Abstract
Oceans are home to much of the world's biodiversity, but we know little about the processes driving speciation in marine ecosystems with few geographical barriers to gene flow. Ecological speciation resulting from divergent natural selection between ecological niches can occur in the face of gene flow. Sister species in the young and ecologically diverse rockfish genus Sebastes coexist in the northeast Pacific, implying that speciation may not require geographical isolation. Here, I use a novel phylogenetic comparative analysis to show that rockfish speciation is instead associated with divergence in habitat depth and depth-associated morphology, consistent with models of parapatric speciation. Using the same analysis, I find no support for alternative hypotheses that speciation involves divergence in diet or life history, or that speciation involves geographic isolation by latitude. These findings support the hypothesis that rockfishes undergo ecological speciation on an environmental gradient.
Keywords: α-niche, β-niche, ecological speciation, parapatric speciation, rockfish
1. Introduction
The scarcity of geographical barriers to gene flow in marine ecosystems poses a challenge to the traditional view of speciation, with its emphasis on geographical isolation [1]. Fully allopatric speciation can occur across barriers such as the Isthmus of Panama [2], but these features are not common enough to explain all marine speciation events [1]. A possible explanation is that ecological speciation—reproductive isolation resulting from divergent natural selection between ecological niches—is widespread in marine taxa [3,4]. Reproductive isolation can arise as a byproduct of ecological divergence, and selection against the production of intermediate phenotypes can favour assortative mating and facilitate speciation. While ecological speciation can occur between strictly allopatric populations, divergent natural selection can also drive speciation in the face of gene flow [5].
Ecological speciation with gene flow may involve divergence in two main aspects of the niche, which have been distinguished in studies of trait-based community assembly [6,7]. Species may diverge between macrohabitats or along environmental gradients (the β-niche), or partition local resources such as food and microhabitats (the α-niche). In the ‘habitat-first’ model, the initial divergence during speciation is between β-niches, with any α-niche divergence coming later [8]. One version of this idea is formalized in models of parapatric speciation along environmental gradients, caused by divergent natural selection and assortative mating [9]. Habitat-first speciation has been inferred from sister-species comparisons in groups including birds [8,10] and Lake Victoria cichlids [11]. An alternative to habitat-first speciation occurs when disruptive selection favours divergence in the α-niche within a habitat. Theoretical models show that sympatric speciation can be driven by disruptive selection on resource-use traits combined with assortative mating based on ecological or marker traits [12,13]. While compelling evidence for ‘within-habitat’ speciation is rare, it comes from α-niche divergence between young sympatric sister species [14].
Sister-species comparisons provide valuable information about modes of niche divergence during recent speciation, but neglect information about earlier speciation events. Molecular phylogenies contain a partial record of such events, allowing the predictions of habitat-first and within-habitat speciation to be tested at the scale of entire clades undergoing adaptive radiation. If certain characters diverge during speciation, the amount of evolutionary change in those traits should be proportional to the number of speciation events, not to the amount of time elapsed [15]. In a clade in which habitat-first speciation predominates, we can predict that traits associated with the β-niche will show this pattern of ‘speciational’ change. Conversely, if within-habitat speciation is common, traits related to the α-niche (e.g. trophic morphology) should exhibit speciational evolution.
(a). Study system
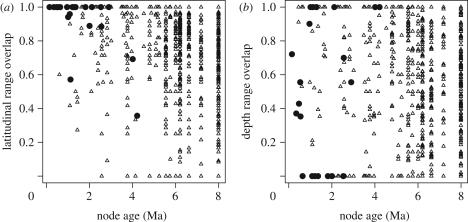
The marine rockfish genus Sebastes originated in the northwest Pacific in the mid-Miocene (ca 8 Ma; [16]) and subsequently diversified to produce over 100 extant species. The centre of rockfish diversity in the northeast Pacific contains at least 66 species, up to 56 of which occur in broad sympatry (i.e. within 1° latitude) off southern California [17]. This considerable diversity in the absence of geographical barriers has sparked interest in the factors promoting speciation in Sebastes [16,18–20]. A recent comprehensive molecular phylogenetic appraisal of the genus [16] indicates that most speciation occurs within oceanic regions. While a few long-distance dispersal events have allowed colonization of new regions, there is little evidence for allopatric speciation between ocean basins or sides of the Pacific Ocean ([16]; electronic supplementary material, figure S1). While rockfishes have a pelagic larval phase and are capable of long-distance dispersal, local recruitment of larvae and site fidelity of adults can permit genetic structure within species (e.g. [21]). Thus, isolation by physical distance might result in allopatric divergence between northern and southern populations [19]. Contrary to the predictions of this hypothesis of allopatric speciation by latitude [22], most close relatives show near-complete latitudinal range overlap (figure 1).
