Abstract
Background
Demethylating agents may alter the expression of genes involved in chemotherapy resistance. We conducted a phase I trial to determine the toxicity and molecular effects of the demethylating agent, decitabine, followed by doxorubicin and cyclophosphamide in children with refractory solid tumors.
Procedure
Stratum A included children with any solid tumor; Stratum B included neuroblastoma patients only. Patients received a 1-hour decitabine infusion for 7 days, followed by doxorubicin (45mg/m2) and cyclophosphamide (1g/m2) on day 7. Pharmacokinetic studies were performed after the first dose of decitabine. Biological studies included methylation and gene expression analyses of caspase-8, MAGE-1 and fetal hemoglobin (HbF), and expression profiling of pre- and post-treatment peripheral blood and bone marrow cells.
Results
The maximum-tolerated dose of decitabine was 5 mg/m2/d for 7 days. Dose-limiting toxicities at 10 mg/m2/d were neutropenia and thrombocytopenia. Decitabine exhibited rapid clearance from plasma. Three of 9 patients in Stratum A and 4/12 patients in Stratum B had stable disease for ≥4 months. Sustained MAGE-1 demethylation and increased HbF expression were observed in the majority of patients post-treatment (12/20 and 14/16 respectively). Caspase-8 promoter demethylation and gene expression were seen in 2/7 bone marrow samples. Differentially expressed genes were identified by microarray analysis.
Conclusion
Low-dose decitabine when combined with doxorubicin/cyclophosphamide has tolerable toxicity in children. However, doses of decitabine capable of producing clinically relevant biologic effects were not well tolerated with this combination. Alternative strategies of combining demethylating agents with non-cytotoxic, biologically targeted agents such as histone deactelyase inhibitors should be explored.
Keywords: demethylation, decitabine, pediatric solid tumor, neuroblastoma
INTRODUCTION
DNA methylation is an epigenetic mechanism used by cells to silence or decrease expression of genes, including tumor suppressors [1–3], and is maintained primarily by the enzyme, DNA methyl transferase 1 (DNMT1) [4]. The cytosine analogues, 5-azacytidine and decitabine (5-aza-2′-deoxycytidine) are potent inhibitors of DNA methylation [5,6]. Their demethylating activity was first exploited by DeSimone et al. who demonstrated increased hemoglobin F levels after treatment in patients with sickle cell disease [7–9]. The doses associated with the demethylating action of decitabine are much lower than those required for cytotoxicity [10–14].
Epigenetic modification of the pro-apoptotic gene, caspase-8, occurs in neuroblastoma [1,15–19], with up to 70% of human neuroblastoma cell lines and 25% of primary tumors lacking caspase-8 expression [15,17,20]. Loss of expression of both CASP8 alleles correlated with methylation of the caspase-8 gene [15], and exposure to 0.1–1.0 μM decitabine restored caspase-8 expression [1,17] and increased susceptibility of neuroblastoma cells to doxorubicin-induced apoptosis [15]. Multiple other genes can also be hypermethylated and silenced in neuroblastoma including RASSF1, TSP-1, and Hsp47 [21–25] and treatment with decitabine has led to significant inhibition of neuroblastoma growth in vivo [23]. Similarly, the cyclin-dependent kinase inhibitor p21WAF1 (CDKN1A), is methylated in primary rhabdomyosarcoma tumors and is reactivated by decitabine treatment [26].
Failure to express genes such as caspase-8 and CDKN1A could be an important mechanism of resistance to chemotherapy in pediatric tumors [17–19,26]. We therefore conducted a phase I study of decitabine administered in combination with cyclophosphamide and doxorubicin in children with relapsed or refractory solid tumors. Cyclophosphamide and doxorubicin are widely used in childhood cancers. Doxorubicin, alone or with decitabine, has been shown to induce apoptosis of neuroblastoma and other cancer cells in vitro [15,27], possibly due to its action as an inhibitor of DNMT1 [1,15,17]. Efficacy has also been reported in clinical trials of decitabine combined with anthracyclines [28,29]. In this study, after determination of the maximum tolerated dose (MTD) of decitabine (Stratum A), we accrued an additional cohort of children with recurrent/refractory neuroblastoma with bone marrow involvement to further delineate the biologic effects of decitabine (Stratum B).
PATIENTS AND METHODS
Trial design
Eligibility
Patients >12 months and ≤21 years of age with recurrent or refractory extracranial solid tumors were eligible. Enrollment on Stratum A required (1) recovery from the acute toxic effects of prior therapy, (2) histopathological verification of malignancy at diagnosis, (3) a Karnofsky or Lansky score ≥50%, and (4) adequate bone marrow function [absolute neutrophil count (ANC) ≥1000/μL, platelet count ≥100,000/mm3, hemoglobin ≥8.0gm/dl], liver function [total serum bilirubin ≤1.5mg/dl, and ALT ≤5 × upper limit of normal for age], renal function (normal serum creatinine for age or GFR≥70ml/min/m2) and cardiac function (shortening fraction of ≥28% or ejection fraction of ≥45%). After determination of the MTD in Stratum A, a cohort of patients with neuroblastoma accessible by bone marrow aspirate were enrolled (Stratum B) if they had the following: ANC ≥750/μL, a platelet count ≥50,000/μL (transfusion independent), and a hemoglobin ≥8.0gm/dl. Exclusion criteria included a prior cumulative anthracycline dose of ≥450mg/m2; pregnancy, lactation or an unwillingness to use an effective contraceptive method; a serious underlying systemic disease or sickle cell anemia; and concomitant use of erythropoietin or hydroxyurea. Informed consent was obtained from a parent or legal guardian, and assent as appropriate, prior to enrollment.
Study Design
Decitabine, supplied by the National Cancer Institute (Bethesda, MD) as a lyophilized powder in 50 mg vials, was reconstituted in 10 ml of sterile water for injection and diluted to a final volume of 25 ml (for patients with a BSA <1.0 m2) or 50 ml (for patients with a BSA ≥1.0 m2). Decitabine was administered intravenously over one hour on Days 0–6. The starting dose of decitabine, based on prior studies demonstrating tolerability of 15–45mg/m2/d in adult patients with myelodysplastic syndrome and hematological malignancies [14,15], was 5 mg/m2/d with planned escalations in 5 mg/m2/d dose increments. Blood and bone marrow samples were collected following the first course of decitabine and prior to the administration of additional chemotherapy on day 7. Dexrazoxane (450 mg/m2, given immediately prior to doxorubicin), doxorubicin (45 mg/m2 IV over <15 minutes), and cyclophosphamide (1 g/m2 IV over 1 hour with appropriate pre- and post-hydration), obtained commercially, were administered on day 7 if the ANC was ≥500/mm3, platelets were ≥50,000/mm3 and renal and liver function met eligibility criteria. G-CSF, 5 mcg/kg/day subcutaneously, was initiated 24 hours following completion of chemotherapy and continued until ANC was >10,000/mm3 post nadir. Courses were repeated every 28 days for up to 12 courses if the patient had at least stable disease and had recovered from the prior course of therapy. A classic 3 + 3 phase I design was used to determine the MTD. Toxicities were graded according to the NCI Common Toxicity Criteria (version 2.0). Tumor response was assessed by Response Evaluation Criteria in Solid Tumors (RECIST) [30].
