Abstract
The genus Clostridium includes major human pathogens and species important to cellulose degradation, the carbon cycle, and biotechnology. Small RNAs (sRNAs) are emerging as crucial regulatory molecules in all organisms, but they have not been investigated in clostridia. Research on sRNAs in clostridia is hindered by the absence of a systematic method to identify sRNA candidates, thus delegating clostridial sRNA research to a hit-and-miss process. Thus, we wanted to develop a method to identify potential sRNAs in the Clostridium genus to open up the field of sRNA research in clostridia. Using comparative genomics analyses combined with predictions of rho-independent terminators and promoters, we predicted sRNAs in 21 clostridial genomes: Clostridium acetobutylicum, C. beijerinckii, C. botulinum (eight strains), C. cellulolyticum, C. difficile, C. kluyveri (two strains), C. novyi, C. perfringens (three strains), C. phytofermentans, C. tetani, and C. thermocellum. Although more than one-third of predicted sRNAs have Shine-Dalgarno (SD) sequences, only one-sixth have a start codon downstream of SD sequences; thus, most of the predicted sRNAs are noncoding RNAs. Quantitative reverse transcription-PCR (Q-RT-PCR) and Northern analysis were employed to test the presence of a randomly chosen set of sRNAs in C. acetobutylicum and several C. botulinum strains, leading to the confirmation of a large fraction of the tested sRNAs. We identified a conserved, novel sRNA which, together with the downstream gene coding for an ATP-binding cassette (ABC) transporter gene, responds to the antibiotic clindamycin. The number of predicted sRNAs correlated with the physiological function of the species (high for pathogens, low for cellulolytic, and intermediate for solventogenic), but not with 16S rRNA-based phylogeny.
IMPORTANCE
Clostridia include major human pathogens and species important to human physiology, cellulose degradation, the carbon cycle, and biotechnology. Small RNAs (sRNAs) are increasingly recognized as crucial regulatory molecules in all organisms, but they remain virtually unexplored in clostridia. We provide the first comprehensive list of computationally identified and experimentally verified small RNAs in the genus Clostridium aiming to accelerate interest in and studies of small RNA molecules in a very important genus. The higher number of sRNAs found in clostridial pathogens suggests a good correlation between the physiological function or niche of the species and the number of predicted and conserved sRNAs. Our list of predicted sRNAs displays a strong enrichment of sRNAs upstream or downstream of ATP-binding cassette (ABC) transporter genes. This, combined with the identification of a conserved sRNA apparently involved in clindamycin resistance, provides a new perspective for future studies of possible regulation of antibiotic resistance genes by sRNAs in bacteria.
INTRODUCTION
Prokaryotic small RNAs (sRNAs) play important regulatory roles in a variety of cellular processes. They are typically 50 to 500 nucleotides (nt) in length and are found on intergenic regions (IRs) (1). Most of these functional RNA molecules normally do not possess a protein-coding function, but some do. They typically act as posttranscriptional regulators by interacting with specific mRNA targets, modulating target stability and/or translation initiation (2). Since the discovery of regulatory sRNA in Escherichia coli (3, 4), several genome-wide methods for sRNA discovery have been developed by combining computational searches with experimental validation of select candidates (5, 6). Using a comparative genomics screen approach, Rivas et al. (7) predicted 275 sRNAs in E. coli, and more than 11 out of the 49 tested candidates were experimentally verified. With the availability of an increasing number of bacterial genome sequences, such strategies have been employed for the discovery of many sRNAs not only in E. coli but also in other prokaryotes (8–13). Most methods that have been developed to predict prokaryotic sRNAs (14–16) employ comparative genomics approaches and have typically been applied to a few genomes in the members of a genus. In addition, most rely solely on comparative genome screening and typically identify highly conserved sRNAs. Promoter and terminator information, which can be important in identifying the length and orientation of sRNAs, is rarely used in such predictions. Two of the most successful predictive tools are the comparative genomics-based computational tools sRNAPredict (17) and SIPHT (18), which combine the positions of various predictive features of sRNAs to predict IR-located sRNAs and which were developed by Linvy et al.
Clostridium is an important prokaryotic genus which includes many species found in soil, nonpathogenic species, and important human and animal pathogens (19). The genus also includes strains of biotechnological importance, including applications in bioremediation (20–22). For example, Clostridium acetobutylicum was used to produce acetone and biobutanol from carbohydrates up to the 1950s using the ABE process (acetone butanol ethanol process), and there has been recent interest in using this species in biofuel production (21, 22), while cellulolytic clostridia are viewed as important industrial organisms for production of chemicals and biofuels from cellulosic feedstocks (23). Little is known about regulatory sRNAs in clostridia. In C. perfringens, a regulatory RNA molecule (VR-RNA) was found to be responsible for the transcriptional regulation of two toxin genes, the collagenase (colA) and alpha-toxin (plc) genes (24). In C. saccharobutylicum P262, expression of the glutamine synthetase gene (glnA) was found to be regulated by an antisense RNA molecule transcribed from downstream and in the direction opposite that of glnA (25–27). A stand-alone S-box and a T-box riboswitch were shown to regulate a sulfur metabolic operon of C. acetobutylicum based on an antisense-RNA mechanism (28). In C. acetobutylicum, a likely synthetic noncoding RNA was found to improve the resistance to the toxicity of butyrate and other carboxylic acids (29). Recently, it was reported that a small noncoding RNA on the pSOL1 megaplasmid of C. acetobutylicum regulates the expression of solvent genes (30).
In this study, we computationally predicted sRNAs in virtually all clostridial genomes sequenced until late 2009 (when the computational work of this study was completed), 21 in total: Clostridium acetobutylicum, C. beijerinckii, C. botulinum (eight strains), C. cellulolyticum, C. difficile, C. kluyveri (two strains), C. novyi, C. perfringens (three strains), C. phytofermentans, C. tetani, and C. thermocellum. The approach integrated and combined genetic features of sequence conservation and notably predictions of transcriptional terminators and clostridial promoters. We show that most predicted clostridial sRNA sequences are well conserved only in the genus Clostridium and cannot be identified in the genomes of species that are phylogenetically close to clostridia like Bacillus subtilis. We experimentally validated a randomly selected set of predicted sRNAs from C. acetobutylicum and three C. botulinum strains. An interesting discovery is a novel sRNA, upstream of an ATP-binding cassette (ABC) transporter gene, which is apparently involved in the antibiotic response to clindamycin and can be identified in several clostridial genomes.
RESULTS
Prediction of sRNAs on clostridial genomes.
Predicted sRNAs on virtually all sequenced clostridial genomes are summarized in Table 1, and a detailed list of predicted sRNA candidates is shown in Table S1 (Excel) in the supplemental material. In total, 113 sRNAs were predicted in Clostridium acetobutylicum, 101 on the chromosome and 12 on the pSOL1 megaplasmid, which carries the essential genes for production of solvent (butanol, acetone, and ethanol) (31), the characteristic stationary-phase trait of solventogenic clostridia. Comparing our prediction to the predicted and annotated RNA sequences in the Rfam database (v9.1), we found that 32 of our predicted sequences had also been predicted by Rfam to be members of known RNA families, which include 16 riboswitches (thiamine pyrophosphate [TPP], flavin mononucleotide [FMN], cobalamin, S-adenosylmethionine [SAM], purine, lysine), 12 upstream leaders (L10_leader, L20_leader, T-box, and ykoK), one sRNA participating in signal recognition particle complex (SRP_bact), one ribozyme (RNaseP_bact_a), and two other sRNAs (6S and tmRNA [named for its dual tRNA and mRNA-like nature]). In Clostridium botulinum, more than 200 sRNAs were predicted in each strain (see Table S1 in the supplemental material). Of these sRNAs, 31 overlap with the Rfam predictions, including 13 riboswitches (TPP, FMN, SAM, purine, and lysine), 16 upstream leaders (L10_leader, L20_leader, T-box, and ykoK), one SRP_bact, and one RNaseP_bact_a.