Figure 1.
Overlap in (a) latitudinal range and (b) depth range compared with time since divergence (node age) for pairs of Sebastes species (sister species denoted by filled symbols). Range overlap is calculated as the range shared by the two species divided by the smaller of the two ranges. In striking contrast to the predictions of allopatric speciation by divergence in latitude [22], recent sister species showed extensive overlap in latitudinal range, which was negatively related to node age (Mantel test, p < 0.001). In contrast, many recently diverged species showed less overlap in depth distribution, with no overall relationship between depth range overlap and node age (p = 0.23).
Ecological divergence over smaller scales may be more important for rockfish speciation than sheer physical distance. Species occupy characteristic depth habitats ranging from the intertidal to more than 600 m [17], and the relatively low overlap of sister species' depth distributions (figure 1) is suggestive of parapatric speciation on a depth gradient [18]. Rockfish species also show extensive α-niche diversity, some feeding primarily on zooplankton and others consuming mostly benthic invertebrates or fish [17]. Coexisting species tend to have limited diet overlap [23] and overdispersed foraging traits [24], suggesting a role for food competition in structuring rockfish assemblages. Within-habitat speciation is possible if these differences evolve in sympatry at the time of speciation. It has been suggested that Sebastes' elaborate courtship rituals and internal fertilization make assortative mating likely [16,25], potentially facilitating either habitat-first or within-habitat speciation in the presence of gene flow [9,12].
I tested these alternative hypotheses by fitting evolutionary models with and without speciational change to rockfish trait data, using a species-level phylogeny of the 66 Sebastes species in the northeast Pacific (electronic supplementary material, figure S1).
2. Material and methods
(a). Rockfish ecological and morphological data
I compiled published estimates of rockfish species' maximum and minimum latitudinal ranges and common adult depth ranges [17]. These were used in calculations of latitudinal and depth overlap (figure 1), while midpoint latitude and square-root-transformed midpoint depth were used as characters in phylogenetic analyses. As a diet (α-niche) axis, I estimated species' mean adult trophic positions (TP) from stable nitrogen isotope ratios (δ15N) for 1–15 individuals from 44 species, sampled in the Santa Barbara Channel and in British Columbia [24]. I calculated TP as TP = (δ15N − δ15Nbase)/3.4 + 2, where δ15Nbase is the average δ15N of a primary consumer (Mytilus californianus) and 3.4 is the average δ15N enrichment per trophic step [26].
I measured morphology of 543 individual rockfish representing all 66 species in the phylogeny (n = 1–46 per species, median 7). I measured total length (TL) and four ecomorphological traits: gill raker number and length, eye width and body depth. I used raw gill raker number data, while other traits were natural-log-transformed. I then adjusted individual values of gill raker length, eye width and body depth to remove the effects of TL. I used the ‘contrast’ program in PHYLIP [27,28] to obtain the phylogenetically corrected reduced major axis regression slopes while accounting for bias owing to both phylogenetic relatedness and intraspecific variation. I then fitted the least squares intercept assuming this slope, and calculated species trait values as mean residuals from this regression line. I used log-transformed published maximum TL as a measure of each species's overall body size [17].
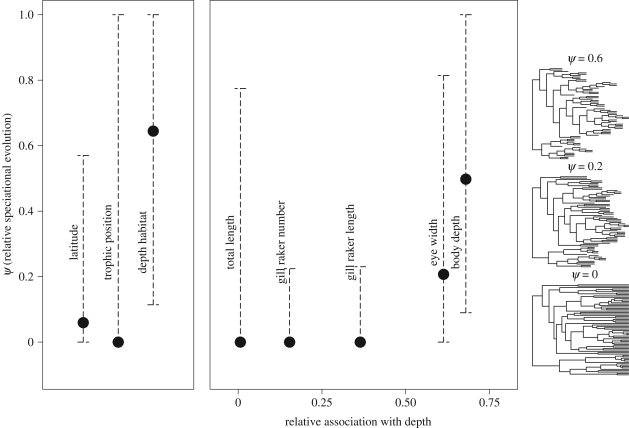
I tested for relationships between morphological traits and ecological variables to identify traits that may adapt species to either different prey types or different depth habitats. I calculated the maximum-likelihood (ML) degree of phylogenetic signal in each morphological trait and niche axis (λsig [29,30]). I pruned the tree of all but the 44 species with TP estimates, then for each character I transformed the tree based on the estimated λsig and calculated phylogenetically independent contrasts [27]. I carried out a multiple regression of the contrasts in each morphological trait (including TL) against contrasts in depth habitat and TP (table 1). I calculated partial r2 values to quantify the association of each trait with depth and with TP, then calculated the relative association with depth as partial 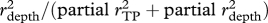 . This metric ranges from 0 to 1 and provides a rough index of the extent to which a trait's evolutionary dynamics should be influenced by a species's α- versus β-niche evolution. I predicted that traits would show a speciational pattern if the niche axis they are more strongly associated with is involved in speciation.
