Abstract
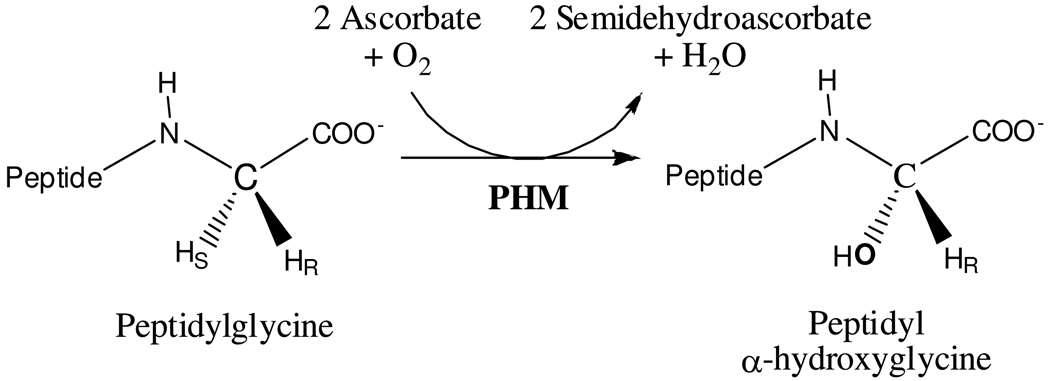
Peptidylglycine α-Hydroxylating Monooxygenase (PHM) catalyzes the stereospecific hydroxylation of the Cα of C-terminal glycine-extended peptides and proteins, the first step in the activation of many peptide hormones, growth factors and neurotransmitters. The crystal structure of the enzyme revealed two non-equivalent Cu sites (CuM and CuH) separated by ~ 11 Å. In the resting state of the enzyme, CuM is coordinated in a distorted tetrahedral geometry by one methionine, two histidines, and a water molecule. The coordination site of the water molecule is the position where external ligands bind. The CuH has a planar T-shaped geometry with three histidines residues and a vacant position that could be potentially occupied by a fourth ligand. Although the catalytic mechanism of PHM and the role of the metals are still being debated, CuM is identified as the metal involved in catalysis while CuH is associated with electron transfer. To further probe the role of the metals, we studied how small molecules such as nitrite (NO2−), azide (N3−) and carbon monoxide (CO) interact with the PHM copper ions. The crystal structure of an oxidized nitrite-soaked PHMcc obtained by 20 hours soaking in mother liquor supplemented with 300 mM NaNO2, shows that nitrite anion coordinates CuM in an asymmetric bidentate fashion. Surprisingly, nitrite does not bind CuH despite the high concentration used in the experiments (nitrite/protein > 1000). Similarly, azide and carbon monoxide coordinate CuM but not CuH in the PHMcc crystal structures obtained by co-crystallization with 40 mM NaN3 and by soaking CO under 3 atm of pressure for 30 minutes. This lack of reactivity at the CuH is also observed in the reduced form of the enzyme: CO binds CuM but not CuH in the structure of PHMcc obtained by exposure of a crystal to 3 atm CO for 15 minutes, in the presence of 5 mM ascorbic acid (reductant). The necessity of CuH to maintain its redox potential in a narrow range compatible to its role as an electron-transfer site seems to explain the lack of coordination of small molecules to CuH; coordination of any external ligand will certainly modify its redox potential.
Introduction
Many peptides such as hormones, growth factors and neurotransmitters require α-amidation of their carboxy-terminus to be fully biologically active.1–3 The α-amidation is carried out in the trans-Golgi network and in secretory granules of neural and endocrine tissues by two sequential reactions. The first reaction is the stereospecific hydroxylation of the Cα of a C-terminal peptidylglycine (see Scheme 1), a reaction catalyzed by Peptidylglycine α-Hydroxylating Monooxygenase (PHM). The second reaction, an N-dealkylation which yields the amidated peptide and glyoxylate, is catalyzed by Peptidyl-α-hydroxyglycine α-Amidating Lyase (PAL).4–5 Although the two enzymes can be expressed and can function independently, in most species they are expressed as a bifunctional single chain enzyme, Peptidylglycine α-Amidating Monooxygenase (PAM).
Scheme 1.
The Stereo-Specific Reaction Catalyzed by PHM
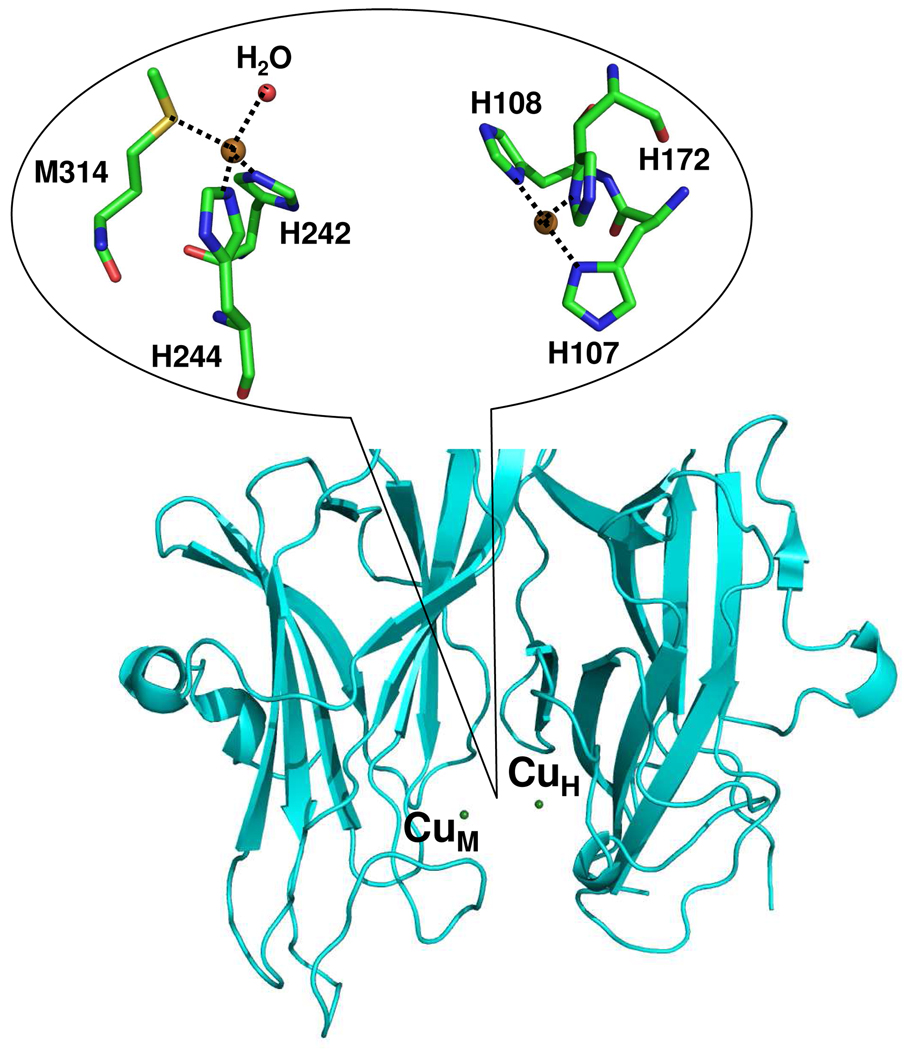
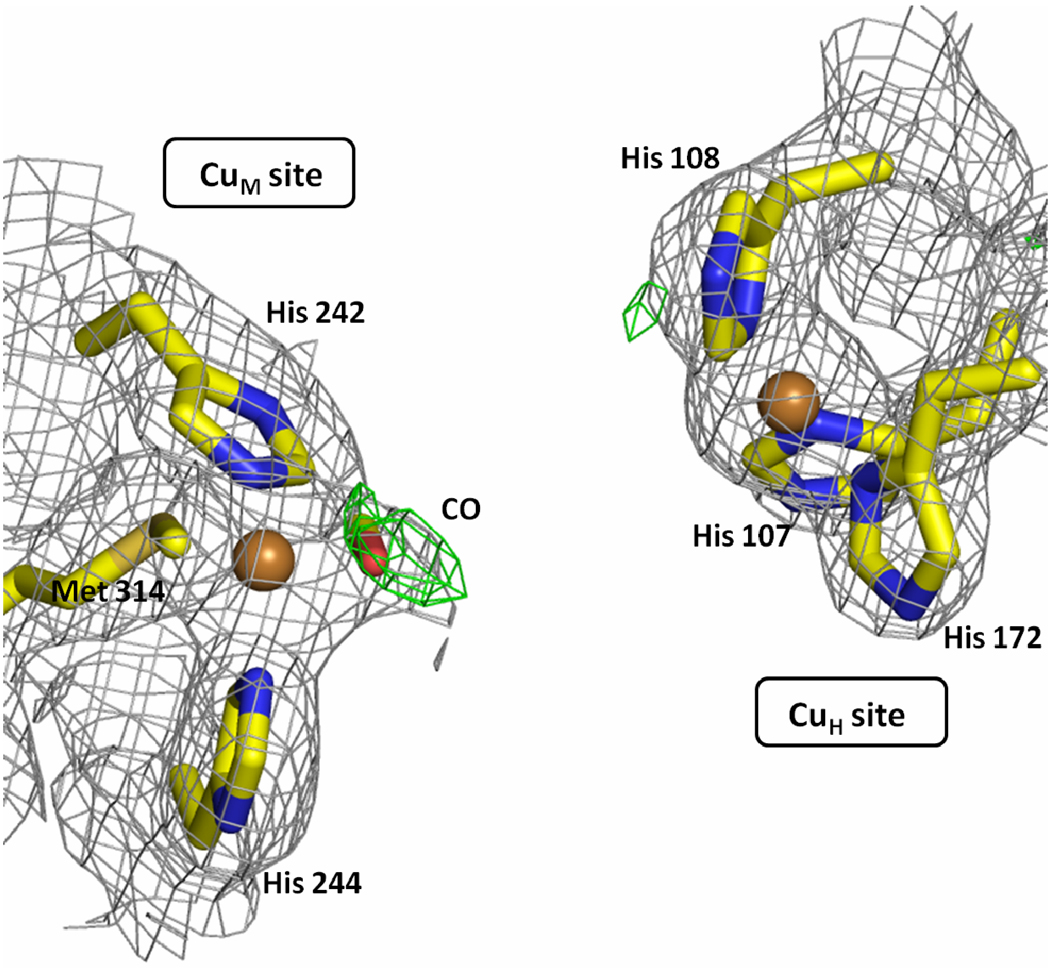
PHM is a binuclear copper protein with two non-equivalent type-2 Cu sites, CuM and CuH, separated by 11 Å.6 It catalyzes the stereospecific Cα hydroxylation of the terminal glycine by incorporating one oxygen atom from dioxygen, using ascorbate as the physiological reductant. In the resting state of the enzyme, CuM has a distorted tetrahedral geometry, with one methionine, two histidines, and a water ligand that can be substituted by an external ligand (Figure 1). CuH has a planar T-shaped geometry with three histidines residues and a vacant ligand position that could potentially bind a fourth ligand (Figure 1). Although the chemical structures of the copper centers CuM and CuH are clearly dissimilar, ligand field Magnetic Circular Dichroism (MCD) and Electron Paramagnetic Resonance (EPR) studies on the resting oxidized PHM did not find any measurable differences between the two Cu sites.7 Interestingly, using nitrite (NO2−) as a metal-ligand, MCD and EPR measurements showed the presence of the two metal centers.7
Figure 1. The structure of CuM and CuH sites of PHMcc.
CuM is the catalytic site and CuH is the electron-transfer site. The two copper centers are separated 11 Å by an inter-domain cleft fully accessible to substrates, solvent and small molecules.