TABLE 1 .
Summary of predicted clostridial sRNAs
| Organism | No. of predicted sRNAs (excludes tRNAs) | Genome size (bp) | No. of genes | Avg IR size (bp) | No. of genes that overlap with Rfam (v9.1) (excludes tRNAs)a |
|---|---|---|---|---|---|
| Pathogenic | |||||
| C. botulinum A ATCC 3502 | 219 | 3,903,260 | 3,590 | 195 | 31 |
| C. botulinum A ATCC 19397 | 233 | 3,863,450 | 3,551 | 198 | 31 |
| C. botulinum A Hall | 231 | 3,760,560 | 3,404 | 210 | 31 |
| C. botulinum A3 Loch Maree | 249 | 4,259,691 | 3,984 | 200 | N/A |
| C. botulinum B1 okra | 246 | 4,107,013 | 3,852 | 209 | N/A |
| C. botulinum B Eklund | 245 | 3,847,969 | 3,520 | 201 | N/A |
| C. botulinum E3 Alaska E43 | 251 | 3,659,644 | 3,256 | 194 | N/A |
| C. botulinum F Langeland | 257 | 4,012,918 | 3,659 | 208 | 31 |
| C. difficile 630 | 264 | 4,298,133 | 3,753 | 194 | 31 |
| C. novyi NT | 119 | 2,547,720 | 2,315 | 146 | 30 |
| C. perfringens 13 | 193 | 3,085,740 | 2,723 | 184 | 26 |
| C. perfringens ATCC 13124 | 181 | 3,256,683 | 2,876 | 183 | 30 |
| C. perfringens SM101 | 131 | 2,921,996 | 2,578 | 204 | 12 |
| C. tetani E88 | 137 | 2,873,333 | 2,432 | 171 | 32 |
| Solventogenic and C. kluyveri | |||||
| C. acetobutylicum ATCC 824 | 113 | 4,132,880 | 3,848 | 153 | 32 |
| C. beijerinckii NCIMB 8052 | 336 | 6,000,632 | 5,020 | 240 | 27 |
| C. kluyveri DSM 555 | 126 | 4,023,800 | 3,913 | 161 | 41 |
| C. kluyveri NBRC 12016 | 136 | 3,955,303 | 3,523 | 186 | N/A |
| Cellulolytic | |||||
| C. cellulolyticum H10 | 45 | 4,068,724 | 3,390 | 159 | N/A |
| C. phytofermentans ISDg | 42 | 4,847,594 | 3,902 | 228 | N/A |
| C. thermocellum ATCC 27405 | 15 | 3,843,301 | 3,189 | 197 | 12 |
N/A, not available.
To examine whether the predicted sRNA sequences could encode small proteins, Shine-Dalgarno (SD) sequences were searched for (32) in the 5′ region of the predicted sequence. The result shows that more than one-third of the predicted sequences have SD sequences near their 5′ end. However, only one-sixth of the predicted sRNA sequences have a start codon following the SD sequences. Even in these latter sequences, some contain many stop codons (e.g., sCAC0610), while others (e.g., sCAC1074 and sCAC2470) have structures similar to known sRNA families (sRNA names explained below in “siRNA nomenclature” in Materials and Methods). Therefore, most of the predicted sRNA sequences do not appear to encode small protein genes.
Experimental validation of predicted sRNAs.
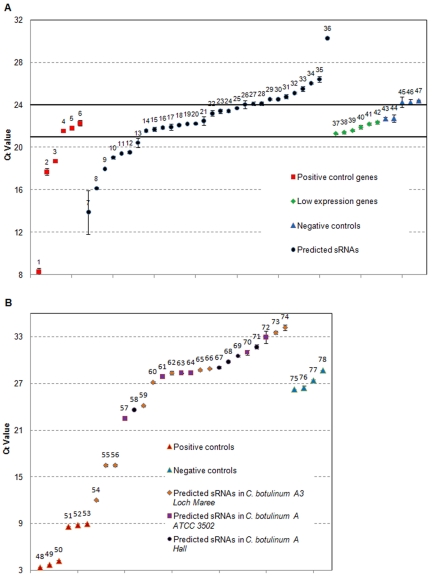
A randomly chosen subset of predicted sRNAs in C. acetobutylicum and C. botulinum (several strains) were examined for expression using quantitative reverse transcription-PCR (Q-RT-PCR) and/or Northern analysis (see Table S2 in the supplemental material). We chose these clostridial species based on the interest of our lab and the broader community in C. acetobutylicum and the desire to also include a major pathogen (C. botulinum) of great interest. Although there have been reports in the literature (10, 33) on the use of Q-RT-PCR for validating sRNAs, there are lingering doubts as to the suitability of Q-RT-PCR as a validation tool. Thirty and 21 of the predicted sRNAs from C. acetobutylicum and C. botulinum, respectively, were tested using Q-RT-PCR (Fig. 1). Selection of predicted sRNAs for validation was based on high GC content of their sequence to facilitate the design of Q-RT-PCR primers. Otherwise the selection of predicted sRNAs was random. We aimed to classify Q-RT-PCR-tested sRNAs in C. acetobutylicum into three groups, “highly expressed,” “possibly expressed,” and “not expressed.” This classification was based on expression profiles of “control genes” that were selected using expression analysis data from a detailed microarray study (34), plus five randomly selected intergenic region (IR) sequences in C. acetobutylicum that do not belong to any annotated or predicted transcripts. According to the cycle threshold (CT) values determined using these “control genes” (Fig. 1), seven sRNA candidates (Fig. 1A, genes 7 to 13) were classified as “highly expressed,” and 13 as “possibly expressed” (Fig. 1A, genes 14 to 27) in an RNA cocktail generated from samples taken at different time points from normal batch cultures, and cultures stressed with either butanol or butyrate (see Materials and Methods and Fig. 1). We used samples from stressed cultures based on the assumption that some sRNAs might be expressed under physiologically relevant stressful conditions, such as butanol and butyrate stress (35, 36). Similarly, four IR sequences in C. botulinum A3 Loch Maree were used as negative controls for testing the expression of predicted C. botulinum sRNAs. Ten candidates from C. botulinum A3 Loch Maree, six candidates from C. botulinum A ATCC 3502, and five candidates from C. botulinum A Hall were tested by Q-RT-PCR using RNA cocktails from normal, unstressed cultures. In total, six sRNAs were found to be highly expressed or possibly expressed (Fig. 1B), four from C. botulinum A3 Loch Maree, one from C. botulinum A ATCC 3502, and one from C. botulinum A Hall.
FIG 1 .