. This metric ranges from 0 to 1 and provides a rough index of the extent to which a trait's evolutionary dynamics should be influenced by a species's α- versus β-niche evolution. I predicted that traits would show a speciational pattern if the niche axis they are more strongly associated with is involved in speciation.
Table 1.
Associations between phylogenetically independent contrasts in four morphological traits and two niche axes: square-root-transformed average adult depth habitat and average trophic position.
| morphological trait | niche axis | slope | standard error | partial r2 | rel. assoc. with deptha |
|---|---|---|---|---|---|
| total length | depth | −0.005 | 0.024 | 0.001 | 0.01 |
| TP | 0.569** | 0.192 | 0.178 | ||
| gill raker number | depth | 1.59 | 0.954 | 0.064 | 0.13 |
| TP | −40.95*** | 7.496 | 0.421 | ||
| gill raker length | depth | 0.207** | 0.059 | 0.233 | 0.37 |
| TP | −2.433*** | 0.461 | 0.405 | ||
| eye width | depth | 0.083* | 0.036 | 0.113 | 0.61 |
| TP | −0.514 | 0.286 | 0.073 | ||
| body depth | depth | −0.078** | 0.025 | 0.191 | 0.68 |
| TP | 0.392 | 0.197 | 0.089 |
*p < 0.05.
**p < 0.01.
***p < 0.001.
aRatio of partial  .
.
(b). Inferring speciational versus gradual evolution
I used a new parameter ψ to quantify the contribution of speciation to a trait's total evolutionary rate. ψ has two advantages over a similar parameter κ [31], which raises branch lengths to an exponent to scale between speciational (κ = 0; all branch lengths equal) and gradual evolution (κ = 1; branches proportional to time). First, ψ is derived from biologically interpretable parameters: rates of evolutionary change over time and at speciation. Second, a limitation of κ is that it neglects any nodes in the phylogeny that are hidden owing to extinction. While we cannot know precisely where the hidden nodes are located in the tree, it is possible to use estimated speciation (λ) and extinction (μ) rates to infer where hidden nodes are most likely to occur [15].
I estimated λ and μ from the distribution of branching times in the phylogeny [15,32,33]. ML estimates were λ = 0.351 (95% CI 0.271–0.447) and μ = 0 (0–0.103). If we assume λ and μ remain constant across the tree, we can estimate the number of hidden speciation events on a branch beginning at time t1 and ending at time t2 as Sh = λα′(t2 − t1). α′ is the probability that a lineage originating at time t left no extant descendants, averaged across the branch [15]. The estimate of no extinction (and thus no hidden speciation events) may be unreliable [34], but results were qualitatively unchanged when I assumed extinction rates up to 75 per cent of the speciation rate (electronic supplementary material, figure S5). I used the (typically non-integer) expected Sh for estimation, though one could also sample integer Sh as part of a Bayesian estimation procedure [15].
I model evolution occurring both gradually (as a Brownian motion process) with rate parameter 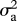 , and as step change at speciation, with change in the trait values of both daughter species drawn from a Gaussian distribution with variance
, and as step change at speciation, with change in the trait values of both daughter species drawn from a Gaussian distribution with variance 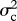 . The rate of speciational evolution is thus
. The rate of speciational evolution is thus 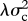 , and the total rate of evolution is
, and the total rate of evolution is
| 2.1 |
The variance of change over a branch is
| 2.2 |
where So is the number of known speciation events affecting that branch and Sh is the expected number of hidden speciation events. So is generally 1 for each branch, although if species present in the phylogeny are missing trait data (e.g. TP in this study), they can be pruned from the tree and deleted nodes accounted for by adding to the So of the affected branches. A simple reparameterization of the above model (equation (2.1)) results in the new parameter 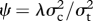 , the fraction of interspecific evolutionary divergence that is due to speciational change. ψ ranges between 0 and 1, and can be compared among traits measured on different scales.