Dose Limiting Toxicity (DLT)
Hematologic DLT was defined as ≥grade 3 neutropenia or thrombocytopenia during decitabine administration, grade 4 neutropenia or grade 4 thrombocytopenia lasting >7 days following the combination of decitabine with cytotoxic chemotherapy, and hematologic toxicity that caused a delay of ≥14 days beyond the planned interval between courses. Non-hematologic DLT was defined as any grade 3 or higher toxicity attributable to decitabine with the exclusion of grade 3 or 4 nausea and vomiting responding to anti-emetics, grade 3 transaminase elevations that returned to grade ≤1 or baseline prior to the next course, grade 3 or greater fever and infection. Determination of the MTD was based upon adverse events with the first treatment cycle.
Pharmacokinetic Analysis
Plasma decitabine concentrations were measured using a specific LC/MS method with a lower limit of quantitation of 1 ng/ml, and intra-, interday coefficient of variations <15% (XenoBiotic Laboratories, Inc. Plainsboro, NJ) [31]. Decitabine plasma concentration-time data were fit by non-linear least squares regression analysis to either a 1 or 2 compartment open model with data weighed by the inverse of the predicted plasma concentration using WinNonlin Pro version 5.2 (Pharsight®). Clearance values were estimated from the end of infusion concentration data assuming that steady-state plasma concentrations were achieved.
Biological Studies
DNA and RNA isolation
Peripheral blood samples (2.5–5 ml) were collected during the first course of therapy into PAX gene tubes (QIAGE®) on days 0, following decitabine treatment but prior to treatment with doxorubicin and cyclophosphamide on day 7, and at the end of the first course of treatment on day 28. Bone marrow samples were collected from Stratum B patients on the same days (Supplemental Table I). RNA was isolated from both blood and bone marrow samples using the Versagene RNA Blood kit (Gentrasystems, Inc.) and genomic DNA was isolated from the flowthrough fraction following phenol extraction and ethanol precipitation. RNA integrity and quality were assayed using the Agilent Technologies 2100 Bioanalyzer Lab-on-a-chip system and DNA integrity was determined by agarose gel electrophoresis and ethidium bromide staining. Only those samples with intact RNA and DNA were used for further analysis.
MAGE-1 and caspase-8 methylation analysis
Analysis of MAGE-1 promoter methylation was performed on peripheral blood samples as described previously [11]. All experiments were performed in triplicate on at least two separate occasions. Statistical significance was determined using the student t-test. For analysis of caspase-8 methylation, genomic DNA samples were treated with bisulfite using the EZ DNA Methylation Kit (ZYMO Research Corp.) and methylation PCR performed as described [15]. Validation studies were performed by mixing known quantities of DNA from the NB13 neuroblastoma cell line that exhibits methylation of both CASP8 alleles and normal peripheral blood in which CASP8 is unmethylated. These studies revealed that this assay was capable of detecting routinely methylated CASP8 if 1% of the input DNA was from NB13 cells (Supplemental Fig. S1).
Hemoglobin F (HbF) and caspase-8 quantitative RT-PCR
cDNA was synthesized from total RNA using random primers (PROMEGA) and Superscript II (Invitrogen). One twenty fifth of the reaction volume was used for each quantitative PCR reaction. The primers and PCR conditions are available on request. All reactions were performed at least 3 times in triplicate in an ABI7900HT thermocycler. Gene expression was normalized to GADPH using the comparative Ct method (2−ΔΔCT)[32]. Statistical analysis was performed using the student t-test.
Affymetrix GeneChip Analysis
RNA samples were processed using standard Affymetrix protocols (Eukaryote Two-Cycle Target Assay). RNA quality was confirmed on an Agilent 2100 Bioanalyzer. Five micrograms of total RNA were processed in the St. Jude Hartwell Center Core Facility using the Affymetrix eukaryote one-cycle target labeling protocol (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Expression analyses were performed using the Affymetrix HG-U133Plus2 GeneChip array which interrogates more than 54,000 transcripts. Signal values and detection calls were generated using the default parameters within the statistical algorithm of the Affymetrix GCOS software version 1.4. Signal values were scaled to a 2% trimmed mean target value of 500. Probeset annotations (December, 2005) were obtained from the Affymetrix website (http://www.affymetrix.com/analysis/index.affx). Signal values were log-start transformed to stabilize variance using STATA 9.2. The transformed signals for each probeset were tested for differences in mean between untreated samples and samples taken on day 7 or 28. In all cases paired t-tests were applied using Partek® Genomics Suite™ 6.2. For each list of P values a correction for multiple testing was also applied [33]. Heat map visualizations were created in Spotfire Decision Site 8.0 (TIBCO).
RESULTS
Clinical Variables
Patient Characteristics
Twenty-three patients were enrolled between March 2004 and May 2006; the primary diagnoses included neuroblastoma (n = 14), osteosarcoma (n = 5), rhabdomyosarcoma (n = 3) and synovial cell sarcoma (n = 1) (Table I). The median age of the treated cohort was 8 years (range 1.8–20.9 years); there were 3 females. All patients had received prior chemotherapy regimens (range, 1–8, median, 3) that included doxorubicin (median cumulative dose, 375 mg/m2, range 120–450 mg/m2) and radiation therapy. Eleven patients were enrolled onto Stratum A, 9 of whom were fully assessable for toxicity. One patient withdrew consent after two doses of decitabine and another had rapid disease progression prior to any study therapy and was taken off study. There were 12 Stratum B patients, all with relapsed or refractory neuroblastoma. Of the 21 patients treated on this study, 11 received only one course of therapy; 3 received 2 courses and 7 received ≥ 5 courses (Table 1).
Table I.