Although the catalytic mechanism of PHM is still being investigated, there is agreement about the main roles of the metals: CuM is the metal involved in catalysis, while CuH is associated with electron transfer.8–10 As mentioned above, these two copper centers can be classified as type-2 on the basis of their structural and spectroscopic properties: (i) both CuM and CuH are coordinated by histidines (CuM also includes a methionine in its coordination sphere) with a vacant position (CuH) or a position occupied by an exogenous ligand as water (CuM) and (ii) the EPR spectrum show typically a large A value (Az = 157 × 10−4 cm−1)7 for the unique copper signal.11 Further, the two Cu(II) signals of the EPR spectrum of the nitrite-perturbed PHM have large A values (Az = 160 × 10−4 cm−1 and 165 × 10−4 cm−1).7 Therefore, there is a question about the type-2 characteristics of CuH, which are typical for catalytic sites, and its postulated role as an electron-transfer site. Single electron transfer reactions are usually carried out by type-1 copper centers (blue copper sites), characterized by a cysteine residue coordinated to the copper.11
To probe further into the role of the metals and to provide additional insight into the results of the MCD and EPR experiments, we studied how small molecules such as nitrite (NO2−), azide (N3−) and carbon monoxide (CO) interact with the copper active sites. Nitrite is the natural substrate of copper-containing nitrite reductases and thus can bind to biological copper sites.12 Nitrite also forms metal complexes with Cu(II) adopting different geometries13–17 and with Cu(I) usually as N-monodentate ligand.18–19 Azide has also been used as a ligand for copper proteins in the oxidized state20–22 and there are several examples in the literature of Cu(II) and Cu(I) complexes with azide.23–24 Carbon monoxide is a well know B-acceptor ligand that can stabilize low oxidation states such as Cu(I) making it a very attractive ligand for reactivity studies of the reduced form of the enzyme. We found that nitrite, azide and carbon monoxide bind CuM but, unexpectedly, do not bind CuH despite the presence of a vacant position in the CuH coordination sphere and the high concentration of the ligands used in the experiments. The lack of coordination of small molecules thus seems to be an essential feature of the CuH site and is probably related to its proposed functional role as an electron-transfer site.
Results and Discussion
ox-PHMcc-nitrite complex
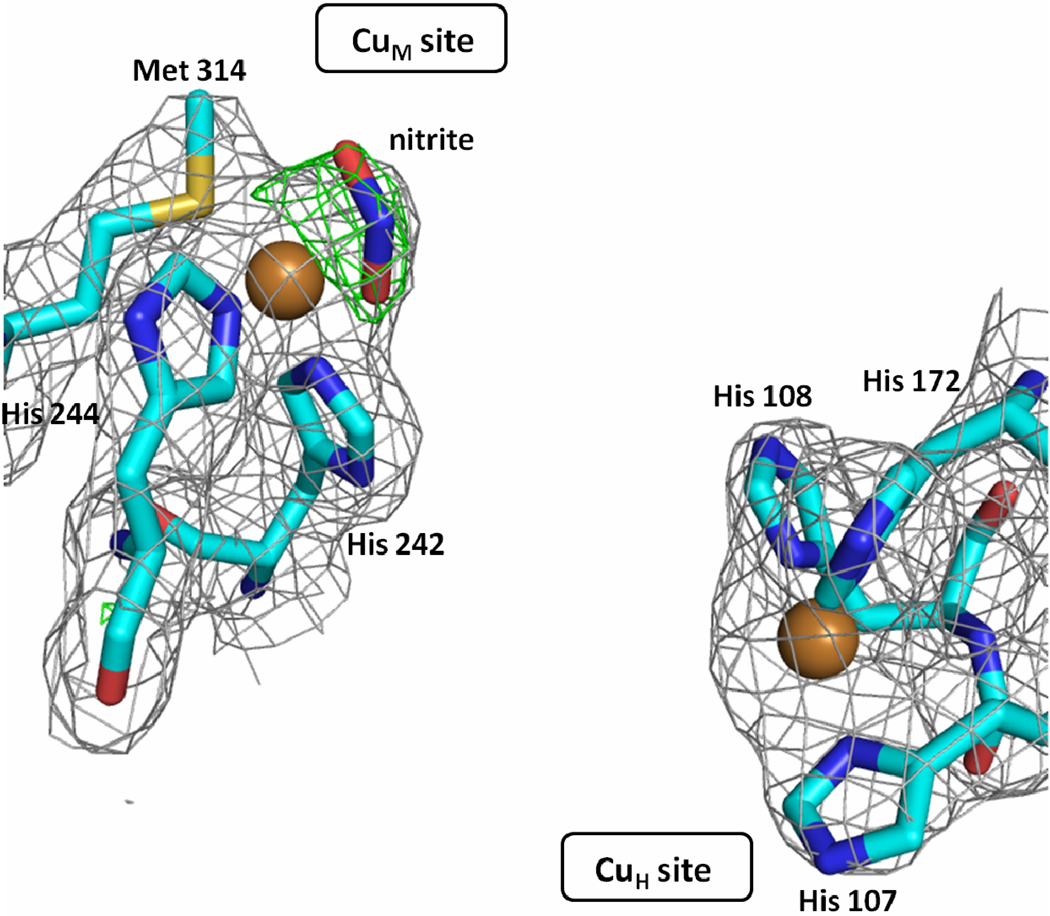
Crystals of the PHMcc-nitrite complex were prepared by soaking protein crystals for 20 hours in mother liquor supplemented with 300 mM NaNO2. The 2.35 Å resolution structure revealed that the nitrite anion coordinates CuM in an asymmetric bidentate fashion with copper-to-nitrite oxygen distances of 1.9 Å and 2.6 Å (Figure 2). It displaces the water ligand found in the oxidized form of the enzyme, with one of its oxygens occupying the approximate position of the oxygen of the water molecule. The CuM environment changes significantly from tetracoordinated N2(His)S(Met)O(water) to pentacoordinated N2(His)S(Met)O2(nitrite). This particular coordination mode of a nitrite ligand has been found in nitrite-soaked crystals of nitrite reductases (CuNRs), enzymes for which nitrite is the natural substrate.25–29 In CuNRs, the catalytic type-2 copper ion is coordinated by three histidine residues, suggesting that the presence of methionine in CuM, at least in the oxidized form of the enzyme, is not playing a critical role in the binding and coordination mode of nitrite. Nitrite has also been found coordinated to the same type-2 Cu of CuNRs as an O-monodentate ligand, but only when substitutions are introduced in residues surrounding the nitrite binding site that render the enzyme inactive.30 In PHM, the CuM site is open, without any neighboring residue that could force nitrite to adopt a different coordination mode. The PHM structure also revealed that the plane defined by the CuM and the two O atoms of nitrite and the plane defined by the N and the two O atoms of nitrite form a 30 degree angle (Figure S1 in Supporting Information). This feature for Cu-(NO2) binding was also observed in several nitrite-soaked CuNRs enzymes.25 However, small molecule compounds having the Cu-(NO2) motif with nitrite bound to Cu as a bidentate ligand, usually exhibit the four atoms (Cu, O1, O2, N) in the same plane. Out of more than 40 structures deposited in the Cambridge Structural Database (CSD),31 only 2 have the nitrite bent out of the plane and even in those cases the angle is small (less than 30 degree).32–33
Figure 2. The structure of CuM and CuH sites of a nitrite-soaked PHMcc.
The nitrite is shown bound to CuM in an asymmetric bidentate fashion (Cu-O1 = 1.9 Å; Cu-O2 = 2.6 Å). The gray mesh shows the sigma weighted 2mFo-DFc (contoured at 1.0 σ) and the green mesh the mFo-DFc omit maps (contoured at 2.8 σ) The omit maps were computed with Refmac5. Carbons are colored cyan, nitrogens blue, oxygens red, sulphur yellow and copper gold.