, the fraction of interspecific evolutionary divergence that is due to speciational change. ψ ranges between 0 and 1, and can be compared among traits measured on different scales.
To calculate the likelihoods of values of ψ and 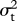 , I first calculated the phylogenetic variance–covariance matrix V for the phylogeny after transforming branch lengths following equation (2.2). The diagonal of V represents the root-to-tip path length of each extant species, while off-diagonal elements represent the path length from the root to the most recent common ancestor of two species. Intraspecific variability or measurement error can be accounted for by adding the squared standard error of species' mean trait values to the diagonal of V [30]. The ancestral state at the root of the tree is estimated as
, I first calculated the phylogenetic variance–covariance matrix V for the phylogeny after transforming branch lengths following equation (2.2). The diagonal of V represents the root-to-tip path length of each extant species, while off-diagonal elements represent the path length from the root to the most recent common ancestor of two species. Intraspecific variability or measurement error can be accounted for by adding the squared standard error of species' mean trait values to the diagonal of V [30]. The ancestral state at the root of the tree is estimated as  , where X is the vector of species means and 1 is a column vector of ones [35]. The likelihood function is the multivariate normal distribution of X, with expectation E(X) (a vector in which each value is
, where X is the vector of species means and 1 is a column vector of ones [35]. The likelihood function is the multivariate normal distribution of X, with expectation E(X) (a vector in which each value is  ) and variance–covariance matrix V. This function is used to calculate the likelihood of ψ and
) and variance–covariance matrix V. This function is used to calculate the likelihood of ψ and 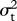 given X, λ, μ,
given X, λ, μ, 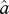 and the tree [30,35].
and the tree [30,35].
I identified the ML estimates of ψ and  for each trait using the ‘subplex’ optimization function in R [36], and estimated approximate confidence intervals on ψ using profile likelihood (for detailed likelihood surfaces see electronic supplementary material, figure S4). Analyses of simulated data indicate that this likelihood estimation procedure can recover known values of ψ with reasonable accuracy (electronic supplementary material, figure S7). I compared evolutionary models using AICc (Akaike's information criterion, corrected for sample size) and Akaike weights, which balance goodness of fit with model complexity [37]. The model in which ψ takes its ML value has three parameters (
for each trait using the ‘subplex’ optimization function in R [36], and estimated approximate confidence intervals on ψ using profile likelihood (for detailed likelihood surfaces see electronic supplementary material, figure S4). Analyses of simulated data indicate that this likelihood estimation procedure can recover known values of ψ with reasonable accuracy (electronic supplementary material, figure S7). I compared evolutionary models using AICc (Akaike's information criterion, corrected for sample size) and Akaike weights, which balance goodness of fit with model complexity [37]. The model in which ψ takes its ML value has three parameters (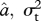 and ψ), while the simpler Brownian motion model (ψ = 0) reduces to two (
and ψ), while the simpler Brownian motion model (ψ = 0) reduces to two ( and
and 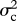 ). To evaluate the sensitivity of this analysis to phylogenetic uncertainty, I estimated ψ for each trait on 100 trees sampled from the posterior distribution.
). To evaluate the sensitivity of this analysis to phylogenetic uncertainty, I estimated ψ for each trait on 100 trees sampled from the posterior distribution.
3. Results
Rockfish species occurring in deeper habitats tended to have larger eyes—consistent with adaptation to low light availability [38]—and smaller body depths. By contrast, species with higher TP tended to be larger, with fewer, shorter gill rakers, consistent with the consumption of larger prey (table 1; electronic supplementary material, figure S3). Thus, although some traits showed some association with both niche axes, eye width and body depth are more related to the β-niche, while total length, gill raker number and gill raker length are more related to the α-niche. There was no tendency for α- or β-niche-associated traits to show greater phylogenetic signal (table 2).
Table 2.
