). The slides were prepared by a standard thin prep processor (Cytec Corporation). Cells were then stained with Papanicolaou stains and sealed with a mounting medium and coverslip. Cells were examined by an expert cytopathologist and non-cancerous-looking cells were analyzed by SL-QPM. For each mouse, about 20 to 30 cells are randomly selected for SL-QPM analysis.
Human Specimens
All studies were performed and all specimens were collected with the approval of the Institutional Review Boards at University of Pittsburgh and University of Washington. Banked frozen mucosal biopsies were obtained from a group of seven patients with ulcerative colitis who underwent colectomy or colonoscopy, four of whom had no evidence of dysplasia, and three of whom were diagnosed with colorectal cancer. Among the three cancer cases, one patient had frankly malignant cells obtained from the cancer site, while the other two patients had abnormal cells from cancer sites, classified as indeterminate by an expert cytopathologist. The cytology slides were made from frozen mucosal biopsies using a touch preparation (TP) method and were subsequently stained by Diff-Quik stains and coversliped. Such methods typically minimize the single-cell thickness variations among different patients for the same type of cell. These slides were reviewed by an experienced cytopathologist who dotted the colon epithelial cells of interest. Approximately 30 to 40 cell nuclei per patient were evaluated for OPL analysis.
Statistical Analysis
We performed the statistical analysis using Microsoft Excel 2007. All the statistical analyses were performed based on the Student’s t test. Two-tailed P values were used for all analyses. The alpha level was assumed to be 0.05. Two-tailed P values of less than 0.05 were considered as statistical significance.
Results
Experiments with APCMin Mouse Model
To explore the ability of nanoscale nuclear architectural characteristics quantified by SL-QPM to identify cancer from cytologically indeterminate cells, we used a well-established animal model of colorectal carcinogenesis—the APCMin mouse model. It is a genetically modified animal model that represents the human condition of familial adenomatous polyposis syndrome in which the adenomatous polyposis coli (APC) gene undergoes a germ-line mutation leading to a truncation in the APC protein and spontaneous development of intestinal adenomas. We analyzed the cytopathologically indistinguishable intestinal epithelial cells from three age-matched wild-type mice and three APCMin mice at 4 to 5 months with the presence of tumors in the small intestine. Figures 3A, 3B show Papanicolaou-stained cytological images of representative intestinal epithelial cells from the APCMin mice compared to those of the wild-type mice obtained from a bright-field microscope and their corresponding OPL maps from the cell nuclei. Although the microscopic cytological images look similar (as confirmed by an expert cytopathologist), the OPL images that characterize the intranuclear distribution of OPL exhibit distinct differences. Based on these OPL maps, three statistical parameters from the nuclei were calculated: the average OPL ⟨OPL⟩ over the 2-D spatial distribution of OPL(x, y); the standard deviation of the OPL σOPL representing a global variation and the entropy EOPL —a measure of randomness, representing a more localized heterogeneity. As shown in Fig. 3C, the nuclear heterogeneity quantified by σOPL and EOPL are significantly increased in approximately 20 to 30 randomly selected intestinal epithelial cells from the APCMin mice compared to those from the wild-type mice (p value < 0.001). However, no statistical difference is found in the average OPL ⟨OPL⟩ (p value = 0.4). These results suggest that the higher nuclear architectural heterogeneity derived from its in situ nanoscale OPL map is associated with carcinogenesis, underlying the potential of this technique in accurate cancer diagnosis at the level of single cell nucleus.
Figure 3.