Q-RT-PCR validation of a select set of predicted sRNAs in Clostridium acetobutylicum and three Clostridium botulinum strains. Cycle threshold (CT) values are shown for genes in four groups: positive-control genes, predicted sRNAs, low-expression genes, and negative-control genes. CT values are averages of 5 or 6 replicates (error bars represent standard deviations). The numbers shown above the symbols (1 to 47) represent specific genes as listed below; the gene name is given first, and the gene number is shown in parentheses after the gene name. (A) Q-RT-PCR test of sRNA expression in C. acetobutylicum using pool CAC1 as described in Text S1 in the supplemental material. Positive-control genes (genes 1 to 6) include the 16S rRNA and genes known to be expressed well on the basis of microarray data (34): 16S (1), 6S (2), CAC0681 (3), CAC2957 (4), CAC1322 (5), and CAC2139 (6). Low- or no-expression genes (under normal culture conditions) (genes 37 to 42) based on microarray data (34) include the following: CAC3313 (37), CAP0060 (38), CAC0428 (39), CAC1094 (40), CAC2614 (41), and CAC2179 (42). The negative controls (genes 43 to 47) include the following intergenic region (IR) sequences (Ig stands for intergenic): IgCAC_1350 (43), IgCAC_232 (44), IgCAC_2999 (45), IgCAC_2630 (46), and IgCAC_2996 (47). The predicted sRNAs (genes 7 to 36) include the following: sCA-P60 (7), sCAC1449 (8), sCAC3821 (9), sCAC137 (10), sCA-P189 (11), sCAC3283 (12), sCAC1645 (13), sCAC646 (14), sCAC1132 (15), sCAC1582 (16), sCAC1760 (17), sCAC2795 (18), sCAC3723 (19), sCAC903 (20), sCAC3340 (21), sCAC3825 (22), sCAC975.1 (23), sCAC1315 (24), sCA-P18 (25), sCAC610 (26), sCAC2819.1 (27), sCAC1313 (28), sCA-P105 (29), sCA-P18.1 (30), sCAC3039 (31), sCAC500 (32), sCAC1594 (33), sCAC2709 (34), sCAC3850 (35), and sCAC349 (36). Notice that the values of the genes expressed well (genes 4 to 6) and the low-expression genes (genes 37 to 42) overlap. Thus, three zones of approximate expression are indicated: expressed (CT ≤ 21), lowly expressed if at all (21 < CT < 24), and probably not expressed (CT ≥ 24). The CT value of 21 was chosen as just below the CT value of the lowest low-expression control genes; the CT value of 24 was chosen based on the subsequent finding (Fig. 3) that sCAC610 represented by gene 26 is highly expressed. (B) Q-RT-PCR test of sRNA expression in C. botulinum A3 Loch Maree,C. botulinum A ATCC 3502, and C. botulinum A Hall using pool CLK, pool CBO, and pool CLC, respectively, as described in Text S2 in the supplemental material . The positive controls include 23S rRNA (genes 48 to 50) and 16S rRNA (genes 51 to 53) in the three C. botulinum strains. The negative controls (genes 75 to 78) include the following intergenic region sequences: IgCLC_2904 (75), IgCLC_2422 (76), IgCLC_2026 (77), and IgCLC_2348 (78). The predicted sRNAs (genes 54 to 74) are sCLK_200 (54), sCLK_3269 (55), sCLK_3642 (56), sCBO3039 (57), sCLC_2889 (58), sCLK_3105 (59), sCLK_3427 (60), sCBO2696 (61), sCLK_3557 (62), sCBO3480 (63), sCBO1976 (64), sCLK_2040 (65), sCLK_2759 (66), sCLC_3353 (67), sCLC_1905 (68), sCLC_2101 (69), sCBO2173 (70), sCLC_3476 (71), sCBO3602 (72), sCLK_1206 (73), and sCLK_3693 (74).
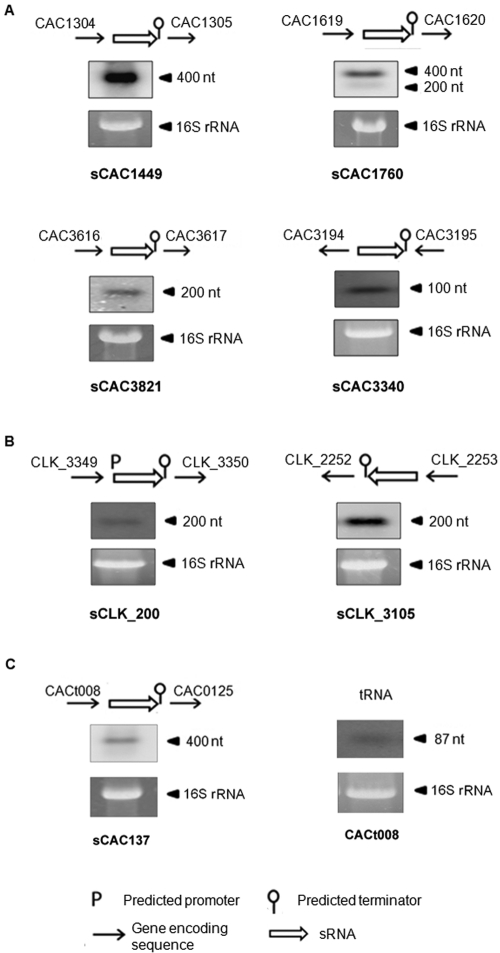
Among the predicted sRNAs which had relatively low CT values in the Q-RT-PCR analysis, we chose the few that have relatively high GC content in order to design good quality probes for Northern analysis. Six sRNAs from C. acetobutylicum (sCAC1449, sCAC1760, sCAC3821, sCAC3340, sCAC137 [Fig. 2A and C] and the sCAC610 sRNA discussed below) and two from C. botulinum (sCLK_200 and sCLK_3105 [Fig. 2B]) were validated using Northern analysis. Considering the accuracy of measuring transcript sizes using Northern blotting, especially at the low size range of 100 to 500 nt, the sizes of sCAC1760, sCAC610 (see below), sCLK_200, and sCLK_3105 were similar to the predicted sizes. An additional 200-nt transcript was also observed when probing for sCAC1760, which is possibly a processed version of the sRNA. The remaining sRNAs (sCAC1449, sCAC3821, and sCAC3340) were found to have different transcript lengths than predicted. The comparative genomics method used here to predict sRNAs can capture only conserved core sequences, but species-dependent variations where sRNAs are trimmed by specific nucleases (37) or additional noncore sequences are added may exist. The observed size (~400 nt), by Northern analysis, of sCAC137 (a putative noncoding RNA of the bacterial signal recognition particle [SRP_bact]) is longer than predicted by us (313 nt) or annotated in the KEGG database (201 nt). To determine the approximate start and stop sites of this sRNA, we carried out Northern analysis of the upstream CACt008 RNA and Q-RT-PCR analysis for the intragenic and downstream regions of this sRNA (see Fig. S1 in the supplemental material). Northern analysis of CACt008 RNA showed a distinct ~87-nt transcript, which is in accordance with the annotated size in KEGG (Fig. 2C). Q-RT-PCR results from both primer set 1 and primer set 2 (Fig. S1) showed very similar CT values (19.82 and 19.66 for primer set 1 and 2, respectively). Taken together, these observations suggest that the 400-nt sequence is a stand-alone transcript transcribed from the IR between CACt008 (tRNA) and CAC0125 (dnaX; DNA polymerase III subunit γ/τ). The IR length between the two genes is similar to the observed length of the sRNA transcript.
FIG 2 .
Northern analysis of predicted sRNAs. Validation of expression for a select set of sRNAs in C. acetobutylicum and C. botulinum by Northern analysis using single-stranded oligonucleotide probes. RNA from pool CAC2 was used for the C. acetobutylicum sRNAs, and RNA from pool CLK was used for the C. botulinum sRNAs. These RNA pools are described in Text S2 in the supplemental material. Ethidium bromide gels of 16S RNA are shown as qualitative and approximate loading controls. (A) Four predicted sRNAs from C. acetobutylicum (sCAC1449, sCAC1760, sCAC3821, and sCAC3340). (B) Two predicted sRNAs from C. botulinum A3 Loch Maree (sCLK_200 and sCLK_3105). (C) The predicted sCAC137 was validated with Northern analysis using a single-stranded oligonucleotide probe. The upstream annotated tRNA, CACt008, was also probed to confirm the tRNA size using a single-stranded oligonucleotide probe.
Experimentally tested sRNAs were computationally classified into Rfam sRNA families (see Tables S3 and S4 in the supplemental material) using GraPPLE as described in Text S1 in the supplemental material.
An sRNA involved in the response to clindamycin and sRNAs near ABC transporter genes.