Two more nitrite anions were also present in the structure. One is located at a protein-nickel-protein crystal contact,34 coordinated to Ni(II) in an asymmetric bidentate fashion (Ni-Onitrite = 1.95 and 2.65 Å), in a manner similar to that observed in CuM. However, in contrast to what was observed for CuM, the Ni, and the N and two O atoms of nitrite are all in the same plane (Figure S2 in Supporting Information). The other nitrite was found also in the periphery of the protein, hydrogen bonded to the -NH of Gly258 (2.8 Å) through one of its oxygens.
Remarkably, CuH does not bind nitrite as evidenced by the lack of density at the fourth coordination site of CuH in a 2Fo-Fc electron density map (Figure 2). This is surprising in view that the concentration of nitrite used in the experiments, 300 mM, was very high (nitrite/protein > 1000). Usually, soaking in a 5 mM NaNO2 solution for 1 hour is enough for binding nitrite to copper(II) in protein crystals.30 There is clearly something intrinsic to the CuH site structure that prevents nitrite binding despite the presence of a vacant coordination position. Azide was similarly found coordinated to CuM but not CuH in the PHMcc-azide crystal structures (see below).
To enable comparisons between spectroscopic and X-ray structural data, PHMcc-nitrite crystals were prepared using the same concentration of sodium nitrite used in the spectroscopic studies carried out by Solomon and co-workers.7 Six transitions were observed in the MCD spectrum of nitrite-perturbed PHMcc [~ 6900 (+), ~ 8900 (−), 10,700 (+), 12,350 (−), 14,200 (+), and 17,000 (−) cm−1], while only two [~12,200 (+) and 16,700 (−) cm−1] are found in the spectrum of the resting PHMcc.7 As the CuH site appears unperturbed in the PHMcc-nitrite structure, the new spectroscopic features found for the nitrite-perturbed PHMcc, the absorptions at ~ 6900 (+), ~ 8900 (−), ~ 10,700 (+) and ~ 12,350 (−), must be assigned to the CuM-(NO2) site of the enzyme. The CuM2+ is in a five-coordinate environment with two histidines, one methionine and nitrite acting as an asymmetric bidentate ligand. The stereochemistry around the copper(II) ion is best described as a distorted square-pyramidal geometry (SPY) rather than trigonal bipyramidal (TBPY). The geometric parameter τ = (β-α)/60 (where β and α are the largest basal angles; τ = 0 corresponds to a perfect SPY while τ = 1 indicates a perfect TBPY) is used to characterize the stereochemistry around the CuM2+ ion.35 The basal plane is defined by the NHis242-SMet-O1nitrite-O2nitrite, where the largest angles are S-Cu-O1 = 152° (β) and N-Cu-O2 = 139° (α). Thus, the value of τ = 0.22 indicates a geometry for CuM better described as a distorted square-pyramid (SPY). The apical position is occupied by the Nε donor of His244. It is interesting to note that all four possible copper(II) d-d transitions are observed in the MCD spectrum of PHM for CuM when it is coordinated by nitrite. Spectral assignments to specific orbital transitions require a more detail ligand field analysis which is beyond the scope of the present work. The difference between the 14,200 (+) (nitrite-perturbed) MCD absorption band assigned to CuH and the band at ~12,200 (+) (native) could be due to the noticeable broadness of the band and the fact that the native ~12,200 (+) band is formed by contributions of both CuH and CuM electronic transitions. Also, binding of ligand to CuM produces in most cases small but significant changes in the histidines coordinating CuH. These involve mainly changes in the conformations of the coordinating histidines in the form of changes in χ2 but sometimes also in χ1. These changes are sometimes as large as 90 degrees. This type of changes will affect in a direct and significant way the interactions between the metal orbitals and those of the ligands and may result in significant spectral changes.
Further spectroscopic characterization was provided by X-band EPR studies.7 The spectrum of PHM shows only one set of Cu(II) hyperfine couplings (gx=2.050, gy=2.060, gz=2.288) making it impossible to distinguish CuM from CuH. The nitrite-PHMcc on the other hand shows two sets of Cu(II) hyperfine couplings in the gz region (2.265 and 2.298), thus distinguishing the two copper sites. The features at gx=2.060, gy=2.060, gz=2.265 can be assigned to the CuIIM-(NO2) site while the ones at gx=2.060, gy=2.060, gz=2.298 to the CuH, because only the CuH was found unperturbed with respect to the native enzyme. The distorted square pyramidal geometry found for CuM-(NO2) is consistent with the EPR spectrum (gz > gx, gy > 2.0) which clearly shows that the ground state orbital for the CuM2+ unpaired electron is d(x2-y2).