(a) Representative cytological images (Papanicolaou stains) and (b) the corresponding OPL maps of cell nuclei (as shown in circles) of intestinal epithelial cells from the wild-type mouse and the APCMin mouse at 4 to 5 months with the development of tumors. Scale bars in the image indicate 5 μm. The colorbar shows the magnitude of the OPL in the cell nucleus in micrometers. Although the cytology images appear similar (cytopathologically indistinguishable), the OPL map of the cell nucleus from the APCMin mice exhibits an increased heterogeneity compared to that from the wild-type mice. (c) The standard deviation of OPL σOPL and the entropy EOPL of the cell nuclei in cytologically normal-appearing intestinal epithelial cells show a statistically significant increase in the APCMin mouse (p value < 0.001), associated with the development of carcinogenesis. The error bar represents the standard error. About 20 to 30 cells were randomly selected from each mouse to conduct SL-QPM measurement and OPL analysis. (Color online only)
Experiments with Colorectal Cancer in Patients with Ulcerative Colitis
To further confirm the significance of in situ nanoscale nuclear architecture in human cells, we analyzed human cytology specimens from a group of seven patients with ulcerative colitis (UC), an inflammatory bowel disease, undergoing colonoscopy or colectomy. Three patients had been diagnosed with colorectal cancer, while four had no evidence of dysplasia or cancer. We analyzed the OPL maps from three groups of colon epithelial cells, selected by an expert cytopathologist: (1) normal cells [i.e., cytologically normal cells from uninvolved tissue site (no active colitis) in the four patients without dysplasia or cancer]; (2) cytologically indeterminate cells obtained at the cancer site (i.e., cells classified by an expert cytologist as being indeterminate in patients with colorectal cancer); and (3) malignant cancer cells obtained at the cancer site (i.e., cytologically classified as malignant cells by cytologist using conventional diagnostic criteria).
Figure 4A presents the representative OPL maps of benign∕normal colonic epithelial cell nuclei in a patient with ulcerative colitis, cell nuclei from a UC patient with concurrent cancer (colonic carcinoma) that was called indeterminate by cytological diagnosis, and malignant cell nuclei that cytologically called malignant from a UC patient with colonic carcinoma. The OPL maps reveal the significantly increased intranuclear heterogeneity and average OPL in the malignant cancer cell.
Figure 4.
(a) Representative cytology images and corresponding OPL maps of a cytologically normal colonic cell from a patient with ulcerative colitis; an indeterminate cell from cancer site, classified by cytology, obtained from a patient with surgically confirmed diagnosis of colonic cancer patient; and a malignant cell from a cancer site, classified by cytology, from a patient with confirmed colonic cancer. The OPL maps show distinct patterns in these three cell groups, with a significantly increased OPL and heterogeneity in cells from cancer patients compared with the normal cells. Scale bars in the image indicate 10 μm. Colorbar shows the magnitude of optical path length in a cell in microns. (b) Scatter plot of the average optical path length ⟨OPL⟩ the intranuclear entropy EOPL and the standard deviation of the OPL σOPL of all the nuclei from a patient with ulcerative colitis without dysplasia or cancer (blue diamond); a colon cancer patient with those cells diagnosed as indeterminate by cytology (purple square); and a colon cancer patient with malignant cells, confirmed by cytology, from a confirmed colon cancer patient with malignant tumor (red triangle). Each point corresponds to a single nucleus. (c) The statistical analysis of EOPL, σOPL, and ⟨OPL⟩ was performed using approximately 30 to 40 cells for each patient. The nuclear heterogeneity derived from EOPL and σOPL is progressively increased from normal cells to cytologically indeterminate cells to frankly malignant cells (Student’s t test, P < 0.0001). The ⟨OPL⟩ in malignant cells from cancer patients is significantly increased compared with that in normal cells (Student’s t test, P < 0.01). However, there was no statistically significant difference in ⟨OPL⟩ between normal and cytologically indeterminate cells.
To illustrate the internuclear variation, Fig. 4B shows a scatter plot of OPL parameters ⟨OPL⟩, σOPL and EOPL for all the cells from one representative patient in each group: (1) normal epithelial cells from an ulcerative colitis (no cancer) patient; (2) cytologically indeterminate cells from a cancer patient; and (3) malignant tumor cells from a cancer patient, with each cell nucleus represented by a point (⟨OPL⟩, σOPL, EOPL). Despite the intrinsic variations among different nuclei, the malignant cells and normal cells are well separated. Importantly, the majority of cytologically indeterminate cells in cancer patients are clearly distinguishable from normal cells from patients without dysplasia. These cells classified as indeterminate by an expert cytolopathologist from a cancer site, resemble the nuclear architectural heterogeneity of the malignant cells.