Results of likelihood analysis of speciational versus gradual evolution. ML estimates of  and ψ are shown for each character. Akaike's information criterion scores (AICc) are shown for the model with (AICψ) and without (AICψ = 0) speciational evolution. The Akaike weight (wψ = 0) is shown for the first model, with weights greater than 0.5 indicating preference for the model with speciational evolution.
and ψ are shown for each character. Akaike's information criterion scores (AICc) are shown for the model with (AICψ) and without (AICψ = 0) speciational evolution. The Akaike weight (wψ = 0) is shown for the first model, with weights greater than 0.5 indicating preference for the model with speciational evolution.
| character | λsig | 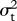 |
ψ | AICψ | AICψ = 0 | wψ = 0 |
|---|---|---|---|---|---|---|
| latitude | 0.77 | 9.36 | 0.059 | 422.4 | 420.8 | 0.31 |
| trophic position | 0.28 | 0.029 | 0 | 40.3 | 38.1 | 0.25 |
| depth habitat | 0.79 | 1.80 | 0.644 | 350.3 | 358.4 | 0.98 |
| total length | 0.94 | 0.045 | 0 | 63.3 | 61.1 | 0.25 |
| gill raker number | 0.87 | 4.63 | 0 | 372.1 | 369.9 | 0.25 |
| gill raker length | 0.78 | 0.022 | 0 | 26.0 | 23.8 | 0.25 |
| eye width | 0.99 | 0.0024 | 0.226 | −106.4 | −105.6 | 0.61 |
| body depth | 0.62 | 0.0013 | 0.565 | −123.7 | −111.2 | 0.99 |
Phylogenetic analysis showed variation among characters in the relative importance of speciational and gradual evolution (figure 2). There was a strong signal of speciational evolution in depth habitat and depth-associated traits, with approximately half of the total evolutionary rate for depth habitat and body depth estimated to occur at speciation. The model with speciational change was strongly preferred by AIC for depth habitat and body depth, and moderately preferred for eye width. Contrary to the predictions of within-habitat speciation, TP and trophic morphology showed no signal of speciational evolution (although confidence intervals were very wide for total length and TP). Latitudinal distribution also showed only a weak signal of speciational change, which was not supported over the simpler model of purely gradual evolution. These results are robust to uncertainty about the phylogeny and the extinction rate (electronic supplementary material, figures S5 and S6).
Figure 2.
Estimated contribution of speciation to the total rate of evolution of each character (ψ). Filled symbols indicate ML estimates of ψ, and error bars indicate the 95 per cent CI (±1.92 log-likelihood units). Morphological traits are arranged on the x-axis by their association with depth habitat relative to TP (table 1). The transformations of the Sebastes tree to the right show the branch lengths implied by three relevant values of ψ (0, 0.2 and 0.6).
4. Discussion
The key result of this study is a strong signal of speciational evolution in the depth habitats of rockfish species and in traits that appear to adapt species to different depths. This finding supports the hypothesis that rockfishes speciate along a depth gradient [18], and argues against several alternative models of speciation. Allopatric speciation between northern and southern populations has been inferred for a possible incipient species pair within S. mystinus [19], but the weak signal of speciational change in latitudinal distribution suggest that fully allopatric speciation is rare in northeast Pacific Sebastes. It has also been proposed that rockfish speciation involves divergence in life history, particularly in maximum lifespan, which varies from 10 to 200 years among species [20]. The lack of a speciational signal in total length (a strong correlate of lifespan) suggests that this is not a general feature of rockfish speciation. Finally, an alternative explanation for a signal of speciational change in traits is that features of the speciation process (e.g. population bottlenecks) lead to abrupt change in many characters [15]. The fact that traits unrelated to depth do not show a signal of speciational change is inconsistent with such a punctuated equilibrium model and indicates a special role for depth habitat in the speciation process.
These findings also draw intriguing connections between the processes of trait evolution, speciation and community assembly. The concepts of the α- and β-niche originated in studies of community assembly, and were developed to describe how species sort themselves within and between habitats, respectively [6,7]. When the assemblages of interest are also members of a clade undergoing adaptive radiation, we can extend this framework to consider how species diversity accumulates via speciation, with the distinct modes of speciation corresponding to the α- and β-niche as described in this manuscript. In the case of Sebastes, the β-niche appears to be involved in speciation, while the α-niche does not.
Some researchers have demonstrated a burst of α-niche evolution early in the diversification of a clade, followed by α-niche conservatism [7,10,39]. By contrast, I found no indication that the α-niche exhibits greater conservatism (i.e. phylogenetic signal) than the β-niche: TP had relatively low phylogenetic signal, while morphological traits more associated with depth versus TP did not differ consistently in phylogenetic signal. This pattern implies that both α- and β-niche evolution have occurred throughout the radiation, but that only β-niche evolution has been concentrated at speciation. Although divergence in TP may not be involved in speciation, α-niche diversification may nonetheless contribute to the species diversity of Sebastes. If species initially isolated by depth habitat later evolve α-niche differences (owing to any combination of adaptation to new resources, competition and genetic drift), they may compete less upon secondary contact and may subsequently be able to coexist [24].