Patient Characteristics
| Accession No. | Diagnosis | Age at enrollment (years) | Gender | Prior chemotherapy regimens | Dose Level (mg/m2) | No. of courses | Best Response |
|---|---|---|---|---|---|---|---|
| Stratum A | |||||||
| 1 | RMS | 14.9 | M | 3 | 5 | 5 | SD |
| 2 | OS | 17.8 | M | 2 | 5 | 1 | PD |
| 3 | NBL | 6.4 | M | 2 | 5 | 5 | SD |
| 4 | RMS | 20.9 | M | 3 | 10 | 9 | SD |
| 5 | OS | 18.3 | M | 4 | 10 | 1 | PD |
| 6 | NBL | 2.9 | F | 3 | 10 | 1 | PD |
| 7* | RMS | 8.5 | M | 2 | 5 | <1 | NA |
| 8 | OS | 20.1 | M | 2 | 5 | 1 | PD |
| 9 | SS | 14.4 | M | 2 | 5 | 1 | PD |
| 10* | OS | 19.5 | M | 3 | 5 | 0 | NA |
| 11 | OS | 8.9 | M | 3 | 5 | 1 | PD |
| Stratum B | |||||||
| 12 | NBL | 14.5 | M | 7 | 5 | 1 | PD |
| 13 | NBL | 4.7 | F | 2 | 5 | 1 | PD |
| 14 | NBL | 1.8 | M | 3 | 5 | 2 | PD |
| 15 | NBL | 8.4 | M | 4 | 5 | 2 | PD |
| 16 | NBL | 9.5 | M | 8 | 5 | 1 | PD |
| 17 | NBL | 20.2 | F | 3 | 5 | 8 | SD |
| 18 | NBL | 13.3 | M | 2 | 5 | 7 | SD |
| 19 | NBL | 9.1 | M | 6 | 5 | 1 | PD |
| 20 | NBL | 13.5 | M | 5 | 5 | 1 | PD |
| 21 | NBL | 5.2 | M | 2 | 5 | 5 | SD |
| 22 | NBL | 5.7 | M | 1 | 5 | 2 | PD |
| 23 | NBL | 14.2 | M | 5 | 5 | 7 | SD |
RMS, Rhabdomyosarcoma; OS, osteosarcoma; NBL, neuroblastoma; SS, synovial sarcoma; SD, stable disease; PD, progressive disease; NA, not applicable;
not evaluable for toxicity.
Toxicity
Of the 9 evaluable patients on Stratum A, one patient experienced grade 3 anemia during the decitabine-only phase. Following administration of cyclophosphamide and doxorubicin, 0/3 patients experienced a DLT at the starting decitabine dose of 5 mg/m2/d. At 10 mg/m2/d, 3/3 patients experienced dose-limiting neutropenia and one, dose-limiting thrombocytopenia. One of these patients continued on therapy at a reduced dose for a total of 9 courses. Three additional patients were enrolled at the 5 mg/m2/d dose level and did not experience DLT, defining 5 mg/m2/d as the MTD of decitabine when administered in combination with cyclophosphamide and doxorubicin. All Stratum B patients were treated at the 5 mg/m2/d MTD. The primary toxicity of the treatment regimen was severe, prolonged but reversible myelosuppression (Table II). Non-hematological DLTs were not observed.
TABLE II.
Toxicities Attributable to Decitabine/Doxorubicin/Cyclophosphamide in Assessable Patients
| Course 1(total, 21 courses) | Courses 2 – 9 (total, 42 courses) | ||||
|---|---|---|---|---|---|
| Maximum grade across course 1 | Maximum grade across courses 2 to 9 | ||||
| Grade 3 | Grade 4 | Grade 3 | Grade 4 | ||
| Toxicity Type | Dose Level | ||||
| Hemoglobin | 5 mg/m2 | 3* | 2 | 3 | |
| 10 mg/m2 | 2 | ||||
| Leukocytes (total WBC) | 5 mg/m2 | 2 | 13 | 1 | 6 |
| 10 mg/m2 | 3 | 1 | |||
| Lymphopenia | 5 mg/m2 | 5 | 3 | 3 | |
| 10 mg/m2 | 1 | ||||
| Neutrophils/granulocytes (ANC/AGC) | 5 mg/m2 | 15 | 8 | ||
| 10 mg/m2 | 3 | 1 | |||
| Platelets | 5 mg/m2 | 3 | 8 | 2 | 5 |
| 10 mg/m2 | 2 | ||||
| Fever (without neutropenia, neutropenia = ANC <1.0 × 10e9/L) | 5 mg/m2 | 1 | |||
| Febrile neutropenia (fever of unknown origin without clinically or microbiologically documented infection) (ANC <1.0 × 10e9/L, fever >=38.5 d | 5 mg/m2 | 7 | 2 | ||
| 10 mg/m2 | 1 | ||||
| Infection (documented clinically or microbiologically) with Grade 3 or 4 neutrophils (ANC <1.0 × 10e9/L) | 5 mg/m2 | 1 | 2 | ||
| 10 mg/m2 | 1 | ||||
| Infection with normal ANC or Grade 1 or 2 neutrophils | 5 mg/m2 | 1 | 1 | ||
| Mucositis/stomatitis | 5 mg/m2 | 1 | |||
| Elevated transaminase (ALT/SGPT) | 5 mg/m2 | 1 | 2 | ||
| Hypophosphatemia | 5 mg/m2 | 1 | |||
| Hypokalemia | 5 mg/m2 | 1 | |||
The numbers refer to number of occurrences of each toxicity type.
Response
There were no objective responses in this heavily pre-treated population. However, stable disease for a median of 5 (range 5–9) courses was noted in 3 of 9 patients on stratum A [rhabdomyosarcoma (n = 2); neuroblastoma (n = 1)], and for a median of 7 (range 5–8) courses in 4 of 12 patients with neuroblastoma enrolled on stratum B (Table I).
Pharmacokinetic Analysis
Seven patients enrolled on stratum A participated in pharmacokinetic studies [5 mg/m2 (n=5) and 10 mg/m2 (n=2)] (Supplemental Fig. S2). Post-infusion data best fit a 2-compartment model for 5 patients and a 1-compartment model for 2 patients. There was substantial interpatient variability in decitabine disposition. The mean (range) half-life of decitabine was 27.9 minutes (18.1–40.2) and clearance, 14.7 L/min/m2 (0.9–31.0) (Table III).
TABLE III.