ox-PHMcc-azide complex
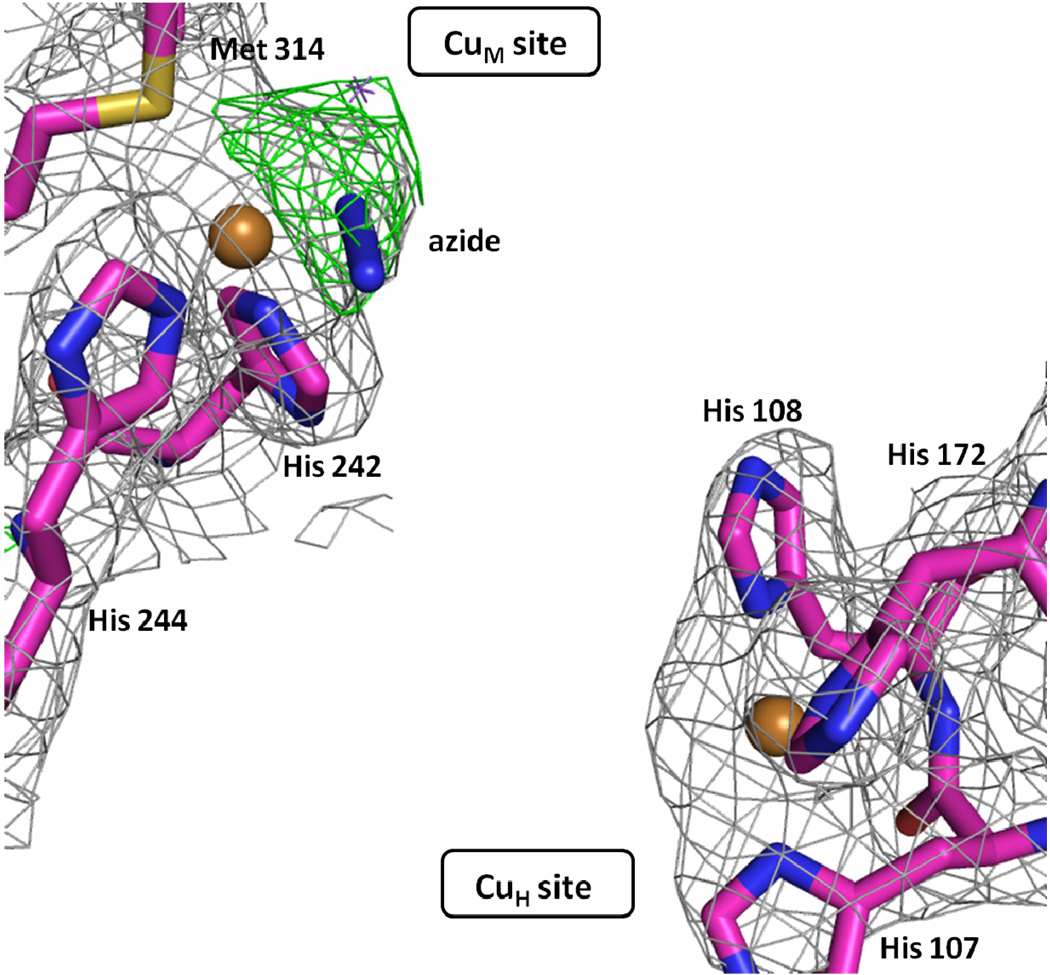
PHMcc crystals soaked in mother liquor supplemented with 350 mM NaN3, the azide concentration used in the spectroscopic studies of Solomon and co-workers,7 were seriously damaged after one hour and completely dissolved a few hours later. Crystals soaked for one hour were tested for X-ray diffraction but the resolution of the data was very low (> 3.5 Å). Crystals that diffracted to an acceptable resolution were prepared by co-crystallization using 40 mM NaN3. Final co-crystallization conditions were 0.5 mM CuSO4, 1.25 mM NiCl2, 100 mM sodium cacodylate pH = 5.5, 5% glycerol, and 40 mM NaN3, at room temperature. After 11 days, data were collected from a crystal that diffracted to 2.4 Å resolution. The structure determined using these data revealed an azide anion coordinated to the CuM (Cu-N = 2.0 Å), replacing the water ligand found in the oxidized state of the native enzyme, or the O2 bound in the reduced form in the presence of substrate, and keeping the same tetrahedral geometry around copper. Kinetic and spectroscopic studies of azide binding to dopamine β-hydroxylase (DBH), a tetrameric PHM analog, were interpreted as indicating that azide binds to a single copper per monomer, in agreement with the crystallographic data on PHM.36
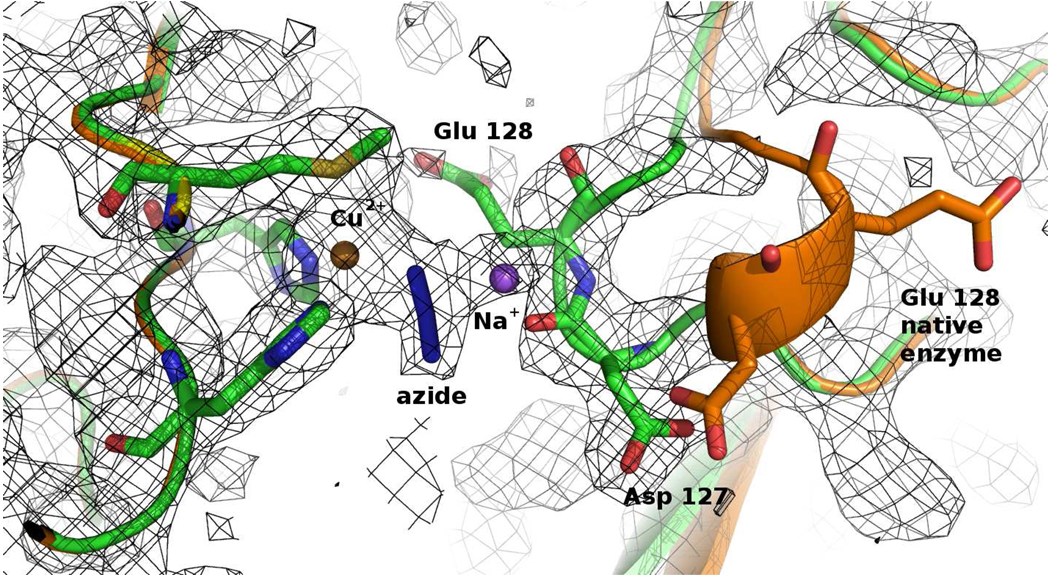
Electron density maps also showed a sodium ion bound to the azide ligand, behaving as a μ(1,1) bridging bidentate ligand connecting the CuM2+ to the Na+ (Figure 3). This Cu2+-:(N3−)-Na+ motif has not been previously observed in proteins and is very rare even in synthetic metal complexes.37 In addition, the loop Asp127-Glu128-Gly129-Thr130 moves closer to the CuM site, in comparison to the free enzyme (Figure 4). The sodium ion binds to the azide, attracts the carboxylate groups of the Asp127 and Glu128, and changes the conformation of the loop. The residues flanking the loop Cys126 and Cys131 are held by disulfide bridges to Cys81 and Cys114, respectively, preventing a large-scale propagation of the new conformation. However, this loop is not moved in PHMcc-azide crystals obtained by soaking (see Supporting Information), suggesting that this conformational change cannot take place within the native crystals upon binding of azide to the CuM site.
Figure 3. Structure of CuM and CuH sites in PHMcc-azide crystals.
The azide is shown bound to CuM in a monodentate fashion (Cu-N = 2.0 Å). The gray mesh shows the sigma weighted 2mFo-DFc (contoured at 1.0 σ) and the green mesh the mFo-DFc omit maps (contoured at 2.8 σ) The omit maps were computed with Refmac5. Carbons are colored magenta, nitrogens blue, oxygens red, sulphur yellow and copper gold.
Figure 4. The loop Asp127 to Thr130 is closer to the CuM site in the PHMcc-azide than in the native enzyme.
The gray mesh represents the 2Fo-Fc electron density contoured at 1.0 σ. Carbons are colored green, nitrogens blue, oxygens red, sulphur yellow, copper gold and sodium purple. The original position of the loop when PHM is crystallized with just 3 mM NaN3 (instead of 40 mM) is shown in orange.