The speciational signal in β-niche evolution reported here suggests that habitat divergence can be an important component of speciation in marine taxa that have little opportunity for strict geographic isolation [1]. Divergence in depth may result from multiple, not mutually exclusive processes. Rockfishes recruit from the plankton to shallow habitats as juveniles, and later undergo ontogenetic migrations to their adult depth habitat. Individuals may recruit by chance to different depth habitats, especially when features such as offshore seamounts lead to a discontinuous depth gradient [18]. Individuals may also recruit to new depth habitats created by sea level fluctuations [16]. This spatial separation may result in individuals reproducing, and populations establishing, in separate habitats. Alternatively, intraspecific variation in traits that affect fitness in different depth habitats may result in divergent selection between shallower and deeper parts of a species's depth range. Sister taxa do not have asymmetric depth range sizes (or latitudinal ranges; electronic supplementary material, figure S2), so parapatric divergence may be more likely than speciation of small, isolated populations [22]. Either selection or behavioural matching of phenotype to habitat may result in a correlation between depth and morphology within species. Migration of suboptimal phenotypes or competition between phenotypes may favour assortative mating, reducing gene flow and ultimately allowing speciation along the depth gradient [9].
However speciation by depth habitat occurs, it will result in sister species experiencing different light environments. This presents a further similarity between Sebastes and the cichlid radiations of Africa's rift lakes [40]. Like cichlids, rockfishes appear to diverge in light environment and perhaps colour [17] during speciation, and to undergo adaptive evolution of visual pigment genes along with habitat shifts [41]. In cichlids, detailed studies have demonstrated that speciation involves sensory drive, the integrated divergence of sexual signals and preferences between environments [11]. Although much work remains to identify the detailed mechanisms involved in rockfish speciation, these similarities suggest that Sebastes is a promising system for future studies of the role of both ecological speciation and sensory drive in marine adaptive radiations.
Acknowledgements
I would like to thank R. Barrett, R. FitzJohn, L. Harmon, N. Kraft, A. Mooers, S. Otto, D. Schluter, J. Shurin, M. Whitlock and three reviewers for comments on this manuscript. F. Bokma, C. Brock and J. Hyde shared files or computer code, and access to rockfish specimens was arranged by M. Love, K. Maslenikov, M. McCrea, H. J. Walker and K. L. Yamanaka. This work was funded by the National Evolutionary Synthesis Center (NESCent) and the National Science and Engineering Research Council of Canada (NSERC).
References
- 1.Palumbi S. R. 1994. Genetic divergence, reproductive isolation, and marine speciation. Ann. Rev. Ecol. Syst. 25, 547–572 10.1146/annurev.es.25.110194.002555 (doi:10.1146/annurev.es.25.110194.002555) [DOI] [Google Scholar]
- 2.Knowlton N., Weigt L. A., Solorzano L. A., Mills D. K., Bermingham E. 1993. Divergence in proteins, mitochondrial DNA, and reproductive compatibility across the Isthmus of Panama. Science 260, 1629–1632 10.1126/science.8503007 (doi:10.1126/science.8503007) [DOI] [PubMed] [Google Scholar]
- 3.Rocha L. A., Robertson D. R., Roman J., Bown B. W. 2005. Ecological speciation in tropical reef fishes. Proc. R. Soc. B 272, 573–579 10.1098/2004.3005 (doi:10.1098/2004.3005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puebla O. 2009. Ecological speciation in marine v. freshwater fishes. J. Fish Biol. 75, 960–996 10.1111/j.1095-8649.2009.02358.x (doi:10.1111/j.1095-8649.2009.02358.x) [DOI] [PubMed] [Google Scholar]