Summary of Pharmacokinetic Parameters for Decitabine
| Patient Accession No. | Dose (mg/m2) | CEOI (ng/ml) | Compartmental Analysis | Cl based on EOI concentration | |||
|---|---|---|---|---|---|---|---|
| t1/2α (min) | t1/2β (min) | Cl (L/min/m2) | Vss (L/m2) | Cl (L/min/m2) | |||
| 1 | 5 | 16.6 | 2.8 | 30.1 | 3.8 | 67.2 | 5.0 |
| 3 | 5 | 4.9 | 2.3 | 26.1 | 15.2 | 333.0 | 17.1 |
| 5 | 10 | 18.4 | 5.8 | 25.5 | 7.8 | 181.0 | 9.1 |
| 6 | 10 | 109.0 | 2.8 | 18.1 | 0.9 | 6.6 | 1.5 |
| 7 | 5 | 2.0 | -- | 29.8 | 31.0 | 1068.0 | 38.8 |
| 8 | 5 | 2.6 | 1.8 | 40.2 | 23.1 | 1065.0 | 32.6 |
| 9 | 5 | 3.0 | -- | 25.2 | 21.1 | 766.0 | 26.6 |
Data fit to a 1-compartment model.
Abbreviations: Cl, clearance; CEOI, concentration at end-of-infusion, Vss, steady state volume of distribution.
Biological Correlative Studies
Blood samples were received from all 21 patients who completed the study. Most blood samples yielded good quality DNA and RNA, although very low amounts of RNA limited the number of assays performed (Supplemental Table I). High quality bone marrow DNA was obtained from 8/9 patients on Stratum B for use in the caspase-8 methylation studies although only 7 were analyzed; one patient was excluded because no pretreatment bone marrow was collected and another patient was excluded because the pretreatment bone marrow was taken from a different site than the Day 7 and Day 28 samples. Isolation of high quality RNA from bone marrow was more problematic; nevertheless, sufficient amounts were obtained to permit analysis of caspase-8 expression from 7/9 patients, HbF expression from 6/9 patients and gene expression analyses at Day 0 and Day 7 from 4 patients and day 28 from 5 patients.
Caspase-8 demethylation
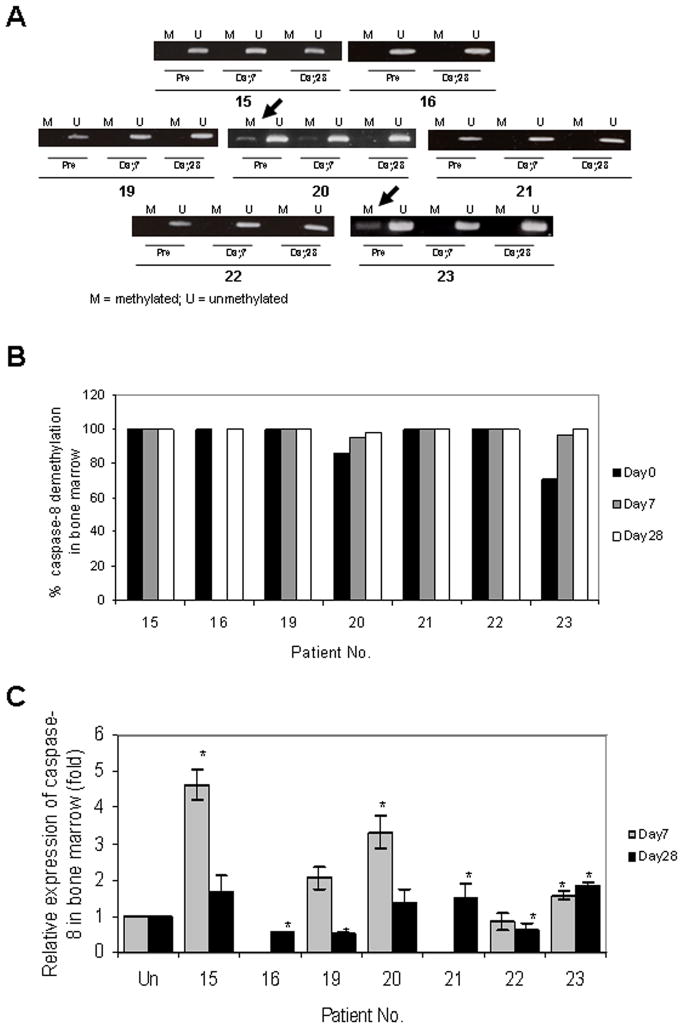
Caspase-8 promoter methylation was not detected in any of 17 pretreatment peripheral blood mononuclear cell (PBMC) samples analyzed, likely due to the low percentage of circulating tumor cells. Partial methylation of caspase-8 was observed in 2/7 pretreatment bone marrow samples (Fig. 1A and 1B; #20 and #23), with complete demethylation in 1 of the 2 (#23) by day 7, and in both samples by day 28. Demethylation correlated with increased caspase-8 mRNA expression in both of these samples on day 7 (Fig. 1C). Increased caspase-8 expression compared to baseline was also observed at day 7 in 2 additional bone marrow samples that did not exhibit methylation prior to treatment (Fig. 1C; #15 and #19). Although caspase-8 expression declined by day 28 in these two samples, a slight elevation in mRNA expression compared to baseline was still observed. Because of significant interpatient variability in pretreatment caspase-8 expression, the primary comparisons were made relative to individual baseline values. All these patients were treated at the 5 mg/m2 decitabine dose level.
Fig. 1.
Changes in caspase-8 gene methylation by methylation PCR (A), quantitative analysis of methylation (B) and mRNA expression (C), in bone marrow following decitabine treatment (day 7) and at the end of course 1 (day 28). Data are normalized to GAPDH and the pretreatment levels set at 1.0 for each patient. Asterisks indicate P-values ≤ 0.05. Insufficient amounts of RNA prevented analysis of Day 7 samples from patients 16 and 21.