The coordination of the CuH site in the structure of the PHMcc-azide crystal is also intriguing. In the free enzyme, CuH has a vacant coordination site that could potentially be occupied by a fourth ligand. However, CuH does not bind azide (Figure 3). This is true not only in the co-crystallized sample at lower azide concentration. Despite the low resolution (3.0 to 3.3 Å) of the X-ray datasets obtained in soaking experiments at (i) 100 mM NaN3 for 1.5 hours, (ii) 50 mM NaN3 for 26 hours and (iii) 50 mM NaN3 for 6 hours, the structures of these crystals all revealed azide bound to CuM but not to CuH (see Table 1 and Supporting Information), consistent with the observations in the higher resolution structure obtained by co-crystallization. Further, the high B factor exhibited for CuH in all datasets (80–105 Å2) in comparison to the protein average B factor (50 Å2) may be indicative of an occupancy lower than 1.0, suggesting that azide may remove CuH after long exposure of crystals to high NaN3 concentration.
Table 1.
Statistics for crystallographic data collection and refinement for oxidized-PHMcc complexes under different conditions
| ox-PHMcc-NO2− 300 mM 20 h soak |
ox-PHMcc-N3− 40 mM Co-crystal |
ox-PHMcc-N3− 100 mM 1.5 h soak |
ox-PHMcc-N3− 50 mM 26 h soak |
ox-PHMcc-CO 3 atm 15 min. soak |
ox-PHMcc- subs-nitrite |
ox-PHMcc- subs-azide |
|
|---|---|---|---|---|---|---|---|
| Unit cell parameters (Å) |
a = 68.6 b = 68.6 c = 80.3 |
a = 68.5 b = 68.6 c = 81.4 |
a = 68.9 b = 69.3 c = 81.5 |
a = 68.7 b = 69.0 c = 81.3 |
a = 69.2 b = 69.7 c = 83.1 |
a = 68.4 b = 68.8 c = 80.0 |
a = 68.1 b = 68.8 c = 79.8 |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Source | X6A | Rotating anode | Rotating anode | Rotating anode | X25 | X4C | X4C |
| Wavelength (Å) | 1.00 | 1.54 | 1.54 | 1.54 | 1.10 | 0.98 | 0.98 |
| Resolution (Å) | 52 – 2.35 | 52 – 2.40 | 53 – 3.05 | 53 – 3.25 | 35 – 2.00 | 52 – 2.70 | 52 – 2.75 |
| Unique reflections | 16,345 | 15,128 | 7,716 | 6,295 | 37,462 | 10,776 | 8,970 |
| Redundancy | 7.1 (6.8) | 5.0 (5.1) | 5.3 (5.4) | 5.6 (5.8) | 5.9 (6.7) | 7.0 (5.6) | 4.8 (3.0) |
| Completeness (%) | 99.7 (100.0) | 99.5 (99.9) | 99.8 (100.0) | 98.7 (99.5) | 97.0 (100.0) | 99.0 (91.4) | 87.3 (49.3) |
| <I>/s<I> | 49.4 (3.7) | 25.2 (3.1) | 22.1 (4.1) | 20.9 (4.3) | 4.6 (1.5) | 35.7 (2.1) | 21.1 (1.7) |
| Rsym (%) | 4.6 (49) | 11.1 (59) | 10.0 (51) | 10.3 (54) | 8.0 (41) | 8.5 (53) | 8.9 (41) |
| Refinement | |||||||
| Rcryst / Rfree (%) | 20 / 24 | 20 / 26 | 20 / 25 | 19 / 24 | 21 / 24 | 22 / 27 | 24 / 30 |
| Stereochemistry | |||||||
| R.M.S. bond lengths (Å) | 0.007 | 0.008 | 0.011 | 0.012 | 0.009 | 0.012 | 0.006 |
| R.M.S. angles (°) | 1.06 | 1.07 | 1.15 | 1.20 | 1.10 | 1.22 | 0.97 |
| Model composition | |||||||
| Amino acids | 308 | 309 | 311 | 310 | 310 | 309 | 307 |
| Cu / Ni / Na | 2 / 1 / - | 2 / 1 / 2 | 2 / 1 / - | 2 / 1 / 1 | 2 / 1 / - | 2 / 1 / - | 2 / 1 / 4 |
| Nitrite | 3 | - | - | - | - | 2 | - |
| Azide | - | 2 | 3 | 3 | - | - | 6 |
| CO | - | - | - | - | 1 | - | - |
| Glycerol | 3 | 3 | 3 | 3 | 4 | 1 | - |
| Water | 168 | 215 | 16 | 12 | 163 | 18 | 18 |
| Total atoms | 2,597 | 2,677 | 2,471 | 2,461 | 2,692 | 2,443 | 2,437 |
| PDB ID code | 3MIB | 3MIC | 3MID | 3MIE | 3MIF | 3MIG | 3MIH |
Numbers in parenthesis correspond to the last resolution shell.
Differences observed between the X-ray structures of PHMcc-azide complexes determined here and the published spectroscopic data (MCD, EPR) obtained on PHMcc solution with 350 mM NaN3,7 may be a consequence of the different NaN3 concentration used in the experiments.
red-PHMcc-CO complex
The ability of metal ions to bind ligands and stabilize their coordination sphere depends, among other factors, on their oxidation state. Both copper ions in PHM alternate between Cu(II) and Cu(I) in the catalytic cycle.38 Thus, the behavior of reduced PHM in the presence of carbon monoxide – a very good ligand for copper(I) – was investigated (see Table 2 for crystallographic data). A PHMcc crystal in 5 mM ascorbic acid was exposed to 3 atm CO for 15 minutes and then analyzed by X-ray crystallography (see experimental section for further details). The structure shows CO bound to CuM as an “end-on, bent” ligand; Cu-C distance is 1.8 Å and CuM-C-O angle is 110° (Figure 5). This geometry is the same geometry adopted by dioxygen in the precatalytic intermediate formed by soaking PHMcc crystals with slow substrate and ascorbate in the presence of dioxygen.34 Again, no electron density is found at the open coordination position of CuH, indicating that Cu(I)H is not reactive against carbon monoxide. A water molecule is found interacting very weakly with the CuH (CuH-Owater = 3.3 Å, not shown in Figure 5). The lack of binding of CO at the CuH site is consistent with the lack of reactivity against molecular oxygen observed for the precatalytic intermediate of PHMcc (see above). In the final refinement, CuH refined to an occupancy of 0.7, a fact that reflects that part of the CuH is displaced under the conditions of the experiment. Binding of CO to CuM and the lack of reactivity of CuH is fully consistent with the previous results of EXAFS and FTIR studies39 as well as with studies of DBH, a PHM analog.40–41
Table 2.
Statistics for crystallographic data collection and refinement for reduced-PHMcc complexes under different conditions
| reduced-PHMcc-nitrite 300 mM |
reduced-PHMcc-azide 50 mM |
reduced-PHMcc-CO 3 atm |
|
|---|---|---|---|
| Unit cell parameters (Å) |
a = 69.0 b = 69.3 c = 81.1 |
a = 68.1 b = 69.1 c = 79.9 |
a = 69.2 b = 68.9 c = 81.7 |
| Space group | P212121 | P212121 | P212121 |
| Source | X4C | X4C | Rotating anode |
| Wavelength | 0.98 | 0.98 | 1.54 |
| Resolution (Å) | 53 – 3.10 | 52 – 3.25 | 53 – 2.15 |
| Unique | 7,468 | 6,287 | 21,690 |
| Redundancy | 7.0 (6.0) | 5.7 (4.6) | 4.5 (4.4) |
| Completeness (%) | 99.9 (99.0) | 99.1 (91.9) | 98.1 (99.4) |
| <I>/s<I> | 23.4 (2.2) | 25.3 (2.2) | 29.7 (2.6) |
| Rsym (%) | 8.1 (57) | 8.8 (47) | 4.6 (66) |
| Refinement | |||
| Rcryst / Rfree (%) | 22 / 28 | 24 / 30 | 21 / 26 |
| Stereochemistry | |||
| R.M.S. bond lengths(Å) | 0.009 | 0.007 | 0.009 |
| R.M.S. angles (°) | 1.11 | 1.06 | 1.11 |
| Model composition | |||
| Amino acids | 306 | 304 | 310 |
| Cu / Ni / Na | 1 / 1 / - | 1 / 1 / - | 2 / 1 / - |
| Nitrite / Azide / CO | 2 / - / - | - / 1 / - | - / - / 2 |
| Acetate | - | - | 1 |
| Glycerol | - | - | 3 |
| Water | 12 | 2 | 153 |
| Total | 2,408 | 2,376 | 2,667 |
| PDB ID code | 3MLK | 3MLL | 3MLJ |
Numbers in parenthesis correspond to the last resolution shell.