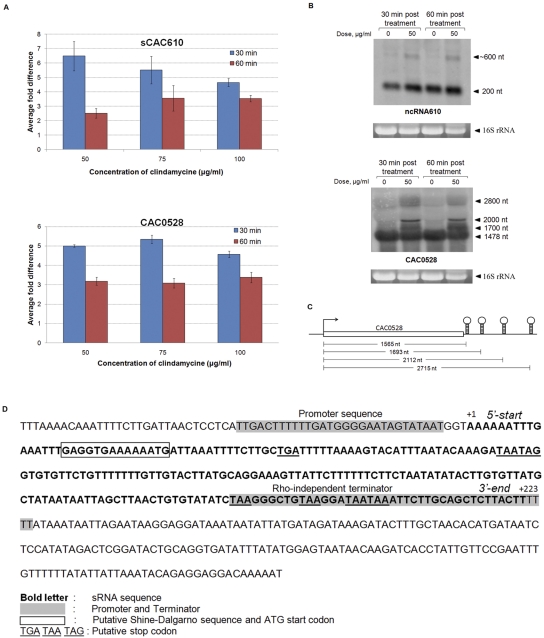
Although it is beyond the scope of this study to examine the functional roles of the predicted sRNAs in clostridia, we carried out a preliminary study on a novel conserved sRNA. We observed that the sCAC610 sRNA (see Table S3 in the supplemental material) of C. acetobutylicum, whose expression was tested by Q-RT-PCR analysis (Fig. 1A, gene 27), was well conserved on most C. botulinum genomes and on the C. beijerinckii genomes (see Fig. S2A in the supplemental material), with a consistent location upstream of putative ABC transporter genes (see Fig. S2B in the supplemental material). The conserved ortholog in C. botulinum A3 Loch Maree, sCLK_3105, was also detected by Northern analysis (Fig. 2B). The distances between these conserved sRNA sequences and the downstream ABC transporter genes were very consistent (~185 bp) (Fig. S2B), thus suggesting a functional relationship between this sRNA and the downstream ABC transporter genes. This potential functional relationship is also supported by the observation that this sRNA sequence does not exist on the genomes of C. botulinum strains B Eklund and E3 Alaska E43, which both lack the ABC transporter gene. Although sCAC610 is conserved only in clostridia, the downstream ABC transporter gene is conserved in many genera (Clostridium, Bacillus, Staphylococcus, Enterococcus, Staphylococcus, Streptococcus, and Lysinibacillus). Furthermore, this gene has been reported to code for a protein (Lsa) that confers low-level resistance to clindamycin in Staphylococcus warneri and Staphylococcus sciuri (38, 39). We therefore hypothesized that the identified sRNA and the downstream ABC transporter gene might be involved in the response of clostridia to clindamycin. To pursue this hypothesis, we investigated whether clindamycin treatment affected the expression of sCAC610 and the putative clindamycin resistance gene (CAC0510) in C. acetobutylicum. The MIC of C. acetobutylicum to clindamycin is 10 µg/ml for solid media, but it is >200 µg/ml for liquid cultures (40). Therefore, we employed a range of clindamycin concentrations up to 100 µg/ml. Expression profiles of sCAC610 and CAC0528 upon clindamycin treatment were assayed by Q-RT-PCR: two biological replicate experiments were carried out, and the data were found to be statistically significant (P < 0.0001). Compared to untreated cells, Q-RT-PCR results showed 4- to 6.5-fold increases in expression of both sCAC610 and CAC0528 after 30 min of 50, 75, and 100 μg/ml clindamycin treatment (Fig. 3A). The differences in expression of sCAC610 and CAC0528 in treated and untreated cells were lower but still significant after 60 min of treatment.
FIG 3 .
Expression of sCAC610 and CAC0528 in response to clindamycin treatment of C. acetobutylicum cultures. A single-stranded oligonucleotide probe was used in Northern analysis of sCAC610, and a double-stranded oligonucleotide probe was used for CAC0528. (A) Relative expression, by Q-RT-PCR analysis, of sCAC610 and CAC0528 upon clindamycin treatment, compared to untreated cells. The cells were treated with vehicle (no-clindamycin control) or with 50, 75, and 100 μg/ml clindamycin for 30 and 60 min. (B) Differential expression of sCAC610 and CAC0528 was confirmed using Northern analysis. The cells were treated with 50 µg/ml clindamycin for 30 and 60 min or not treated with clindamycin (0 µg/ml). Ethidium bromide gels of 16S RNA are shown as qualitative loading controls. (C) Predicted rho-independent terminators downstream of CAC0528. (D) DNA sequence of the entire intergenic region upstream of CAC0528. The sCAC610 transcript determined by the 5′ and 3′ RACE reactions is shown in bold type.
Northern analysis (Fig. 3B) showed a strong, ca. 200-nt transcript (despite the rather high CT value of 24 in Fig. 1A) for sCAC610. There were increased levels of sCAC610 in samples treated with clindamycin after 30 and 60 minutes of treatment compared to untreated samples (Fig. 3B), and interestingly, an uncharacterized 600-nt transcript hybridizing to the probe used was also observed to be differentially expressed upon clindamycin treatment (Fig. 3B). 5′ rapid amplification of cDNA ends (5′ RACE) and 3′ RACE sequencing revealed a transcript length of 223 nt, which is consistent with the prediction and Northern analysis. Though computational analysis found a promoter sequence, a Shine-Dalgarno box, and an AUG start codon near the 5′ end of the transcript, the sCAC610 sequence contains several stop codons in the transcript, and therefore, it is unlikely that sCAC610 codes for a small protein. sCAC610 was predicted to have a stable secondary structure (see Fig. S3 in the supplemental material), which is often observed in many prokaryotic noncoding RNAs. These lines of evidence suggest that sCAC610 is a noncoding RNA.
Northern analysis of the downstream gene (CAC0528) coding for the ABC transporter gene showed increased expression of several transcripts in cells treated with clindamycin. Although the expected 1,470-nt transcript for CAC0528 did not show differential expression upon clindamycin treatment, three other transcripts (of ca. 1,700 nt, 2,000 nt, and 2,800 nt) hybridizing to the probe were apparently induced by clindamycin treatment (Fig. 3B). The ~2,000-nt transcript appeared to be the most significantly and specifically induced transcript upon clindamycin treatment. Multiple rho-independent terminators were identified by TransTermHP after the annotated CAC0528 coding region (Fig. 3C); this suggests that there may be multiple transcripts expressed upon clindamycin treatment. All observed transcript sizes (Fig. 3B) are close to those predicted (1,700 nt, 2,110 nt, and 2,730 nt) based on the alternate transcriptional terminators. However, these extended transcripts contain many stop codons, and thus are unlikely to code for different proteins. We confirmed the presence of several longer transcripts by 3′ RACE analysis using probes outside the CAC0528 open reading frame (ORF) (data not shown). The mechanism by which this sRNA may be impacting the expression of CAC0528 remains to be investigated. Most sRNAs impact mRNA expression by an antisense mechanism. We did not find antisense pairing sequences between sCAC610 and the coding part of the downstream CAC0528 mRNA. We did notice, however, a good set of pairing sequences (see Fig. S3 in the supplemental material) that could affect the stability of the loop structures that correspond to the first three putative rho-independent terminators shown in Fig. 3C.
sCAC610 is not the only sRNA predicted to be near ABC transporter genes. In fact, we found sRNA enrichment in intergenic regions upstream or downstream of other ABC transporter genes in the list of predicted clostridial sRNAs (highlighted lines in Table S1 in the supplemental material). For example, for C. botulinum A3 Loch Maree, we predicted 25 sRNAs near ABC transporter genes. Taking into account the number of annotated ABC transporter genes (214), the number of predicted sRNAs (249), and the total number of genes (3,984) on C. botulinum A3 Loch Maree genome, Fisher’s exact test gave a significant P value of <0.005. Significant enrichment was also observed for C. botulinum E3-Alaska-E43 (P value of <0.01), C. difficile 630 (P value of <0.02), C. botulinum A Hall (P value of <0.04), and C. botulinum A ATCC 19397 (P value of <0.05). The other annotated clostridial genomes, with the exceptions of C. cellulolyticum H10 and C. thermocellum ATCC 27405, also contain many sRNA/ABC transporter pairs, though not as many as in the strains above.