MAGE-1 and HbF methylation
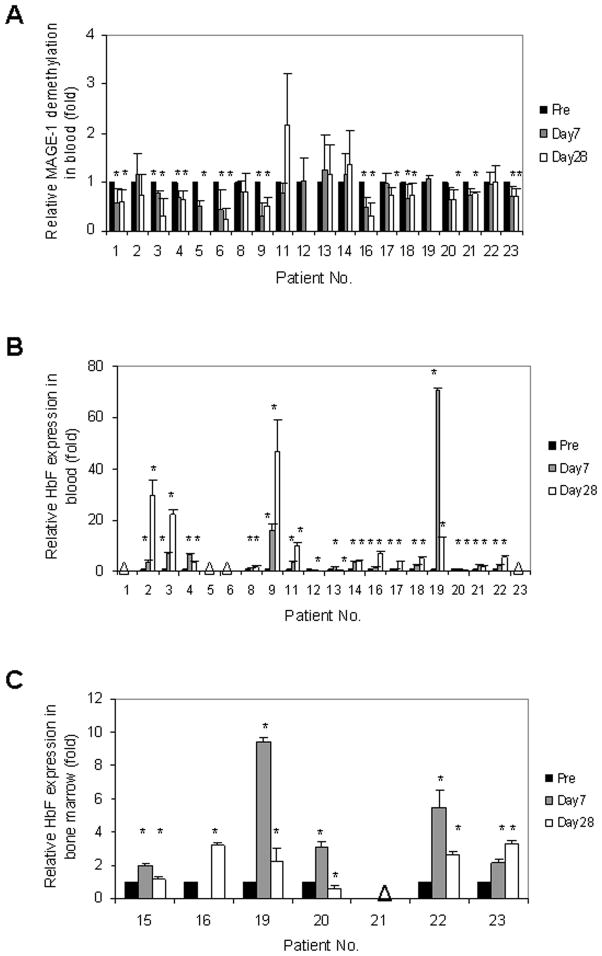
Exploratory studies were performed to assess MAGE-1 and HbF methylation status in peripheral blood as potential biomarkers of demethylation in children (Fig. 2). Sufficient high quality DNA was obtained to allow MAGE-1 methylation analysis of 20/21 day 0 and 7 and 18/21 day 28 patient blood samples. In our study all patients retained at least one MAGE-1 allele based on the fact that PCR products were obtained using primers recognizing both the promoter regions and the MAGE-1 mRNA product (Fig. 2A). Demethylation (24–80% decrease in methylation) of the MAGE-1 promoter following decitabine was detected in the PBMC of 12/20 patient samples on day 7. All but one remained demethylated on day 28 (P<0.05) (Fig. 2A).
Fig. 2.
Biomarker analyses of demethylation and increased gene expression. Changes in MAGE-1 promoter methylation in peripheral blood mononuclear cells (PBMC) following decitabine treatment (day 7), and cytotoxic chemotherapy (day 28) (A). The ratio of methylated to unmethylated MAGE-1 promoter sequences is shown. Quantitative PCR analysis of HbF RNA expression in PBMCs (B), and in bone marrow samples (C) from Stratum B patients. Data were normalized to GAPDH expression levels and day 0 levels set at 1.0 for each patient. Asterisks indicate P-values ≤ 0.05.
Of the 21 patients enrolled in this study, peripheral blood samples from 16 were analyzable for HbF mRNA expression by qRT-PCR. Of these, 14 exhibited increased HbF mRNA expression (Fig. 2B). Ten out of the 14 patient samples had increased HbF expression on day 7 and further increases on day 28, and 4 exhibited increased HbF expression on day 7 which declined somewhat by day 28. HbF mRNA expression also increased in bone marrow samples on either day 7 or day 28 in all Stratum B patients with sufficient amount of RNA for analysis (5/6 on day 7 and 5/7 on day 28) (Fig. 2C). However, elevated HbF protein levels were not detected by standard electrophoresis. There was no association between disease progression and HbF mRNA expression or MAGE-1 promoter demethylation.
Gene Expression Analysis
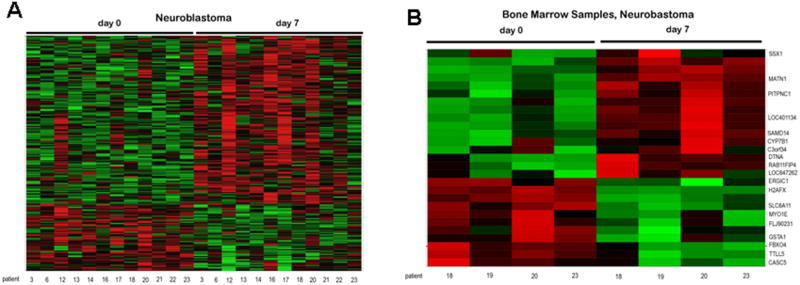
To determine the global effects of treatment on gene expression we performed exploratory microarray expression analysis on PBMCs of neuroblastoma patients (n=12) on days 0, 7, and 28 (Fig. 3A). As expected, given the large number of tests and the relatively small heterogeneous sample size, no gene was statistically significant at the 0.05 level. Therefore, the unadjusted P-values and log ratios were used to determine relative rank and best performing probe sets. Comparison of each of the pretreatment samples with day 7 (decitabine only) and day 28 (decitabine plus doxorubicin and cyclophosphamide) samples revealed multiple genes with greater than two-fold expression changes and unadjusted P-values ≤ 0.005 (Supplemental Table II). Of interest, only a small number of differentially expressed genes (<10%) were common to both day 7 and day 28. More genes were upregulated on day 7, while on day 28 more genes were downregulated. Gene ontology analysis was used to rank groups of transcripts with similar functions (Supplemental Table III). The neuroblastoma day 7 patient PBMC samples were enriched in genes with roles in synaptogenesis, B cell activation, antigen processing and peptide presentation and negative regulation of cell growth, whereas the day 28 samples also included genes involved in biogenic amine catabolism, regulation of interferon gamma, calcium mediated signaling and regulation of cdc42 GTPase activity. Microarray expression analysis was also performed on bone marrow samples from 4 out of 9 patients with neuroblastoma (Fig. 3B). While the quantity of bone marrow was not sufficient to isolate pure tumor cell populations, we ascertained that these samples did contain tumor cells based on the presence GD2 synthase, MYCN and tyrosine hydroxylase (TH) gene expression, none of which are seen in normal bone marrow [34] (Supplemental Fig. S3). Despite the lack of statistical significance when accounting for the large number of probesets, the annotations of the highest ranking probe sets provided a preliminary indication of the biological processes that may be regulated by decitabine (Supplemental Table IV).
Fig. 3.
Heat maps of differential changes in gene expression in peripheral blood mononuclear cells (A), and bone marrows (B), of patients with neuroblastoma. Z-scores are shown across the rows. Data is colored so that a z-score of zero is black, a score of 2 is red and a score of −2 is green.
DISCUSSION
The aims of this study were to pilot the use of low-dose decitabine in children with relapsed or refractory solid tumors and to study the pharmacokinetics, toxicity and drug exposures associated with biological and clinical response. This information will inform the design of future studies combining demethylating agents and cytotoxics. In addition, our study illustrates the challenges and logistics of pediatric patient sample collection, transport, quality control and analysis of samples for biological targets and correlates.
The MTD of decitabine, when administered for 7 days in combination with doxorubicin and cyclophosphamide, was 5 mg/m2/d. As in adult patients [11,14,35], neutropenia and thrombocytopenia were the dose-limiting toxicities. Non-myelosuppression-related toxicity occurs rarely in adults and includes nausea, vomiting, diarrhea, and fatigue [36], all of which were also seen in our patients.