Figure 5. The structure of CuM and CuH sites of the reduced-PHMcc-CO complex.
The CO is shown bound to CuM in a bent configuration (Cu-C = 1.8 Å; Cu-C-O = 110°). The gray mesh shows the sigma weighted 2mFo-DFc (contoured at 1.0 σ) and the green mesh the mFo-DFc omit maps (contoured at 2.8 σ) The omit maps were computed with Refmac5. Carbons are colored yellow, nitrogens blue, oxygens red, sulfur yellow-orange and copper gold.
ox-PHMcc-CO complex
Although CO is not as good a ligand for Cu(II) as for Cu(I), CO-soaking experiments were carried out with the oxidized form of the enzyme to further probe the reactivity of Cu(II)H and to compare it with the structure of the reduced PHMcc-CO. The structure revealed that CO coordinates Cu(II)M with the same geometry observed in the complex with the reduced form of the enzyme. However, in addition to its regular conformation, the loop spanning residues 127–130 is found also in an alternative conformation (occupancy=0.5), similar to that found in the ox-PHMcc-azide structure obtained by co-crystallization (see above). In the CO case, the carboxylate of Glu128 of the new conformation coordinates CuM as a monodentate ligand. Even though pressure unfolding usually requires much higher pressures (e.g. on the order of 10,000 psi)42 than that used in these investigations (50 psi), one tentative explanation for this double conformation is that it was caused by pressure.
red-PHMcc-nitrite and red-PHMcc-azide complexes
The behavior of reduced PHM in the presence of high concentrations of nitrite and azide was also investigated (see Table 2 for crystallographic data). As in the oxidized state of the enzyme, nitrite and azide bind Cu(I)M (Figure S4 in Supporting Information). Surprisingly, Cu(I)H is completely removed from its site in the presence of the high concentrations of nitrite or azide (300 mM NaNO2 or 40 mM NaN3). Also, in the azide complex, the Cu(I)M site is only partially occupied; the structure was refined with half occupancy for the Cu(I)M-azide moiety. The low resolution of these crystals (> 3 Å) in comparison with the typical resolution of reduced PHM crystals (around 2 Å) is probably related to structural deterioration caused by the loss of CuH.
Binding experiments in the presence of substrate
We carried out experiments to investigate whether the presence of substrate has any effect at the copper centers that could modify their chemical behavior towards molecules such as nitrite or azide. Our experiments had to be carried out in the oxidized state of the enzyme because CuH is removed from the protein in the reduced state (see above). Using the substrate (N-α-acetyl-3,5-diiodotyrosylglycine, Ac-DiI-YG) and the experimental conditions reported previously,8,43 crystals of PHM-substrate were prepared by soaking for one hour native crystals of PHMcc in mother liquor (ML) supplemented with 1 mM substrate, followed by soaking in ML supplemented with 1mM substrate and sodium nitrite or sodium azide (see Table 1 for crystallographic data). The X-ray structures did not show bound substrate in either case but, as found in the experiments without substrate, CuM binds nitrite or azide while CuH did not (Figure S5 in Supporting Information). Binding of nitrite or azide and/or high concentration of sodium nitrite or sodium azide in the media seem to prevent substrate binding to PHM.
Why does CuH NOT bind small molecules?
We have shown that under the conditions used azide, nitrite and carbon monoxide do not bind to CuH. We have previously shown that CuH does not bind dioxygen under conditions in which it binds to CuM. Blackburn and coworkers showed that isocyanides bind to CuM but not to CuH.44 They also showed that in DBH, CO binds to a single copper per monomer.40 All these results point to CuH as a metal site with unusual reactivity.
The catalytic α-hydroxylation reaction uses dioxygen as the oxygen source. The two electrons required for the dioxygen reduction are provided by ascorbate via one-electron reduction of CuM and CuH. In this manner, PHM starts its catalytic cycle in the CuIM…CuIH stage. The substrate binds close to the CuM site and a reactive oxygen species bound to CuM is responsible for substrate hydrogen abstraction. In this scenario, CuH is an electron-transfer site, providing the second electron required to complete the catalytic cycle. The driving force for this step is given by the difference in redox potential between CuH and that of the acceptor species, which includes an oxygen-bound CuM. The lack of coordination of small molecules to CuH is probably related to the necessity for CuH to maintain its redox potential in a narrow range to maintain its function as an electron donor. It is known that alterations in the coordination sphere (number and type of ligand donor atoms) and stereochemistry at the metal center can produce great differences in the potential at which electron-transfer reactions occur.45 The change from three histidines T-shape geometry to, for instance, three histidines and one small ligand (e.g. monodentate nitrite, azide or carbon monoxide) square planar geometry would surely change Cu(I)/(II)H redox potential. Were this site accessible to small molecule binding, metabolites present in the physiological environment would change the enzyme kinetics.
Binding of O2 to Cu(I)H would be particularly detrimental as it could be reduced to superoxide and released. The X-ray structure of reduced PHMcc with dioxygen bound to the CuM in the presence of a slow-substrate, revealed that CuH does not bind dioxygen.34 This is essentially the same reactivity we observe with carbon monoxide. These results are consistent with the proposal presented here: the architecture of the CuH site prevents small molecule binding to avoid any redox potential change.
From a chemical point of view it is very difficult to understand why very good copper ligands such as nitrite, azide or carbon monoxide do not bind CuH, since there is a vacant position on its coordination sphere. Apparently the bis-histidines His107–His108, in concert with His172 -all three histidines binding through the Nδ donor- form a particular geometry and electronic structure for CuH that prevents binding at the fourth vacant position. One characteristic of the CuH coordination that may account for its lack of reactivity is its unusual geometry: the three histidines are in a shape that we described in general as a T-configuration. However, the coordination of CuH is more unusual than that. The three His-Cu-His angles in the 13 complexes (10 reported in this manuscript) are the following: H107-CuH-H108 ranges between 138.6 and 151.4 degrees; H107-CuH-H172 between 107.2 and 111.9; and H108-CuH-H172 between 94.3 and 110.9. This arrangement is somewhere between a square planar with one empty position and a planar trigonal. It is possible that this coordination, imposed by the protein environment, prevents binding to a fourth site. In addition, in all three cases the histidines coordinate the copper with their Nδ the most positionally restricted of the two histidine nitrogens, making it more difficult for the coordination to rearrange to accommodate a fourth ligand. A detailed quantum mechanical analysis of CuH could provide insights on this matter.
The metal coordination in outer-sphere electron-transfer sites is found fully occupied with ligands provided by the protein structure. Type-1 copper sites in blue copper proteins, which almost exclusively function as electron-transfer molecules, are coordinated by two His and one Cys in a distorted trigonal plane with an axial protein residue (usually Met) in an overall distorted tetrahedral geometry; no vacant/open position is present in their coordination sphere.46–47 The binuclear CuA center as in cytochrome c oxidase is another example.48 Recently, a red copper protein with a copper site with an open position in the reduced form (a water molecule occupies that position in the oxidized form) has been spectroscopically and structurally characterized.49–50 It had been proposed that this red copper protein (absorption band at 390 nm), isolated from Nitrosomonas europaea, might be involved in electron-transfer because it shares significant sequence homology to blue copper proteins and has a typical Cu-thiolate bond. However, later investigations suggest that electron-transfer is accomplished by an inner-sphere mechanism.51
The same feature (absence of an open coordination position) is observed for iron electron-transfer proteins that work via an outer-sphere mechanism. From the simple tetrahedral iron-sulfur center such as is found in rubredoxin to the very complex iron-sulfur clusters as in other ferredoxins,52 and even the six-coordinated heme in cytochromes,53 in all cases, efficient outer-sphere electron-transfer is guaranteed with a fully saturated coordination sphere for the metal site. The only exception is the heme at cytochrome c’ which has an open coordination position at the active site;54 however its physiological role as an electron-transfer site is still unclear.55–56
In summary, all very well known outer-sphere electron-transfer metal-sites have the attribute to be fully saturated in their coordination sphere by ligands provided by the protein structure. Surprisingly, the CuH outer-sphere electron-transfer site in PHM has an apparent open coordination position that remains empty at high concentrations of small anions/molecules such as nitrite, azide and carbon monoxide. Something essential with the typical architecture of CuH creates an electronic structure for CuH that impedes the binding of an extra-ligand. This property ensures CuH maintains its redox potential in a narrow range compatible to its role as an efficient electron-transfer in the catalytic cycle of PHM.