Most predicted clostridial sRNAs are not conserved in bacilli or other genera.
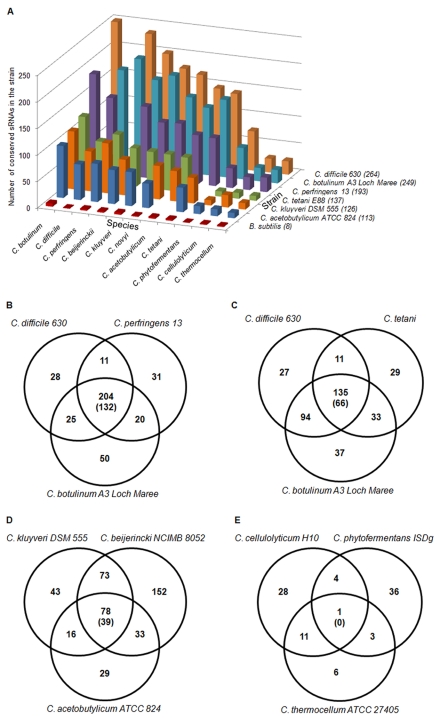
To date, most known sRNAs are highly conserved in several genera. We suspected that predictions based on closely related genomes might impact the ability to predict sRNAs and therefore, that current methods of prediction might underestimate the number of sRNAs. We therefore examined conservation of sRNAs in different clostridial genomes and in clostridial genomes and genomes from other genera. BLAST analysis of the 113 predicted sRNAs in C. acetobutylicum ATCC 824 against prokaryotic chromosomes in the NCBI genome database (E value of <0.001) found only 38 conserved sequences in nonclostridial organisms. However, of the 32 Rfam annotated sequences we predicted in C. acetobutylicum ATCC 824, 25 were included in these 38 conserved sequences and most of these 25 sequences are riboswitches such as SAM, T-box, or TPP, which function by binding to small target molecules. We believe that this is because the sRNAs covered by Rfam usually belong to common sRNA families, which are conserved in multiple genera. In the aforementioned 38 conserved sequences, only 17 were conserved in bacilli, which are members of the phylum Firmicutes and phylogenetically closely related to clostridia. Using our approach, we also examined sRNA predictions in B. subtilis; conserved IRs between B. subtilis and clostridial genomes rather than between the genomes of different bacilli were used as the search space. Surprisingly, we predicted that only eight sRNAs were conserved in B. subtilis and clostridia (Fig. 4A), all of which have been annotated in the Rfam database. Thus, the great majority of predicted clostridial sRNA sequences are not conserved in bacilli or across more distantly related genera.
FIG 4 .
Conservation of predicted sRNAs in different clostridial species. (A) The number of predicted sRNAs in representative strains that are conserved in other clostridial species which includes all substrains. The number in parentheses next to the strain name is the total number of predicted sRNAs in each strain. (B to E) sRNA conservation between different clostridial species. Because an sRNA sequence may have multiple conserved sequences in another organism, the number of sRNAs conserved between two organisms could vary. The common number of sRNA sequences between the three species in each Venn diagram is the maximum number of conserved sRNAs in the three species, and the number in parentheses is the minimum number of conserved sRNAs in the three species.
Further analysis of conservation of predicted sRNAs among various species within the Clostridium genus showed much better conservation among functionally related clostridia, such as among pathogenic species (Fig. 4). For example, among the three pathogens (C. difficile 630, C. botulinum A3 Loch Maree, and C. perfringens 13) shown in the Venn diagram in Fig. 4B, 204 (maximum number) (minimum number, 132; see the legend to Fig. 4) total sRNA sequences are conserved. This preservation is quite high for any combination of two or three clostridial species (data not shown) except for cellulolytic clostridia (Fig. 4E), for which we have predicted a much lower number of sRNAs. Based on the data shown in Fig. 4 and other data (not shown), about half of the predicted clostridial sRNAs are conserved among most (but not all) clostridial strains. One would then conclude that including even a few clostridial genomes in the prediction effort would generate a large number of sRNA predictions. This is in contrast to what would have been predicted by employing less-related genomes, as our computational results discussed above have shown. Still, the larger the number of clostridial genomes included in the prediction method, the larger the number of predicted sRNAs.
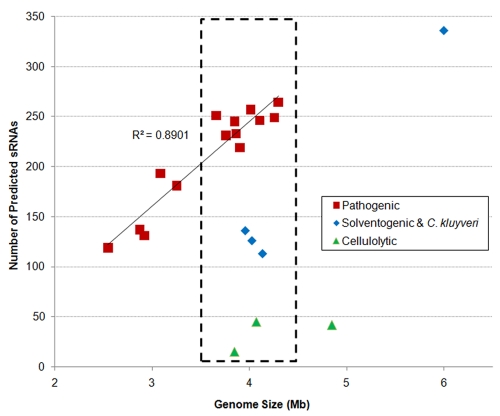
Biological function and then genome size, but not phylogenetic closeness, may correlate with the number of sRNAs on a genome; there are more sRNAs in pathogenic clostridia.
We did not find a correlation between the number of conserved sRNA sequences and the 16S rRNA-based phylogenetic distance in clostridial genomes (see Fig. S4 in the supplemental material). This is best demonstrated by Clostridium difficile. The phylogenetic distance between C. difficile and other pathogenic clostridia is larger than the phylogenetic distance between strains in the nonpathogenic clostridial categories in this study (41). However, a much larger number of sRNAs was still predicted in C. difficile than in other nonpathogenic strains with similar genome size. We noticed that there is a good correlation between the physiological function or niche of the species and the number of predicted and conserved sRNAs. Given the similar genome sizes, we predicted a much larger number of sRNAs for pathogenic clostridia than for cellulolytic clostridia and solventogenic clostridia (which we grouped functionally with C. kluyveri) (Table 1 and Fig. 5). This indicates a good correlation between the physiological function or niche of the species and the number of predicted and conserved sRNAs. The larger number of predicted sRNAs in pathogenic clostridial strains than in other strains did not arise from the difference in genomic GC content or gene densities either. The genomes of pathogenic clostridia have GC contents of ~29%, the solventogenic and C. kluyveri genomes have GC contents of ~30%, and the cellulolytic clostidrial genomes have higher GC contents of ~35% to 40%. Higher GC content may result in more predicted terminators and thus affect the number of predictable sRNAs. Our predictions go against this possible impact of the GC content: pathogenic clostridia have the most predicted sRNAs, and cellulolytic the least. The gene densities in different clostridial genomes are quite close to each other (less than 5% difference). Though the average IR size varies with species (Table 1), it is unlikely that the IR size is the cause of the differences in the number of predicted sRNAs. For example, the genome size and average IR size of C. thermocellum ATCC 27405 are similar to those of C. botulinum strains, but there are many more predicted sRNAs for C. botulinum strains.
FIG 5 .
Number of predicted sRNAs versus genome size. The number of predicted sRNAs for each clostridial species is plotted against the size of its genome. The number of predicted sRNAs in pathogenic clostridial strains varies linearly with genome size (R2 = 0.8901). For species with similar genome sizes (shown within the dashed-line box), the number of predicted sRNAs varies systematically with the type (grouped based on physiological niche) of clostridial species: very low in cellulolytic species, high for pathogenic species, and intermediate for solventogenic and C. kluyveri (this is statistically significant; the t test comparing any two of the three clostridial types gives P values of <0.005).