Similar to studies in adult patients [37–39], there was wide interpatient variability in decitabine drug disposition. Drug clearance in children was greater than that reported in adults [37]. However, direct comparisons with other trials should be made cautiously as non-compartmental methods using the trapezoidal rule tend to underestimate area under the curve during the infusion and hence lead to an overestimate of clearance.
The biologic correlates of the approach of using a demethylating agent were analyzed using several methods: caspase 8 methylation status and expression, MAGE-1 methylation and HbF expression. Observed changes after decitabine exposure included demethylation of the caspase 8 gene and an increase in mRNA expression albeit in a limited number of bone marrow samples. The limited methylation noted could be attributed to alterations in normal bone marrow cells or in the proportion of tumor cells, or deletion of one or both of the caspase-8 alleles in a subset of samples, although homozygous or heterozygous loss of the CASP8 gene is extremely rare [15]. It is also possible that in primary tumors, as opposed to cell lines, caspase-8 expression is regulated by multiple mechanisms, only one of which may be promoter hypermethylation. This was previously noted in a study where hypermethylation-associated CD44 receptor silencing was found in neuroblastoma cell lines but not in primary tumor samples [22].
Similarly, MAGE-1 methylation was examined in PBMC as a potential marker of tumor methylation. The MAGE-1 promoter is normally methylated in somatic tissues [40]. The percentage of patients who exhibited significant demethylation of MAGE-1 in PBMCs during the first course of treatment was lower in our study than in a previous study in adults using 2 mg/m2/d of decitabine infused continuously over 7 days (60% vs. 100%) [10]. Others have found that a 10 mg/m2/d decitabine dose level was required to attain plasma peak concentrations associated with biologic effects [41–43], suggesting that in our study of heavily pre-treated patients optimal concentrations of drug might not have been achieved. Nonetheless, the duration of MAGE-1 demethylation in our study appears to be similar to that observed in adult studies [10,11,35,44]. MAGE-1 hypomethylation was retained, and in a few patients increased slightly, on day 28 as noted previously [11,45]. Although this is consistent with the possibility that decitabine-induced demethylation is prolonged after exposure and that even low levels of decitabine can alter methylation to some extent, the administration of cytotoxic chemotherapy may have impacted these results. We are not aware of any published literature regarding epigenetic modifications induced by dexrazoxane.
We also examined changes in HbF expression as a biomarker of tumor demethylation. Increased expression of HbF mRNA was found in 88% of blood samples and 100% of bone marrow samples, although by HbF electrophoresis, no change in protein levels was observed. These differences likely reflect the difference in the sensitivity of the two methodologies; a recent adult study showed an increase in HbF protein levels by western blotting following decitabine [46]. These data suggest that MAGE-1 and HbF could potentially serve as biomarkers of methylation status, especially because such an analysis is relatively easily performed on DNA (MAGE-1) and RNA (HbF) isolated from peripheral blood cells.
Similar to other reports [35] preliminary microarray analyses of global changes in gene expression revealed significant interpatient heterogeneity. Other studies have reported that interpatient heterogeneity can result from differences in patient age, sex, ethnicity, time of sample collection and the proportions of the various cell types in the blood and bone marrow [47,48]. Since all of our patients had treatments that could alter the relative proportions of the various cell types in peripheral blood, and also because their bone marrow samples most likely would exhibit differences in the proportions of neuroblasts and stromal cells, the interpatient heterogeneity is not especially surprising. Despite this variation, sets of genes with differential expression could be identified at day 7 and 28. Only 10% of the genes whose expression was altered in response to decitabine (i.e., day 7) exhibited changes in expression on day 28, which could be due to the fact that a different set of genes responds to cytotoxic agents. In our study, increases in gene expression were more common after decitabine, while decreases were more commonly seen after treatment with cytotoxic agents. While analysis of which genes and pathways are affected by these agents is clearly more important than the number of genes with altered expression, the fact that the cellular response to these drugs differed so dramatically is nonetheless potentially interesting and may reflect differences in the mechanism of action of these agents. Our data are consistent with the proposed effect of decitabine: reactivation of epigenetically silenced genes by irreversibly binding DNMT1 and depleting intracellular DNMT1 activity. This was also noted in a study that analyzed the gene expression profile of a neuroblastoma cell line following exposure to decitabine; of 44 genes that were differentially expressed (≥ 3-fold), 27 were upregulated, among them Hsp47, and 17 were downregulated following treatment [24]. Further studies with additional patients and quantitative analyses will be required to validate these data and eliminate false positives.
Most responses to low-dose decitabine have been observed in patients with myelodysplastic syndrome [36,49] with limited evidence of activity in patients with solid tumors [11,38,39]. In adults with hematopoietic malignancies, 15 mg/m2 administered over one hour daily, 5 days a week, for 2 weeks induced the highest response rate [14,36]. Two of 9 adults with refractory solid tumors treated with continuous infusion decitabine at 2 mg/m2/day for 7 days had stable disease [11]. Similar results were seen in patients with ovarian cancer treated with a cytotoxic agent following decitabine [46]. The limited clinical responses seen in our study may reflect the fact that we were not able to achieve drug exposures similar to those that have been observed in other clinical studies. It has been reported that the higher the dose of demethylating agent, the more marked the epigenetic change [50]. Failure to achieve an adequate concentration to induce both biological and clinical response could have been due to several reasons including the inability to dose escalate due to the heavy pre-treatment of patients, the additional cytotoxicity incurred with cyclophosphamide and doxorubicin, the dosing schedule employed, the rapid plasma clearance of decitabine, and the use of single agent epigenetic therapy. Of course, we cannot exclude the possibility that decitabine may be inactive at any exposure. The use of alternative cytotoxic agents may allow further dose escalation of decitabine to achieve concentrations that would result in clinical and biologic response. In addition, targeting multiple points of the apoptotic pathway and/or the use of other agents that induce more extensive chromatin modifications may be necessary to induce response.
In summary, when administered in combination with cyclophosphamide and doxorubicin, the MTD of low-dose decitabine is 5 mg/m2/day in pediatric patients with relapsed solid tumors, with significant myelosuppression being the primary toxicity observed. At this dose, increased caspase-8 expression was found in a limited number of tumor-containing bone marrow samples. Sustained demethylation and overexpression of biomarkers MAGE-1 and HbF in peripheral blood was maintained for 4 weeks in the majority of samples tested and MAGE-1 could be used as a potential biomarker to assess demethylation status of the tumor. Since aberrant DNA methylation is often complemented by other epigenetic events that alter chromatin structure such as histone deacetylation, a combination of these two epigenetic modifications with chemotherapeutic agents is an attractive strategy for future investigation. Lastly, the methylation profiles of tumors should be explored to determine whether they correlate with response to chemotherapy.