Experimental Section
Preparation of crystals and soaking experiments
Stably transfected Chinese Hamster Ovary (CHO) cell lines secreting PHMcc (residues 42 to 356) were constructed using the pCIS vector system57 and purified as described previously.6 The native protein was crystallized by the hanging-drop diffusion method at 293 K. Thus, PHMcc (1 µl; concentration = 16 mg/ml) was mixed with an equal volume of mother liquor (0.1–0.5 mM CuSO4, 1.0–1.25 mM NiCl2, 100 mM sodium cacodylate pH = 5.5, 3.08 mM NaN3, 5% glycerol) and crystals appeared in 3–4 days. As reported before, nickel(II) ion is incorporated into PHMcc structure as a crystal contact site.
Oxidized PHMcc-nitrite crystals
Nitrite-soaked PHMcc crystals were obtained by placing native PHMcc crystals in mother liquor supplemented with 300 mM NaNO2 for 20 hours at room temperature. The crystals were then transferred to fresh mother liquor supplemented with 300 mM NaNO2 and 30 % glycerol as a cryo-protectant, prior to flash-freezing. Data collection and refinement statistics are reported in Table 1.
Oxidized PHMcc-azide crystals
ox-PHM-azide crystals were obtained by co-crystallization at 293 K, using the same methodology as for crystallizing native PHMcc but the mother liquor contained 40 mM NaN3 instead of 3.08 mM. Crystals appear in 3–4 days.
Oxidized PHMcc-CO crystals
A pressure chamber built by Kas Kumar was used for CO soaking into PHM (unpublished work). The reservoir of the pressure chamber was filled with mother liquor containing 25% of glycerol and saturated with CO. Native PHMcc crystals were soaked into the CO-containing cryoprotectant solution for less than a minute and transferred with a loop to the pressure chamber. The chamber was purged several times with low pressure of CO, then gradually filled with 50 psi CO and finally maintained at this pressure during 30 minutes. After bringing back the chamber to atmospheric pressure, the crystal was rapidly removed and cryo-frozen in liquid nitrogen.
Oxidized PHMcc-nitrite crystals in the presence of substrate
PHMcc crystals, grown as described above, were first soaked in mother liquor supplemented with 1 mM substrate (Ac-3,5-diI-YG) for 1 hour at room temeprature. Then the crystals were soaked in a solution containing 300 mM NaNO2, 1 mM substrate and the rest of the mother liquor components, for 14 hours at room temperature.
Oxidized PHMcc-azide crystals in the presence of substrate
Crystals of PHMcc complexed with substrate were obtained as described above (1 hour soaking in mother liquor with 1 mM substrate). Then the crystals were soaked in a solution containing 40 mM NaN3, 1 mM substrate and the rest of the mother liquor components, for 8 hours at room temperature.
Reduced PHMcc-CO crystals
Reduced CO-soaked PHMcc crystals were prepared following the same pressure chamber and protocol described for the oxidized CO-soaked PHMcc crystals (see above) but adding 5 mM ascorbic acid to the cryoprotectant solution and the crystal was in the pressure chamber filled with 50 psi CO for 15 minutes.
Reduced PHMcc-nitrite crystals
PHMcc crystals, grown as described above, were reduced with ascorbic acid by soaking native crystals for 1 hour in mother liquor with 5 mM ascorbic acid (1.85 mM NiCl2, 100 mM sodium cacodylate pH = 5.5, 3.08 mM NaN3, 5% glycerol). The reduced crystals were then soaked in a solution containing 300 mM NaNO2, 5 mM ascorbic acid and the rest of the mother liquor components, for 13 hours at room temperature.
Reduced PHMcc-azide crystals
Reduced PHMcc crystals were obtained as described above, then soaked in a solution containing 40 mM NaN3, 5 mM ascorbic acid in mother liquor components, for 6 hours at room temperature.
Data collection, structures determination and refinement
Diffraction data were collected on single frozen crystals, either at a home source (Rigaku RU-200 rotating anode and R-AXIS IV image plate detector) or at beamline X6A, X4C or X25 of the National Synchrotron Light Source at Brookhaven National Laboratory. Frames were processed with HKL2000 software package.58 All crystals belong to the orthorhombic space group P212121. All structures were determined by molecular replacement with the program AMoRe59 using the coordinates of the native enzyme (1PHM.pdb) as the search model. The models were built interactively with program O60 and refined using REFMAC 5.0 as implemented in CCP4 suite programs.61–62 Anisotropic refinement using translation, libration and screw rotation (TLS) of rigid bodies was carried out using four groups (residues 46–65; 66–185; 186–302 and 303–354) or each domain (46–197 and 198–354) as TLS group.63–64 Solvent molecules were added automatically using program ARP/WARP62,65 and visually inspected with program O.60 Refinement was monitored by calculating Rfree values using 5% of the reflections set aside for cross-validation. Crystallographic data collection and refinement statistics are summarized in Tables 1 and 2.
Supplementary Material
Acknowledgments
Mario Bianchet and the staff at beamlines X6A, X4C and X25 of the National Synchrotron Light Source, Brookhaven National Laboratory, are gratefully acknowledged for assistance during synchrotron data collection. This work was supported by a National Science Foundation grant MCB-0450465 (L.M.A. and S.T.P.) and National Institutes of Health grant DK032949 (E.A.E.).
Footnotes
Accession Numbers
Atomic coordinates and structure factors have been submitted to the Protein Data Bank; PDB ID codes are indicated in Tables 1 and 2.
Supporting Information
Supporting Information Available: Description of the material included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Eipper BA, Stoffers DA, Mains RE. Annu Rev Neurosci. 1992;15:57. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 2.Merkler DJ. Enzyme Microb Technol. 1994;16:450. doi: 10.1016/0141-0229(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 3.Prigge ST, Mains RE, Eipper BA, Amzel LM. Cellular and Molecular Life Sciences. 2000;57:1236. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolhekar AS, Bell J, Shiozaki EN, Jin L, Keutmann HT, Hand TA, Mains RE, Eipper BA. Biochemistry-Us. 2002;41:12384. doi: 10.1021/bi0260280. [DOI] [PubMed] [Google Scholar]
- 5.Chufan EE, De M, Eipper BA, Mains RE, Amzel LM. Structure. 2009;17:965. doi: 10.1016/j.str.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Science. 1997;278:1300. doi: 10.1126/science.278.5341.1300. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Bell J, Eipper BA, Solomon EI. Biochemistry-Us. 2004;43:5735. doi: 10.1021/bi0362830. [DOI] [PubMed] [Google Scholar]
- 8.Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Nat Struct Biol. 1999;6:976. doi: 10.1038/13351. [DOI] [PubMed] [Google Scholar]
- 9.Klinman JP. J Biol Chem. 2006;281:3013. doi: 10.1074/jbc.R500011200. [DOI] [PubMed] [Google Scholar]
- 10.Crespo A, Marti MA, Roitberg AE, Amzel LM, Estrin DA. J Am Chem Soc. 2006;128:12817. doi: 10.1021/ja062876x. [DOI] [PubMed] [Google Scholar]