The much larger number of predicted sRNAs in pathogenic clostridial genomes did not result from using a larger number of pathogenic genomes. When we repeated the prediction by keeping only three randomly chosen pathogenic genomes of different species, for every combination of three pathogenic genomes, we still predicted more than 95% of the sRNAs we predicted when using all pathogenic genomes. However, we found a correlation between the number of predicted sRNAs and sequence conservation of the genomic IRs. In pathogenic clostridial genomes, the number of conserved IRs found by BLAST is much higher than those in solventogenic, C. kluyveri, and cellulolytic clostridial genomes. For example, using BLAST analysis, we found 644 nonoverlapping conserved sequences when we compared the C. botulinum A 3502 IRs against those in C. difficile 630. In contrast, we found only 167 nonoverlapping conserved sequences when we compared the C. acetobutylicum ATCC 824 IRs against those in C. kluyveri DSM 555. This ratio (644/167) is considerably higher than the corresponding ratio of predicted sRNAs (219/113). This would suggest that higher conservation of IR sequences does not translate to a proportionally higher number of predicted sRNAs. Thus, it is not logical to argue that the higher degree of IR conservation among pathogenic genomes per se is the reason for the significantly larger number of predicted sRNAs on pathogenic genomes. For pathogenic clostridia, the number of predicted sRNAs correlated well with genome size (Fig. 5), and the same may be true for solventogenic clostridia and the physiologically related C. kluyveri (Fig. 5). The large number (336) of sRNAs predicted for C. beijerinckii makes the point that prediction of a large number of sRNAs for a genome does not necessarily require a large number of functionally related genomes. This may then argue for the interpretation that the low number of sRNAs predicted on cellulolytic genomes is not due to the small number (3) of such genomes included in the analysis and would support the hypothesis that the number of sRNAs that are coded on a genome is indeed related to the physiology of the organism.
DISCUSSION
We reported the prediction of a large number of sRNAs on most sequenced clostridial genomes. In addition to the pairwise genome comparisons, we also used promoter and terminator information in predicting clostridial sRNAs. Computationally predicted promoters and terminators provide another layer of information for sRNA predictions. The predicted sRNAs included most previously predicted (Rfam database) sRNAs, but, significantly, identified novel sRNAs that are not well conserved in other prokaryotes.
Many predicted sRNA sequences were tested using Q-RT-PCR, and a subset of these sequences were tested by Northern analysis. In the laboratory, we tested Clostridium acetobutylicum RNA samples from cells growing under normal culture conditions, butanol stress, and butyrate stress. For Clostridium botulinum, we tested only samples from cells growing under normal culture conditions. Thus, because of the limited growth conditions we tested, the inability to experimentally verify the presence of some predicted sRNAs does not necessarily mean that these predicted sRNAs do not exist, as they could be transcribed at extremely low levels or not transcribed at all under the culture conditions employed in this study. Overall, our experience is that using Q-RT-PCR (as in Fig. 1) as the first screen for predicted sRNAs seems to be sound and useful.
We have also shown that the primary sequences of most predicted sRNAs are conserved only within the genus Clostridium. Our analysis shows that the number of predicted sRNAs in B. subtilis that are conserved in clostridial genomes is limited to only eight sRNAs, a much smaller number than might have been expected for two closely related genera. This may be due to the different physiological niches of organisms in the two classes/genera, but it may also be due to the fact that bacilli evolved much later than clostridia, after the “great oxidation” event (19), and suggests a relatively low conservation of many sRNAs even in evolutionarily close genera. Within the genus Clostridium, our analysis shows that many sRNAs are well conserved in several clostridial species and that conservation is higher in physiologically related species, such as among pathogens (Fig. 4). This means that the algorithm would have predicted a large number of sRNAs even if only three pathogenic clostridial gnomes were included in the analysis and that the small number of sRNAs predicted among cellulolytic clostridial genomes would not dramatically increase by including additional cellulolytic genomes. What would be the reason for pathogens to carry genes encoding more sRNAs? Perhaps the reason is that the niche of these organisms as human or animal pathogens requires larger flexibility in adapting to changing host environments to give them a survival advantage and perhaps their pathogenic potency. It has been reported that the expression of some sRNAs is induced by the host environment (42, 43). What does the low number of predicted sRNAs suggest for cellulolytic clostridial species? Perhaps that their environmental niche is very predictable and secure in terms of nutrient resources and that they have relatively little competition from other organisms.
Few sRNAs impacting drug resistance have been discovered. These sRNAs include micF, whose transcriptional activation is associated with resistance to multiple antibiotics (44, 45), and an sRNA which confers drug resistance to spectinomycin in E. coli (46). An ABC transporter gene has been reported to be trans-regulated by an sRNA which folds as a pseudoknot and binds Hfq protein in E. coli, Salmonella, and Shigella (47). It has also been reported that sRNAs are encoded within pathogenicity islands in Staphylococcus aureus (48), but the study did not examine whether these sRNAs are involved in regulating drug resistance or not. Here, the discovery of the clindamycin-responsive sRNA (sCAC610) in C. acetobutylicum, which is strongly conserved in other clostridial genomes (C. beijerinckii and several strains of C. botulinum), suggests that this sRNA plays an important role in drug resistance. We do not have evidence that sCAC610 works with the putative RNA chaperone Hfq protein (coded by CAC1834) in C. acetobutylicum. However, we observed that sCAC610 displays a consistent location and distance to its downstream putative ABC transporter gene in all the genomes in which it was identified. It is noteworthy that the ABC transporter gene CAC0528 is conserved in many bacteria other than clostridia. In contrast, sCAC610 is a unique sequence found only in the C. botulinum, C. acetobutylicum, and C. beijerinckii genomes. The enrichment of predicted sRNAs in upstream and downstream regions of other ABC transporter genes suggests that sRNAs could be broadly involved in cis-regulation of efflux pump activity. Since it is well established that efflux pumps regulate clinically relevant resistance to antibiotics (49, 50), our findings provide a new perspective for future research into mechanisms responsible for antibiotic resistance. Finally, our study did not predict any sRNA within the pathogenicity islands of pathogenic clostridial genomes.
MATERIALS AND METHODS
Computational identification of sRNAs encoded on intergenic regions.
The sequences of all 21 clostridial genomes (Table 1) were downloaded from NCBI (ftp://ftp.ncbi.nih.gov/). tRNAs, rRNAs, previously annotated small RNAs (sRNAs), and riboswitches were downloaded from the Rfam database (http://www.sanger.ac.uk/Software/Rfam/) (51). Genetic features used in the prediction of sRNAs include conserved intergenic regions (IRs), rho-independent terminators, and predicted promoters targeted by clostridial sigma factors σA, σD, σE, and σF. The computed data for these features were prepared as described below. IR sequence conservation data were prepared by pairwise genome comparisons. For sRNA predictions, to avoid predicting obvious conservation between strains of the same species, the genomes of all other clostridial species were used as its comparative partner genomes. For example, the Clostridium botulinum strains were not compared to each other, but to all other strains. Because sRNAs are usually 50 to 550 nt in length, we were interested only in IR sequences longer than 50 nt. We identified the conserved IRs between the target genome and its partner genome with WU BLAST 2.0 (52). A BLAST E value cutoff of 1 × 10−10 was applied, which assesses the significance of an alignment. Though a number of repeat regions are found in the IRs of prokaryotic genomes, we did not remove such repeats because these repeats may carry functional sequences. Putative IR rho-independent transcription terminators were predicted with TransTerm (53) and RNAMotif (54). For TransTerm, we used only those rho-independent terminators that had a confidence of 96% or higher in the terminator prediction. For RNAMotif, we used the descriptor file included in the sRNAPredict2 (55) package. Clostridial promoters targeted by σA, σD, σE, and σF were predicted using a hidden Markov model (56). After collecting the data described above for each genome, we applied sRNAPredict2 (55) to combine the computed features and predict sRNAs on each genome. The following criteria were used in the prediction: (i) every sRNA candidate must have a putative terminator no more than 20 nt downstream of its 3′ end; (ii) every sRNA candidate must be conserved and predicted on at least two clostridial genomes of different species. We also predicted sRNAs on the B. subtilis genome using the approach described above, except that the prediction examined conservation of IR sequences between B. subtilis and clostridial genomes rather than between different bacillus genomes.