- 5.Schluter D. 2001. Ecological speciation. Trends Ecol. Evol. 16, 372–380 10.1016/S0169-5347(01)02198-X (doi:10.1016/S0169-5347(01)02198-X) [DOI] [PubMed] [Google Scholar]
- 6.Pickett S. T. A., Bazzaz F. A. 1978. Organization of an assemblage of early successional species on a soil moisture gradient. Ecology 59, 1248–1255 10.2307/1938238 (doi:10.2307/1938238) [DOI] [Google Scholar]
- 7.Ackerly D. D., Schwilk D. W., Webb C. O. 2006. Niche evolution and adaptive radiation: testing the order of trait divergence. Ecology 87, S50–S61 [DOI] [PubMed] [Google Scholar]
- 8.Diamond J. M. 1986. Evolution of ecological segregation in the New Guinea montane avifauna. In Community ecology (eds Diamond J. M., Case T. J.), pp. 98–125 Cambridge, MA: Harper and Row [Google Scholar]
- 9.Doebeli M., Dieckmann U. 2003. Speciation along environmental gradients. Nature 421, 259–264 10.1038/nature01274 (doi:10.1038/nature01274) [DOI] [PubMed] [Google Scholar]
- 10.Richman A. D., Price T. 1992. Evolution of ecological differences in the Old World leaf warblers. Nature 355, 817–821 10.1038/355817a0 (doi:10.1038/355817a0) [DOI] [PubMed] [Google Scholar]
- 11.Seehausen O., et al. 2008. Speciation through sensory drive in cichlid fish. Nature 455, 620–626 10.1038/nature07285 (doi:10.1038/nature07285) [DOI] [PubMed] [Google Scholar]
- 12.Dieckmann U., Doebeli M. 1999. On the origin of species by sympatric speciation. Nature 400, 354–357 10.1038/22521 (doi:10.1038/22521) [DOI] [PubMed] [Google Scholar]
- 13.Van Doorn G. S., Edelaar P., Weissing F. J. 2009. On the origin of species by natural and sexual selection. Science 326, 1704–1707 10.1126/science.1181661 (doi:10.1126/science.1181661) [DOI] [PubMed] [Google Scholar]
- 14.Barluenga M., Stölting K. N., Salzburger W., Muschick M., Meyer A. 2006. Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature 439, 719–723 10.1038/nature04325 (doi:10.1038/nature04325) [DOI] [PubMed] [Google Scholar]
- 15.Bokma F. 2008. Detection of ‘punctuated equilibrium’ by Bayesian estimation of speciation and extinction rates, ancestral character states, and rates of anagenetic and cladogenetic evolution on a molecular phylogeny. Evolution 62, 2718–2726 10.1111/j.1558-5646.2008.00492.x (doi:10.1111/j.1558-5646.2008.00492.x) [DOI] [PubMed] [Google Scholar]
- 16.Hyde J. R., Vetter R. D. 2007. The origin, evolution, an diversification of rockfishes of the genus Sebastes (Cuvier). Mol. Phylogenet. Evol. 44, 790–811 10.1016/j.ympev.2006.12.026 (doi:10.1016/j.ympev.2006.12.026) [DOI] [PubMed] [Google Scholar]
- 17.Love M. S., Yaklovich M., Thorsteinson L. 2002. The rockfishes of the northeast Pacific. Berkeley, CA: University of California Press [Google Scholar]
- 18.Hyde J. R., Kimbrell C. A., Budrick J. E., Lynn E. A., Vetter R. D. 2008. Cryptic speciation in the vermillion rockfish (Sebates miniatus) and the role of bathymetry in the speciation process. Mol. Ecol. 17, 1122–1136 10.1111/j.1365-294X.2007.03653.x (doi:10.1111/j.1365-294X.2007.03653.x) [DOI] [PubMed] [Google Scholar]
- 19.Burford M. O. 2009. Demographic history, geographical distribution and reproductive isolation of distinct lineages of blue rockfish (Sebastes mystinus), a marine fish with a high dispersal potential. J. Evol. Biol. 22, 1471–1486 10.1111/j.1420-9101.2009.01760.x (doi:10.1111/j.1420-9101.2009.01760.x) [DOI] [PubMed] [Google Scholar]
- 20.Mangel M., Kindsvater H. K., Bonsall M. B. 2007. Evolutionary analysis of life span, competition, and adaptive radiation, motivated by the Pacific rockfishes (Sebastes). Evolution 61, 1208–1224 10.1111/j.1558-5646.2007.00094.x (doi:10.1111/j.1558-5646.2007.00094.x) [DOI] [PubMed] [Google Scholar]
- 21.Johansson M. L., Banks M. A., Glunt K. D., Hassel-Finnegan H. M., Buonaccorsi V. P. 2008. Influence of habitat discontinuity, geographical distance, and oceanography on fine-scale population genetic structure of copper rockfish (Sebastes caurinus). Mol. Ecol. 17, 3051–3061 10.1111/j.1365-294X.2008.03814.x (doi:10.1111/j.1365-294X.2008.03814.x) [DOI] [PubMed] [Google Scholar]