Supplementary Material
Relative sensitivity of caspase-8 methylation was determined by mixing unmethylated DNA from normal PBMCs and NB13 neuroblastoma cell line DNA in varying ratios prior to performing the methylated PCR reaction. These studies showed a direct relationship between the relative density of the PCR product and the percentage of unmethylated or methylated DNA (A). PCR products from these studies were analyzed by gel electrophoresis to verify specific amplification (B).
Post-infusion plasma profiles o patients treated with a 1-hour infusion of 5 mg/m2 (filled symbol) or 10 mg/m2 (open symbol) decitabine. Filled circles represent actual plasma concentrations. Lines illustrate fitted data.
The presence of neuroblastoma cells in bone marrow samples was assessed by quantitative reverse-transcriptase PCR using primers that specifically detected (A) GD2-synthase, (B) MYCN and (C) tyrosine hydroxylase (TH). Data is shown relative to the level of expression in peripheral blood samples from normal controls.
Acknowledgments
NIH/NINDS K08NS047983, ASCO and COG YIA (REG). Trial supported by National Cancer Institute Grants U01 CA97452 and NCRR M01 RR00188.
We acknowledge the outstanding assistance of Elizabeth O’Connor and Dori Greenwald of the COG Phase 1 Committee and that of all the CRAs of the participating institutions. We appreciate the help of Cheryl Medeiros-Nancarrow with preparation of the protocol. We also thank Dr. Tanya Tekautz for assistance during the preparation of the protocol, and Dr. Geoff Neale from the Hartwell Center at St. Jude Children’s Research Hospital for helpful advice on microarray analysis.
Footnotes
Previous Presentations: Presented in part at the 41st and 43rd Annual Meetings of the American Society of Clinical Oncology, 2005 and 2007.
Disclaimers: no conflict of interest.
AUTHORS DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
We dedicate this article to the memory of Dr Vincent Kidd on whose research this clinical trial was based.
References
- 1.Fulda S, Kufer MU, Meyer E, et al. Sensitization for death receptor- or drug-induced apoptosis by re-expression of caspase-8 through demethylation or gene transfer. Oncogene. 2001;20(41):5865–5877. doi: 10.1038/sj.onc.1204750. [DOI] [PubMed] [Google Scholar]
- 2.Razin A, Riggs AD. DNA methylation and gene function. Science (New York, NY) 1980;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 3.Merlo A, Herman JG, Mao L, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1(7):686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 4.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annual review of biochemistry. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 5.Brown R, Plumb JA. Demethylation of DNA by decitabine in cancer chemotherapy. Expert review of anticancer therapy. 2004;4(4):501–510. doi: 10.1586/14737140.4.4.501. [DOI] [PubMed] [Google Scholar]
- 6.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer research. 1998;58(1):95–101. [PubMed] [Google Scholar]
- 7.DeSimone J, Heller P, Hall L, et al. 5-Azacytidine stimulates fetal hemoglobin synthesis in anemic baboons. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(14):4428–4431. doi: 10.1073/pnas.79.14.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley TJ, DeSimone J, Noguchi CT, et al. 5-Azacytidine increases gamma-globin synthesis and reduces the proportion of dense cells in patients with sickle cell anemia. Blood. 1983;62(2):370–380. [PubMed] [Google Scholar]
- 9.DeSimone J, Koshy M, Dorn L, et al. Maintenance of elevated fetal hemoglobin levels by decitabine during dose interval treatment of sickle cell anemia. Blood. 2002;99(11):3905–3908. doi: 10.1182/blood.v99.11.3905. [DOI] [PubMed] [Google Scholar]
- 10.Oki Y, Aoki E, Issa JP. Decitabine--bedside to bench. Critical reviews in oncology/hematology. 2007;61(2):140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Samlowski WE, Leachman SA, Wade M, et al. Evaluation of a 7-day continuous intravenous infusion of decitabine: inhibition of promoter-specific and global genomic DNA methylation. J Clin Oncol. 2005;23(17):3897–3905. doi: 10.1200/JCO.2005.06.118. [DOI] [PubMed] [Google Scholar]
- 12.Yang AS, Doshi KD, Choi SW, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer research. 2006;66(10):5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 13.Daskalakis M, Nguyen TT, Nguyen C, et al. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. Blood. 2002;100(8):2957–2964. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 14.Issa JP, Garcia-Manero G, Giles FJ, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103(5):1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 15.Teitz T, Wei T, Valentine MB, et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nature Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 16.Teitz T, Lahti JM, Kidd VJ. Aggressive childhood neuroblastomas do not express caspase-8: an important component of programmed cell death. Journal of molecular medicine (Berlin, Germany) 2001;79(8):428–436. doi: 10.1007/s001090100233. [DOI] [PubMed] [Google Scholar]
- 17.Eggert A, Grotzer MA, Zuzak TJ, et al. Resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in neuroblastoma cells correlates with a loss of caspase-8 expression. Cancer research. 2001;61(4):1314–1319. [PubMed] [Google Scholar]
- 18.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, et al. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer research. 2000;60(16):4315–4319. [PubMed] [Google Scholar]
- 19.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, et al. Loss of caspase-8 expression in neuroblastoma is related to malignancy and resistance to TRAIL-induced apoptosis. Medical and pediatric oncology. 2000;35(6):608–611. doi: 10.1002/1096-911x(20001201)35:6<608::aid-mpo25>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 20.Takita J, Yang HW, Chen YY, et al. Allelic imbalance on chromosome 2q and alterations of the caspase 8 gene in neuroblastoma. Oncogene. 2001;20(32):4424–4432. doi: 10.1038/sj.onc.1204521. [DOI] [PubMed] [Google Scholar]
- 21.Astuti D, Agathanggelou A, Honorio S, et al. RASSF1A promoter region CpG island hypermethylation in phaeochromocytomas and neuroblastoma tumours. Oncogene. 2001;20(51):7573–7577. doi: 10.1038/sj.onc.1204968. [DOI] [PubMed] [Google Scholar]
- 22.Yan P, Muhlethaler A, Bourloud KB, et al. Hypermethylation-mediated regulation of CD44 gene expression in human neuroblastoma. Genes Chromosomes Cancer. 2003;36(2):129–138. doi: 10.1002/gcc.10150. [DOI] [PubMed] [Google Scholar]