Shine-Dalgarno (SD) sequences were predicted with the program free_align described in reference 32. This program simulates the binding between mRNAs and single-stranded 16S rRNA 3′ tail and identifies SD sequences by the position of the lowest ΔG° value. The ∆G° was calculated between the 16S 3′-tail sequence 5′-GAUCACCUCCUUUCU-3′ and the 5′ end (+1 bp to +45 bp) of the predicted sRNA sequences. If ΔG° is less than −3.4535 kcal/mol, the transcript was assumed to have an SD sequence. Translation start codons (AUG, GUG, and UUG) were then searched for within 20 bp downstream of the predicted SD sequence.
Predicted sRNAs chosen for experimental validation were further classified into functionally known sRNA families using GraPPLE. The sRNA family with the largest probability was reported as the family that each sRNA belongs to. sCAC610 was also analyzed for riboswitch elements using several online computational tools (57–59).
sRNA nomenclature.
A predicted sRNA on a genome is indicated by an initial lowercase “s,” followed by the three-letter (capitalized) genome identification (ID) used in the KEGG database, and ending with a number that indicates its genomic location, specifically, its genomic left coordinate (in kb units) plus one, regardless of its orientation. For example, the C. acetobutylicum sRNA that starts from chromosome coordinate 3,500 is named sCAC4; the sRNA that starts from the C. acetobutylicum pSOL1 megaplasmid coordinate 600 is designated sCA-P1.
Strains and growth conditions.
C. acetobutylicum was grown as reported previously (34). C. botulinum strains were grown in Eric Johnson’s lab (University of Wisconsin, Madison). C. botulinum strains A ATCC 3502 and A Hall were grown in type A toxin production medium (60). C. botulinum strain A3 Loch Maree was grown in MM medium (61, 62).
Cultures to collect cell samples for RNA isolation.
In order to validate predicted sRNAs, we collected cells for RNA extraction as follows. For Clostridium acetobutylicum, cultures were grown under three conditions: no stress, butanol stress (0.5% [vol/vol]), and butyric acid stress (0.35% [vol/vol]). These cultures were meant to provide RNA that might include a good fraction of the sRNAs coded on the C. acetobutylicum genome, based on the assumption that some of the sRNAs are expressed under some physiologically encountered stress, such as metabolite (butanol or butyrate) stress. A flask of 250 ml of clostridial growth medium (CGM) was inoculated with 10 ml (4% vol/vol) of preculture at an A600 of 0.6 to 0.8. This culture was grown to an A600 of ~0.6 and then used to inoculate 12 subcultures of 50 ml each with a 10% (vol/vol) inoculum. Four of these subcultures were allowed to grow unstressed, four were stressed with butyrate, and four were stressed with butanol. For butyrate stress, 175 µl of butyric acid was added at an A600 of 0.8, and for butanol stress, 250 µl of n-butanol was added at an A600 of 0.8. For the unstressed cultures, RNA samples were taken at 6 h (exponential phase), 12 h (transition phase), 18 h (early stationary phase), and 30 h (late stationary phase). For the stressed cultures (butanol and butyrate), RNA samples were taken 30 min and 1 h after stress.
To examine the effect of clindamycin on C. acetobutylicum, two biological replicate 250-ml flask cultures were inoculated with a 4% (vol/vol) preculture at an A600 of ~0.6 and grown to an A600 of ~1.0. Aliquots of 10 ml of culture were then transferred to duplicate individual flasks containing the following concentrations of clindamycin (Sigma-Aldrich): 0, 50, 75, and 100 µg/ml. RNA samples were taken 30 min and 1 h after stress.
C. botulinum strains were grown in 500-ml static flasks without stress with a 1% (vol/vol) inoculum as described previously (63). C. botulinum cultures were sampled at 4, 5, 6, 8, 10, 12, and 24 hours during growth. Culture aliquots were flash-frozen and stored in liquid nitrogen until RNA preparation.
RNA sampling and isolation.
For details on RNA sampling and isolation, see Text S2 in the supplemental material.
RNA pools.
For information on RNA pools, see Text S2 in the supplemental material.
cDNA generation and Q-RT-PCR analysis.
For details on cDNA generation and Q-RT-PCR analysis, see Text S2 in the supplemental material.
Northern analysis.
Northern blots using a single-stranded oligoprobe were performed as described previously (29). Oligonucleotide sequences used are listed in Table S2 in the supplemental material. In each lane of the Northern blot, 20 µg of total RNA was loaded and electrophoretically resolved on a 1.2% denaturing morpholinepropanesulfonic acid (MOPS)-agarose gel. RNA markers, 0.1 to 1 kb and 0.2 to 10 kb (USB, Cleveland, OH) were used as size standards. Northern blotting using double-stranded DNA (dsDNA) probes was performed as described previously (29) with a few modifications. Unincorporated radiolabeled [α-32P]dCTP was removed using Illustra G-50 spin columns (GE, Piscataway, NJ). Prehybridization and hybridization were carried out at 42°C with ULTRAhyb hybridization buffers (Ambion).
RACE reactions (5′ RACE and 3′ RACE).
For details on rapid amplification of cDNA ends (RACE), see Text S2 in the supplemental material.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We thank Marite Bradshaw and Eric A. Johnson of the Department of Bacteriology and the Food Research Institute at the University of Wisconsin (Madison) for preparing and donating C. botulinum RNA for this research.
This work was supported by National Science Foundation (United States) grant CBET-0853490.
Footnotes
Citation Chen, Y., D. C. Indurthi, S. W. Jones, and E. T. Papoutsakis. 2011. Small RNAs in the genus Clostridium. mBio 2(1):e00340-10. doi:10.1128/mBio.00340-10.