Figure 4C shows the analysis of three statistical parameters from all cells in seven patients (approximately 30 to 40 cells from each patient). The heterogeneity parameters—σOPL and EOPL —were significantly increased in cytologically malignant cancer cells (p value < 0.0001) compared with those in noncancerous cells. The same heterogeneity parameters can also distinguish cytologically indeterminate cells in cancer patients from those in patients without dysplasia with a high level of statistical significance (p value < 0.0001). Although ⟨OPL⟩ is significantly increased in malignant cancer cells (p value < 0.01), ⟨OPL⟩ does not present any statistical difference in cytologically indeterminate cells obtained at the cancer site when compared with normal-appearing epithelial colonic cells obtained from patients without cancer or dysplasia. These results suggest the importance of the in situ nanoscale nuclear heterogeneity in detecting subtle changes in tumor malignancy.
Discussion
Here we presented a novel method to investigate in situ nuclear architectural characteristics with a simple and novel optical instrument—SL-QPM. SL-QPM provides an ultrasensitive measurement of subtle alterations of nuclear structure, represented by a spatial map of the OPL of a single cell nucleus with a sensitivity of less than a nanometer. Importantly, SL-QPM can be directly used on the original cytology specimens prepared with the standard clinical protocol without any special processing, such as fluorescence staining or coverslip removal. We demonstrated the importance of nanoscale nuclear architectural characteristics in cancer diagnosis by identifying cancer patients or carcinogenesis from otherwise cytologically indeterminate or normal-appearing cells. We found that the increased heterogeneity of cell nuclear architecture quantified by the nanoscale-sensitive OPL map, accompanies the development of carcinogenesis or malignancy, with better sensitivity than standard cytopathology, in both the animal model and human cytology specimens from patients with colorectal cancer. These nanoscale nuclear architectural heterogeneity characteristics could be used as potential diagnostic markers for subtle changes of malignancy, especially when only a small amount of human cell or tissue samples are available. The simplicity of this optical instrument and its applicability to clinical specimens make this technique easily translatable to basic research and clinical care settings.
SL-QPM quantifies the subtle alterations of nuclear architecture in situ with a high sensitivity, through the spatial mapping of the nanoscale-sensitive OPL within a single cell nucleus. The architectural characteristics of each nucleus are described with three OPL statistical parameters: the average OPL of the individual nucleus ⟨OPL⟩, the standard deviation of the OPL σOPL , and the entropy EOPL , derived from the texture analysis of the OPL map. The σOPL of the cell nucleus characterizes a global intranuclear heterogeneity; while EOPL quantifies the randomness in the nuclear architecture, representing a local heterogeneity.
The in situ nanoscale-sensitive nuclear architectural heterogeneity parameters—σOPL and EOPL —have shown a high level of sensitivity in detecting carcinogenesis in the animal model and identifying colorectal malignancy in cells that are characterized by pathologists as cytologically indeterminate. Such significance is even more pronounced in distinguishing frankly malignant cells from benign cells. Interestingly, the cells from cancer patients (regardless of their cytological diagnosis) also exhibit a higher level of internuclear heterogeneity (i.e., pleomorphism), evidenced by the wider spread of σOPL and EOPL in cell nuclei from colorectal cancer patients, as shown in Fig. 4B. On the other hand, the average OPL of the nucleus ⟨OPL⟩ also indicates certain capability of discriminating frankly benign and malignant cells, but by itself does not demonstrate higher sensitivity than conventional cytopathology.
The OPL, as the key parameter to characterize the nuclear architecture is essentially the product of the refractive index and the physical thickness [OPL(x, y) = ⟨n(x, y)⟩ L(x, y)]. Due to the standardized or automated clinical cytology slide preparation (e.g., ThinPrep processor), most cytology specimens have a monolayer of cells with consistent physical thickness for the same cell type. If we assume the same physical thickness of each cell, the OPL quantifies the changes in refractive index or the mass density (i.e., macromolecular concentration) of the nuclear components17, 18 such as chromatin and the nuclear matrix. Indeed, the increased nuclear density (i.e., hyperchromasia) in cancer cells has been well documented as one of the important pathological criteria for cancer diagnosis.19 Our quantitative measurement of increased average OPL (i.e., ⟨OPL⟩) or nuclear density in frankly malignant cells is in good agreement with this pathologic diagnostic criteria. On the other hand, the spatial heterogeneity of cell nuclear density quantified by σOPL and EOPL , exhibit a progressive increase in benign cells, indeterminate cells from cancer sites, and frankly malignant cells from cancer patients. Importantly, our results underscore the significance of in situ nanoscale intranuclear heterogeneity as a highly sensitive diagnostic characteristic for cancer, which cannot be obtained with any other techniques for nuclear or DNA analysis, such as flow cytometry.20 Although the specific biological events responsible for the increased nuclear architectural heterogeneity in cancer cells are not identified, these subtle changes in the nuclear structure are likely the results of complex genetic and molecular events from multiple molecular pathways, such as chromatin clumping, nucleolar alterations, and genetic instability.1, 2, 21
This technique should not be considered as advanced image analysis of conventional digital microscopic images. The OPL map provides quantitative nanoscale information about the nuclear architecture that may be otherwise undetectable with conventional optical microscopy. For example, we performed a similar texture analysis on conventional bright-field cytology images and found that the heterogeneity parameters (i.e., standard deviation σ and entropy E) cannot distinguish cytologically normal-appearing epithelial cell nuclei from the wild-type and APCMin mice (P value = 0.30 and 0.29, respectively), underlining the importance of the nanoscale OPL map.