- 11.Wijma HJ, MacPherson I, Farver O, Tocheva EI, Pecht I, Verbeet MP, Murphy ME, Canters GW. J Am Chem Soc. 2007;129:519. doi: 10.1021/ja064763j. [DOI] [PubMed] [Google Scholar]
- 12.MacPherson IS, Murphy MEP. Cellular and Molecular Life Sciences. 2007;64:2887. doi: 10.1007/s00018-007-7310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehnert N, Cornelissen U, Neese F, Ono T, Noguchi Y, Okamoto K, Fujisawa K. Inorganic Chemistry. 2007;46:3916. doi: 10.1021/ic0619355. [DOI] [PubMed] [Google Scholar]
- 14.Karlin KD, Tyeklár Z. Bioinorganic chemistry of copper. New York: Chapman & Hall; 1993. [Google Scholar]
- 15.Tolman WB. Inorganic Chemistry. 1991;30:4877. [Google Scholar]
- 16.Ruggiero CE, Carrier SM, Tolman WB. Angew Chem Int Edit. 1994;33:895. [Google Scholar]
- 17.Walsh A, Walsh B, Murphy B, Hathaway BJ. Acta Crystallogr B. 1981;37:1512. [Google Scholar]
- 18.Kujime M, Izumi C, Tomura M, Hada M, Fujii H. Journal of the American Chemical Society. 2008;130:6088. doi: 10.1021/ja075575b. [DOI] [PubMed] [Google Scholar]
- 19.Halfen JA, Mahapatra S, Wilkinson EC, Gengenbach A, Young VG, Jr, Que L, Tolman WB. J. Am. Chem. Soc. 1996;118:763. [Google Scholar]
- 20.Tocheva EI, Eltis LD, Murphy MEP. Biochemistry-Us. 2008;47:4452. doi: 10.1021/bi7020537. [DOI] [PubMed] [Google Scholar]
- 21.Rogers MS, Tyler EM, Akyumani N, Kurtis CR, Spooner RK, Deacon SE, Tamber S, Firbank SJ, Mahmoud K, Knowles PF, Phillips SEV, McPherson MJ, Dooley DM. Biochemistry-Us. 2007;46:4606. doi: 10.1021/bi062139d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai LC, Bonander N, Harata K, Karlsson G, Vanngard T, Langer V, Sjolin L. Acta Crystallogr D. 1996;52:950. doi: 10.1107/S0907444996004982. [DOI] [PubMed] [Google Scholar]
- 23.Casella L, Gullotti M, Pallanza G, Buga M. Inorganic Chemistry. 1991;30:221. [Google Scholar]
- 24.Pettinari C. Polyhedron. 2001;20:2755. [Google Scholar]
- 25.Tocheva EI, Rosell FI, Mauk AG, Murphy ME. Science. 2004;304:867. doi: 10.1126/science.1095109. [DOI] [PubMed] [Google Scholar]
- 26.Dodd FE, Van Beeumen J, Eady RR, Hasnain SS. J Mol Biol. 1998;282:369. doi: 10.1006/jmbi.1998.2007. [DOI] [PubMed] [Google Scholar]
- 27.Murphy MEP, Turley S, Adman ET. Journal of Biological Chemistry. 1997;272:28455. doi: 10.1074/jbc.272.45.28455. [DOI] [PubMed] [Google Scholar]
- 28.Antonyuk SV, Strange RW, Sawers G, Eady RR, Hasnain SS. Proc Natl Acad Sci U S A. 2005;102:12041. doi: 10.1073/pnas.0504207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson F, Pistorius A, Farkas D, De Grip W, Hansson O, Sjolin L, Neutze R. J Biol Chem. 2007;282:6347. doi: 10.1074/jbc.M605746200. [DOI] [PubMed] [Google Scholar]
- 30.Boulanger MJ, Murphy ME. Protein Sci. 2003;12:248. doi: 10.1110/ps.0224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen FH. Acta Crystallogr B. 2002;58:380. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 32.Chattopadhyay S, Drew MGB, Ghosh A. Inorg Chim Acta. 2006;359:4519. [Google Scholar]
- 33.Shim YB, Choi SN, Lee SJ, Kang SK, Lee YM. Acta Crystallogr E. 2004;60:M1573. [Google Scholar]
- 34.Prigge ST, Eipper BA, Mains RE, Amzel LM. Science (Washington, DC, United States) 2004;304:864. doi: 10.1126/science.1094583. [DOI] [PubMed] [Google Scholar]
- 35.Addison AW, Rao TN, Reedijk J, van Rijn J, Verschoor GC. J. Chem. Soc. Dalton Trans. 1984:1349. [Google Scholar]
- 36.Blackburn NJ, Collison D, Sutton J, Mabbs FE. Biochem J. 1984;220:447. doi: 10.1042/bj2200447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goher MAS, Mautner FA. Polyhedron. 1995;14:1439. [Google Scholar]
- 38.Freeman JC, Villafranca JJ, Merkler DJ. J. Am. Chem. Soc. 1993;115:4923. [Google Scholar]
- 39.Jaron S, Blackburn NJ. Biochemistry-Us. 1999;38:15086. doi: 10.1021/bi991341w. [DOI] [PubMed] [Google Scholar]
- 40.Blackburn NJ, Pettingill TM, Seagraves KS, Shigeta RT. J Biol Chem. 1990;265:15383. [PubMed] [Google Scholar]
- 41.Pettingill TM, Strange RW, Blackburn NJ. J Biol Chem. 1991;266:16996. [PubMed] [Google Scholar]
- 42.Panick G, Malessa R, Winter R, Rapp G, Frye KJ, Royer CA. Journal of Molecular Biology. 1998;275:389. doi: 10.1006/jmbi.1997.1454. [DOI] [PubMed] [Google Scholar]
- 43.Siebert XPhD. Johns Hopkins School of Medicine. 2005 [Google Scholar]
- 44.Rhames FC, Murthy NN, Karlin KD, Blackburn NJ. J Biol Inorg Chem. 2001;6:567. doi: 10.1007/s007750100233. [DOI] [PubMed] [Google Scholar]
- 45.Lippard SJ, Berg JM. Principles of bioinorganic chemistry. Mill Valley, Calif: University Science Books; 1994. [Google Scholar]
- 46.Solomon EI, Szilagyi RK, DeBeer George S, Basumallick L. Chem Rev. 2004;104:419. doi: 10.1021/cr0206317. [DOI] [PubMed] [Google Scholar]
- 47.Sakurai T, Kataoka K. Cell Mol Life Sci. 2007;64:2642. doi: 10.1007/s00018-007-7183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim E, Chufan EE, Kamaraj K, Karlin KD. Chemical Reviews (Washington, DC, United States) 2004;104:1077. doi: 10.1021/cr0206162. [DOI] [PubMed] [Google Scholar]
- 49.Basumallick L, Sarangi R, DeBeer George S, Elmore B, Hooper AB, Hedman B, Hodgson KO, Solomon EI. J Am Chem Soc. 2005;127:3531. doi: 10.1021/ja044412+. [DOI] [PubMed] [Google Scholar]
- 50.Lieberman RL, Arciero DM, Hooper AB, Rosenzweig AC. Biochemistry-Us. 2001;40:5674. doi: 10.1021/bi0102611. [DOI] [PubMed] [Google Scholar]
- 51.Solomon EI. Inorg Chem. 2006;45:8012. doi: 10.1021/ic060450d. [DOI] [PubMed] [Google Scholar]
- 52.Venkateswara Rao P, Holm RH. Chem Rev. 2004;104:527. doi: 10.1021/cr020615+. [DOI] [PubMed] [Google Scholar]
- 53.Walker FA. Chem Rev. 2004;104:589. doi: 10.1021/cr020634j. [DOI] [PubMed] [Google Scholar]
- 54.Benini S, Rypniewski WR, Wilson KS, Ciurli S. J Inorg Biochem. 2008;102:1322. doi: 10.1016/j.jinorgbio.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 55.Meyer TE, Cheddar G, Bartsch RG, Getzoff ED, Cusanovich MA, Tollin G. Biochemistry-Us. 1986;25:1383. doi: 10.1021/bi00354a029. [DOI] [PubMed] [Google Scholar]
- 56.Mayburd AL, Kassner RJ. Biochemistry-Us. 2002;41:11582. doi: 10.1021/bi020058l. [DOI] [PubMed] [Google Scholar]
- 57.Kolhekar AS, Keutmann HT, Mains RE, Quon AS, Eipper BA. Biochemistry-Us. 1997;36:10901. doi: 10.1021/bi9708747. [DOI] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. Methods in Enzymology. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.Navaza J. Acta Crystallogr D Biol Crystallogr. 2001;57:1367. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 60.Jones TA, Kjeldgaard M. Methods Enzymol. 1997;277:173. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 61.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 62.Acta Crystallogr D Biol Crystallogr. 1994;50:760. [Google Scholar]
- 63.Painter J, Merritt EA. J Appl Crystallogr. 2006;39:109. [Google Scholar]
- 64.Winn MD, Murshudov GN, Papiz MZ. Methods Enzymol. 2003;374:300. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 65.Lamzin VS, Wilson KS. Method Enzymol. 1997;277:269. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.