REFERENCES
- 1. Gottesman S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303–328 [DOI] [PubMed] [Google Scholar]
- 2. Waters L. S., Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersen J., Delihas N., Ikenaka K., Green P. J., Pines O., Ilercil O., Inouye M. 1987. The isolation and characterization of RNA coded by the micF gene in Escherichia coli. Nucleic Acids Res. 15:2089–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizuno T., Chou M. Y., Inouye M. 1984. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. U. S. A. 81:1966–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E. G., Margalit H., Altuvia S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941–950 [DOI] [PubMed] [Google Scholar]
- 6. Wassarman K. M., Repoila F., Rosenow C., Storz G., Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rivas E., Klein R. J., Jones T. A., Eddy S. R. 2001. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 11:1369–1373 [DOI] [PubMed] [Google Scholar]
- 8. Mandin P., Repoila F., Vergassola M., Geissmann T., Cossart P. 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 35:962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. del Val C., Rivas E., Torres-Quesada O., Toro N., Jimenez-Zurdo J. I. 2007. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol. Microbiol. 66:1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Panek J., Bobek J., Mikulik K., Basler M., Vohradsky J. 2008. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics 9:217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Axmann I. M., Kensche P., Vogel J., Kohl S., Herzel H., Hess W. R. 2005. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 6:R73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Livny J., Brencic A., Lory S., Waldor M. K. 2006. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 34:3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Voss B., Georg J., Schon V., Ude S., Hess W. R. 2009. Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genomics 10:123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kulkarni R. V., Kulkarni P. R. 2007. Computational approaches for the discovery of bacterial small RNAs. Methods 43:131–139 [DOI] [PubMed] [Google Scholar]
- 15. Machado-Lima A., del Portillo H. A., Durham A. M. 2008. Computational methods in noncoding RNA research. J. Math. Biol. 56:15–49 [DOI] [PubMed] [Google Scholar]
- 16. Washietl S., Hofacker I. L., Stadler P. F. 2005. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 102:2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Livny J., Fogel M. A., Davis B. M., Waldor M. K. 2005. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res. 33:4096–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Livny J., Teonadi H., Livny M., Waldor M. K. 2008. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS One 3:e3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paredes C. J., Alsaker K. V., Papoutsakis E. T. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 20. Kopke M., Held C., Hujer S., Liesegang H., Wiezer A., Wollherr A., Ehrenreich A., Liebl W., Gottschalk G., Dürre P. 2010. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U. S. A. 107:13087–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papoutsakis E. T. 2008. Engineering solventogenic clostridia. Curr. Opin. Biotechnol. 19:420–429 [DOI] [PubMed] [Google Scholar]
- 22. Dürre P. 2008. Fermentative butanol production—bulk chemical and biofuel, p. 353–362 In Wiegel J., Maier R. J., Adams M. W. W., Incredible anaerobes: from physiology to genomics to fuels. Blackwell Publishing, Oxford, United Kingdom: [DOI] [PubMed] [Google Scholar]
- 23. Demain A. L., Newcomb M., Wu J. H. D. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shimizu T., Yaguchi H., Ohtani K., Banu S., Hayashi H. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 43:257–265 [DOI] [PubMed] [Google Scholar]
- 25. Fierro-Monti I. P., Reid S. J., Woods D. R. 1992. Differential expression of a Clostridium acetobutylicum antisense RNA: implications for regulation of glutamine synthetase. J. Bacteriol. 174:7642–7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Janssen P. J., Jones D. T., Woods D. R. 1990. Studies on Clostridium acetobutylicum glnA promoters and antisense RNA. Mol. Microbiol. 4:1575–1583 [PubMed] [Google Scholar]
- 27. Woods D. R., Reid S. J. 1995. Regulation of nitrogen metabolism, starch utilisation and the beta-hbd-adh1 gene cluster in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:299–306 [DOI] [PubMed] [Google Scholar]
- 28. Andre G., Even S., Putzer H., Burguiere P., Croux C., Danchin A., Martin-Verstraete I., Soutourina O. 2008. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 36:5955–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Borden J. R., Jones S. W., Indurthi D., Chen Y., Papoutsakis E. T. 2010. A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing. Metab. Eng. 12:268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schiel B., Nold N., Dürre P. 2010. Identification of a small noncoding RNA in Clostridium acetobutylicum, abstr. GRV09, p. 135. Programme for the 3rd Joint Conference of the German Society for Hygiene and Microbiology (Jahrestagung der Deutschen Gesellschaft für Hygiene und Mikorobiologie [DGHM]) and the Association for General and Applied Microbiology (Jahrestagung der Vereinigung für Allgemeine und Angewandte Mikrobiologie [VAAM]), 28 to 31 March 2010. , Hannover, Germany [Google Scholar]
- 31. Cornillot E., Nair R. V., Papoutsakis E. T., Soucaille P. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Starmer J., Stomp A., Vouk M., Bitzer D. 2006. Predicting Shine-Dalgarno sequence locations exposes genome annotation errors. PLoS Comput. Biol. 2:e57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiao B., Li W., Guo G., Li B., Liu Z., Jia K., Guo Y., Mao X., Zou Q. 2009. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Curr. Microbiol. 58:258–263 [DOI] [PubMed] [Google Scholar]
- 34. Jones S. W., Paredes C. J., Tracy B., Cheng N., Sillers R., Senger R. S., Papoutsakis E. T. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 9:R114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tummala S. B., Junne S. G., Paredes C. J., Papoutsakis E. T. 2003. Transcriptional analysis of product-concentration driven changes in cellular programs of recombinant Clostridium acetobutylicum strains. Biotechnol. Bioeng. 84:842–854 [DOI] [PubMed] [Google Scholar]
- 36. Alsaker K. V., Paredes C., Papoutsakis E. T. 2010. Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol. Bioeng. 105:1131–1147 [DOI] [PubMed] [Google Scholar]
- 37. Reichenbach B., Gopel Y., Gorke B. 2009. Dual control by perfectly overlapping sigma 54- and sigma 70-promoters adjusts small RNA GlmY expression to different environmental signals. Mol. Microbiol. 74:1054–1070 [DOI] [PubMed] [Google Scholar]
- 38. Kehrenberg C., Aarestrup F. M., Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kehrenberg C., Ojo K. K., Schwarz S. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 54:936–939 [DOI] [PubMed] [Google Scholar]
- 40. Mermelstein L. D., Papoutsakis E. T. 1993. Evaluation of macrolide and lincosamide antibiotics for plasmid maintenance in low pH Clostridium acetobutylicum ATCC 824 fermentations. FEMS Microbiol. Lett. 113:71–75 [DOI] [PubMed] [Google Scholar]
- 41. Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K. H., Ludwig W., Glockner F. O., Rossello-Mora R. 2008. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241–250 [DOI] [PubMed] [Google Scholar]
- 42. Padalon-Brauch G., Hershberg R., Elgrably-Weiss M., Baruch K., Rosenshine I., Margalit H., Altuvia S. 2008. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 36:1913–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vercruysse M., Fauvart M., Cloots L., Engelen K., Thijs I. M., Marchal K., Michiels J. 2010. Genome-wide detection of predicted non-coding RNAs in Rhizobium etli expressed during free-living and host-associated growth using a high-resolution tiling array. BMC Genomics 11:53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller P. F., Sulavik M. C. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441–448 [DOI] [PubMed] [Google Scholar]
- 45. Nikaido H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382–388 [DOI] [PubMed] [Google Scholar]
- 46. Zimmerman J. M., Maher L. J., III 2002. In vivo selection of spectinomycin-binding RNAs. Nucleic Acids Res. 30:5425–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Antal M., Bordeau V., Douchin V., Felden B. 2005. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 280:7901–7908 [DOI] [PubMed] [Google Scholar]
- 48. Pichon C., Felden B. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U. S. A. 102:14249–14254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nishino K. 2008. Role of drug efflux pumps in bacterial multidrug resistance and virulence. Analysis to identify novel drug targets and counteract multidrug resistance and virulence. Jpn. J. Antibiot. 61:105–113 (In Japanese) [PubMed] [Google Scholar]
- 50. Piddock L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629–636 [DOI] [PubMed] [Google Scholar]
- 51. Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S. R., Bateman A. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33:D121–D124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ermolaeva M. D., Khalak H. G., White O., Smith H. O., Salzberg S. L. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27–33 [DOI] [PubMed] [Google Scholar]
- 54. Macke T. J., Ecker D. J., Gutell R. R., Gautheret D., Case D. A., Sampath R. 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 29:4724–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Livny J. 2007. Efficient annotation of bacterial genomes for small, noncoding RNAs using the integrative computational tool sRNAPredict2. Methods Mol. Biol. 395:475–488 [DOI] [PubMed] [Google Scholar]
- 56. Paredes C. J., Rigoutsos I., Papoutsakis E. T. 2004. Transcriptional organization of the Clostridium acetobutylicum genome. Nucleic Acids Res. 32:1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bengert P., Dandekar T. 2004. Riboswitch finder—a tool for identification of riboswitch RNAs. Nucleic Acids Res. 32:W154–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Abreu-Goodger C., Merino E. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 33:W690–W692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang T. H., Huang H. D., Wu L. C., Yeh C. T., Liu B. J., Horng J. T. 2009. Computational identification of riboswitches based on RNA conserved functional sequences and conformations. RNA 15:1426–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schantz E. J., Johnson E. A. 1992. Properties and use of botulinum toxin and other microbial neurotoxins in medicine. Microbiol. Rev. 56:80–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Demain A. L., Gerson D. F., Fang A. 2005. Effective levels of tetanus toxin can be made in a production medium totally lacking both animal (e.g., brain heart infusion) and dairy proteins or digests (e.g., casein hydrolysates). Vaccine 23:5420–5423 [DOI] [PubMed] [Google Scholar]
- 62. Mueller J. H., Miller P. A. 1954. Variable factors influencing the production of tetanus toxin. J. Bacteriol. 67:271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bradshaw M., Dineen S. S., Maks N. D., Johnson E. A. 2004. Regulation of neurotoxin complex expression in Clostridium botulinum strains 62A, Hall A-hyper, and NCTC 2916. Anaerobe 10:321–333 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.