Due to the limited sample size, the diagnostic utility of this technique for various cancers must be further validated with a larger patient population. The relevance of increased nuclear architectural heterogeneity parameters to the development of carcinogenesis or malignancy was supported by the following evidence. First, the increased nuclear heterogeneity parameters (σOPL and EOPL) were consistently presented in both animal model and human cytology specimens of colorectal cancer. Second, the progressive increase of these parameters in benign cells, indeterminate cells from cancer patients and frankly malignant cells implies the capability of SL-QPM of detecting the subtle cytologically undetectable “transitional” alterations in cell nuclei in cancer patients. Third, the nuclear heterogeneity parameters from cytologically indeterminate or normal cells from cancer subjects resemble those from frankly malignant cells, distinct from normal cells from their benign counterparts, indicating that these cytological indeterminate cells from cancer subjects may share common biological events with the frankly malignant cells.
We do not suggest that these nanoscale nuclear architectural features should replace existing pathological diagnostic procedures or criteria, but instead can enhance the accurate detection of malignancy that would otherwise be missed by conventional cytopathology. Our proposed approach is practical and can rapidly be introduced into clinical practice since no special sample preparation is required and would only be used in those cases that conventional cytopathology does not make a definitive diagnosis. Although the cytology specimens from colorectal cancer were studied, this approach can be applied to a wide variety of tumor types and tissue. Ongoing studies aimed at expanding our tumor types are being conducted. From the basic science perspective, this technique may provide a new capability for elucidating the mechanism of malignancy and correlating functional and molecular parameters with malignancy-associated structural changes. Such work could ultimately advance the discovery of new therapeutic targets and provide novel approaches to monitoring the effects of treatment or preventive strategies.
Acknowledgments
We acknowledge Dr. Christopher J. Bakkenist for valuable discussions. This work is supported by research grants from the National Institute of Health (R21CA138370 and R21CA152935), James F. Walsh Foundation, the University of Pittsburgh Medical Center (Y.L., R.B.), and the National Energy Technological Laboratory Research Participation Program (P.W.) sponsored by the U.S. Department of Energy and administered by the Oak Ridge Institute for Science and Education.