- 23.Yang QW, Liu S, Tian Y, et al. Methylation-associated silencing of the thrombospondin-1 gene in human neuroblastoma. Cancer research. 2003;63(19):6299–6310. [PubMed] [Google Scholar]
- 24.Yang Q, Liu S, Tian Y, et al. Methylation-associated silencing of the heat shock protein 47 gene in human neuroblastoma. Cancer research. 2004;64(13):4531–4538. doi: 10.1158/0008-5472.CAN-04-0956. [DOI] [PubMed] [Google Scholar]
- 25.van Noesel MM, van Bezouw S, Voute PA, et al. Clustering of hypermethylated genes in neuroblastoma. Genes Chromosomes Cancer. 2003;38(3):226–233. doi: 10.1002/gcc.10278. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, He L, Savell VH, et al. Inhibition of the interferon-gamma/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21WAF1 promoter region. Cancer research. 2000;60(12):3290–3298. [PubMed] [Google Scholar]
- 27.Yokochi T, Robertson KD. Doxorubicin inhibits DNMT1, resulting in conditional apoptosis. Molecular pharmacology. 2004;66(6):1415–1420. doi: 10.1124/mol.104.002634. [DOI] [PubMed] [Google Scholar]
- 28.Schwartsmann G, Fernandes MS, Schaan MD, et al. Decitabine (5-Aza-2′-deoxycytidine; DAC) plus daunorubicin as a first line treatment in patients with acute myeloid leukemia: preliminary observations. Leukemia. 1997;11 (Suppl 1):S28–31. [PubMed] [Google Scholar]
- 29.Willemze R, Archimbaud E, Muus P. Preliminary results with 5-aza-2′-deoxycytidine (DAC)-containing chemotherapy in patients with relapsed or refractory acute leukemia. The EORTC Leukemia Cooperative Group. Leukemia. 1993;7 (Suppl 1):49–50. [PubMed] [Google Scholar]
- 30.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors [see comments] J Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 31.Gu Z, Zhou G. Report. XenoBiotic Laboratories, Inc. (XBL); 2002. May 23, Analytical Method Development and Validation of Decitabine in Human Plasma; pp. 1–74. [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung IY, Cheung NK. Quantitation of marrow disease in neuroblastoma by real-time reverse transcription-PCR. Clin Cancer Res. 2001;7(6):1698–1705. [PubMed] [Google Scholar]
- 35.Schrump DS, Fischette MR, Nguyen DM, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res. 2006;12(19):5777–5785. doi: 10.1158/1078-0432.CCR-06-0669. [DOI] [PubMed] [Google Scholar]
- 36.Issa JP, Gharibyan V, Cortes J, et al. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol. 2005;23(17):3948–3956. doi: 10.1200/JCO.2005.11.981. [DOI] [PubMed] [Google Scholar]
- 37.Cashen AF, Shah AK, Todt L, et al. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer chemotherapy and pharmacology. 2008;61(5):759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- 38.Momparler RL, Bouffard DY, Momparler LF, et al. Pilot phase I-II study on 5-aza-2′-deoxycytidine (Decitabine) in patients with metastatic lung cancer. Anti-cancer drugs. 1997;8(4):358–368. doi: 10.1097/00001813-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 39.van Groeningen CJ, Leyva A, O’Brien AM, et al. Phase I and pharmacokinetic study of 5-aza-2′-deoxycytidine (NSC 127716) in cancer patients. Cancer research. 1986;46(9):4831–4836. [PubMed] [Google Scholar]
- 40.De Smet C, Lurquin C, Lethe B, et al. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Molecular and cellular biology. 1999;19(11):7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thibault A, Figg WD, Bergan RC, et al. A phase II study of 5-aza-2′deoxycytidine (decitabine) in hormone independent metastatic (D2) prostate cancer. Tumori. 1998;84(1):87–89. doi: 10.1177/030089169808400120. [DOI] [PubMed] [Google Scholar]
- 42.Aparicio A, Eads CA, Leong LA, et al. Phase I trial of continuous infusion 5-aza-2′-deoxycytidine. Cancer chemotherapy and pharmacology. 2003;51(3):231–239. doi: 10.1007/s00280-002-0563-y. [DOI] [PubMed] [Google Scholar]
- 43.Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 44.Oki Y, Kantarjian HM, Gharibyan V, et al. Phase II study of low-dose decitabine in combination with imatinib mesylate in patients with accelerated or myeloid blastic phase of chronic myelogenous leukemia. Cancer. 2007;109(5):899–906. doi: 10.1002/cncr.22470. [DOI] [PubMed] [Google Scholar]
- 45.De Smet C, De Backer O, Faraoni I, et al. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(14):7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Appleton K, Mackay HJ, Judson I, et al. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol. 2007;25(29):4603–4609. doi: 10.1200/JCO.2007.10.8688. [DOI] [PubMed] [Google Scholar]
- 47.Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100(4):1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan HP, Di Liao C, Fu BY, et al. Interindividual and interethnic variation in genomewide gene expression: insights into the biological variation of gene expression and clinical implications. Clin Chem. 2009;55(4):774–785. doi: 10.1373/clinchem.2008.119107. [DOI] [PubMed] [Google Scholar]
- 49.Wijermans P, Lubbert M, Verhoef G, et al. Low-dose 5-aza-2′-deoxycytidine, a DNA hypomethylating agent, for the treatment of high-risk myelodysplastic syndrome: a multicenter phase II study in elderly patients. J Clin Oncol. 2000;18(5):956–962. doi: 10.1200/JCO.2000.18.5.956. [DOI] [PubMed] [Google Scholar]
- 50.Lemaire M, Chabot GG, Raynal NJ, et al. Importance of dose-schedule of 5-aza-2′-deoxycytidine for epigenetic therapy of cancer. BMC Cancer. 2008;8:128. doi: 10.1186/1471-2407-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative sensitivity of caspase-8 methylation was determined by mixing unmethylated DNA from normal PBMCs and NB13 neuroblastoma cell line DNA in varying ratios prior to performing the methylated PCR reaction. These studies showed a direct relationship between the relative density of the PCR product and the percentage of unmethylated or methylated DNA (A). PCR products from these studies were analyzed by gel electrophoresis to verify specific amplification (B).
Post-infusion plasma profiles o patients treated with a 1-hour infusion of 5 mg/m2 (filled symbol) or 10 mg/m2 (open symbol) decitabine. Filled circles represent actual plasma concentrations. Lines illustrate fitted data.
The presence of neuroblastoma cells in bone marrow samples was assessed by quantitative reverse-transcriptase PCR using primers that specifically detected (A) GD2-synthase, (B) MYCN and (C) tyrosine hydroxylase (TH). Data is shown relative to the level of expression in peripheral blood samples from normal controls.