- 22.Barraclough T. G., Vogler A. P. 2000. Detecting the geographical pattern of speciation from species-level phylogenies. Am. Nat. 155, 419–434 10.1086/303332 (doi:10.1086/303332) [DOI] [PubMed] [Google Scholar]
- 23.Hallacher L. E., Roberts D. A. 1985. Differential utilization of space and food by the inshore rockfishes (Scorpaenidae: Sebastes) of Carmel Bay, California. Environ. Biol. Fish. 12, 91–110 10.1007/BF00002762 (doi:10.1007/BF00002762) [DOI] [Google Scholar]
- 24.Ingram T., Shurin J. B. 2009. Trait-based assembly and phylogenetic structure in northeast Pacific rockfish assemblages. Ecology 90, 2444–2453 10.1890/08-1841.1 (doi:10.1890/08-1841.1) [DOI] [PubMed] [Google Scholar]
- 25.Helvey M. 1982. First observations of courtship behavior in rockfish, genus Sebastes. Copeia 1982, 763–770 10.2307/1444084 (doi:10.2307/1444084) [DOI] [Google Scholar]
- 26.Post D. M. 2002. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology 83, 703–718 10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2 (doi:10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2) [DOI] [Google Scholar]
- 27.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15 10.1086/284325 (doi:10.1086/284325) [DOI] [Google Scholar]
- 28.Felsenstein J. 2008. Comparative methods with sampling error and within-species variation: contrasts revisited and revised. Am. Nat. 171, 713–725 10.1086/587525 (doi:10.1086/587525) [DOI] [PubMed] [Google Scholar]
- 29.Freckleton R. P., Harvey P. H., Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 160, 712–726 10.1086/343873 (doi:10.1086/343873) [DOI] [PubMed] [Google Scholar]
- 30.Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131 10.1093/bioinformatics/btm538 (doi:10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 31.Pagel M. 1997. Inferring evolutionary processes from phylogenies. Zool. Scripta 26, 331–348 10.1111/j.1463-6409.1997.tb00423.x (doi:10.1111/j.1463-6409.1997.tb00423.x) [DOI] [Google Scholar]
- 32.Nee S., Holmes E. C., May R. M., Harvey P. H. 1994. Extinction rates can be estimated from molecular phylogenies. Phil. Trans. R. Soc. Lond. B 344, 77–82 10.1098/rstb.1994.0054 (doi:10.1098/rstb.1994.0054) [DOI] [PubMed] [Google Scholar]
- 33.Rabosky D. L. 2006. LASER: a maximum-likelihood toolkit for detecting temporal shifts in diversification rates. Evol. Bioinform. Online 2, 257–260 [PMC free article] [PubMed] [Google Scholar]
- 34.Rabosky D. L. 2010. Extinction rates should not be estimated from molecular phylogenies. Evolution 64, 1816–1824 [DOI] [PubMed] [Google Scholar]
- 35.O'Meara B. C., Ané C., Sanderson M. J., Wainwright P. C. 2006. Testing for different rates of continuous trait evolution using likelihood. Evolution 60, 922–933 [PubMed] [Google Scholar]
- 36.R Development Core Team 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 37.Anderson D. R., Burnham K. P., Thompson W. L. 2000. Null hypothesis testing: problems, prevalence, and an alternative. J. Wildl. Manage. 64, 912–923 [Google Scholar]
- 38.Warrant E., Locket N. A. 2004. Vision in the deep sea. Biol. Rev. 79, 671–712 10.1017/S1464793103006420 (doi:10.1017/S1464793103006420) [DOI] [PubMed] [Google Scholar]
- 39.Glor R. E., Kolbe J. T., Powell R., Larson A., Losos J. B. 2003. Phylogenetic analysis of ecological and morphological diversification in Hispaniolan trunk-ground anoles (Anolis cybotes group). Evolution 57, 2383–2397 [DOI] [PubMed] [Google Scholar]
- 40.Johns G. C., Avise J. C. 1998. Tests for ancient species flocks based on molecular phylogenetic appraisals of Sebastes rockfishes and other marine fishes. Evolution 52, 1135–1146 10.2307/2411243 (doi:10.2307/2411243) [DOI] [PubMed] [Google Scholar]
- 41.Sivasundar A., Palumbi S. R. 2010. Parallel amino acid replacements in the rhodopsins of the rockfishes (Sebastes spp.) associated with shifts in habitat depth. J. Evol. Biol. 23, 1159–1169 10.1111/j.1420-9101.2010.01977.x (doi:10.1111/j.1420-9101.2010.01977.x) [DOI] [PubMed] [Google Scholar]