References
- Nickerson J. A., “Nuclear dreams: the malignant alteration of nuclear architecture,” J. Cell. Biochem. 70(2), 172–180 (1998). 10.1002/(SICI)1097-4644(19980801)70:2 172::AID-JCB3 3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- Zink D., Fischer A. H., and Nickerson J. A., “Nuclear structure in cancer cells,” Nature Rev. Cancer 4(9), 677–687 (2004). 10.1038/nrc1430 [DOI] [PubMed] [Google Scholar]
- Underwood J. C. E. and Crocker J., Pathology of the Nucleus, Springer-Verlag, Berlin; New York: (1990). [Google Scholar]
- Konety B. R. and Getzenberg R. H., “Nuclear structural proteins as biomarkers of cancer,” J. Cell. Biochem. Suppl. 32–33, 183–191 (1999). 10.1002/(SICI)1097-4644(1999)75:32+ 183::AID-JCB22 3.0.CO;2-A [DOI] [PubMed] [Google Scholar]
- Ahlquist D. A., “Molecular detection of colorectal neoplasia,” Gastroenterology 138(6), 2127–2139 (2010). 10.1053/j.gastro.2010.01.055 [DOI] [PubMed] [Google Scholar]
- Ferraro P., Alferi D., De Nicola S., De Petrocellis L., Finizio A., and Pierattini G., “Quantitative phase-contrast microscopy by a lateral shear approach to digital holographic image reconstruction,” Opt. Lett. 31(10), 1405–1407 (2006). 10.1364/OL.31.001405 [DOI] [PubMed] [Google Scholar]
- Joo C., Akkin T., Cense B., Park B. H., and de Boer J. F., “Spectral-domain optical coherence phase microscopy for quantitative phase-contrast imaging,” Opt. Lett. 30(16), 2131–2133 (2005). 10.1364/OL.30.002131 [DOI] [PubMed] [Google Scholar]
- Rylander C. G., Dave D. P., Akkin T., Milner T. E., Diller K. R., and Welch A. J., “Quantitative phase-contrast imaging of cells with phase-sensitive optical coherence microscopy,” Opt. Lett. 29(13), 1509–1511 (2004). 10.1364/OL.29.001509 [DOI] [PubMed] [Google Scholar]
- Shaked N. T., Rinehart M. T., and Wax A., “Dual-interference-channel quantitative-phase microscopy of live cell dynamics,” Opt. Lett. 34(6), 767–769 (2009). 10.1364/OL.34.000767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Rao B., Yu L., and Chen Z., “High-dynamic-range quantitative phase imaging with spectral domain phase microscopy,” Opt. Lett. 34(21), 3442–3444 (2009). 10.1364/OL.34.003442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G., Ikeda T., Dasari R. R., and Feld M. S., “Diffraction phase microscopy for quantifying cell structure and dynamics,” Opt. Lett. 31(6), 775–777 (2006). 10.1364/OL.31.000775 [DOI] [PubMed] [Google Scholar]
- Sarunic M. V., Weinberg S., and Izatt J. A., “Full-field swept-source phase microscopy,” Opt. Lett. 31(10), 1462–1464 (2006). 10.1364/OL.31.001462 [DOI] [PubMed] [Google Scholar]
- Barty A., Nugent K. A., Paganin D., and Roberts A., “Quantitative optical phase microscopy,” Opt. Lett. 23(11), 817–819 (1998). 10.1364/OL.23.000817 [DOI] [PubMed] [Google Scholar]
- Wang P., Bista R., Bhargava R., Brand R. E., and Liu Y., “Spatial-domain low-coherence quantitative phase microscopy for cancer diagnosis,” Opt. Lett. 35(17), 2840–2842 (2010). 10.1364/OL.35.002840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li X., Kim Y. L., and Backman V., “Elastic backscattering spectroscopic microscopy,” Opt. Lett. 30(18), 2445–2447 (2005). 10.1364/OL.30.002445 [DOI] [PubMed] [Google Scholar]
- Gonzalez R. C., Woods R. E., and Eddins S. L., Digital Image Processing Using MATLAB, Pearson/Prentice Hall, Upper Saddle River, NJ: (2004). [Google Scholar]
- Barer R. and Tkaczyk S., “Refractive index of concentrated protein solutions,” Nature 173(4409), 821–822 (1954). 10.1038/173821b0 [DOI] [PubMed] [Google Scholar]
- Kékicheff P., Laughlin R. G., and Munyon R. L., “Diffusive interfacial transport: a new approach to concentrated protein solution studies,” Langmuir 17(16), 4693–4696 (2001). 10.1021/la010226v [DOI] [Google Scholar]
- Orell S. R., Sterrett G. F., and Whitaker D., Fine Needle Aspiration Cytology, Elsevier Churchill Livingstone, Edinburgh; New York: (2005). [Google Scholar]
- Sampedro A., Salas-Bustamante A., Lopez-Artimez M., Garcia-Muniz J. L., and Urdiales G., “Cell cycle flow cytometric analysis in the diagnosis and management of colorectal carcinoma,” Anal. Quant. Cytol. Histol. 21(4), 347–352 (1999). [PubMed] [Google Scholar]
- Coffey D. S., “Self-organization, complexity and chaos: the new biology for medicine,” Nature Med. 4(8), 882–885 (1998). 10.1038/nm0898-882 [DOI] [PubMed] [Google Scholar]