Abstract
The discrepancies between the published QM/MM studies (Schöneboom, J. C.; Cohen, S.; Lin, H.; Shaik, S.; Thiel, W. J. Am. Chem. Soc. 2004, 126, 4017 / Guallar, V.; Friesner, R. A. J. Am. Chem. Soc. 2004, 126, 8501) on H-abstraction of camphor in P450cam have largely been resolved. The crystallographic water molecule 903 situated near the oxo atom of Compound I acts as a catalyst for H-abstraction, lowering the barrier by about 4 kcal/mol. Spin density at the A-propionate side chain of heme can occur in the case of incomplete screening, but has no major effect on the computed barrier.
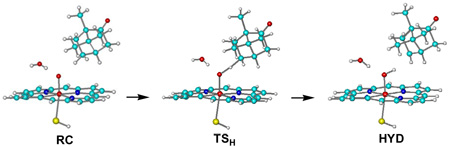
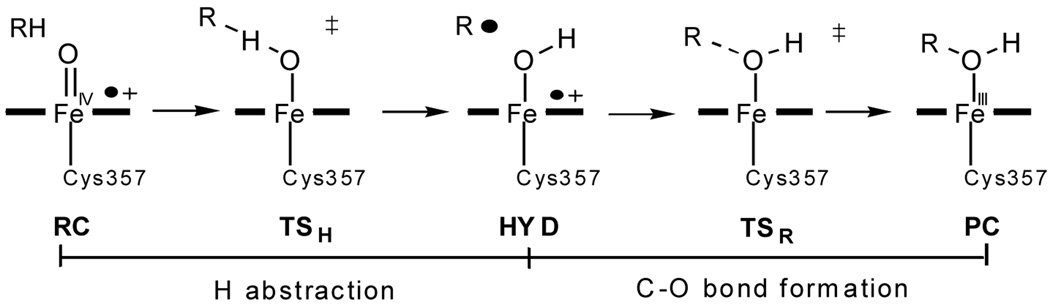
Cytochrome P450 monooxygenases catalyze regio- and stereoselective C–H bond hydroxylation in bioorganisms.1 The primary reactive species of P450 enzymes is deemed to be an elusive high-valence oxyferryl-porphyrin π-cation species called Compound I (Cpd I). In the consensus rebound mechanism (see Scheme 1),1 an initial rate-determining hydrogen abstraction from the substrate yields an iron-hydroxo intermediate radical pair HYD via a transition structure TSH which then recombines to give the ferric-alcohol complex PC. P450cam hydroxylates camphor at the C5 position leading to 5-exo-hydroxycamphor as the sole product.
Scheme 1.
Rebound mechanism of C–H hydroxylation by P450
Combined quantum mechanical/molecular mechanical (QM/MM) studies on biohydroxylation of camphor by Cpd I of P450cam have been performed by two groups (Shaik/Thiel [S/T]2 and Guallar/Friesner [G/F]3,4) recently. These studies followed the same general strategy but differed in scope, system setup, and QM/MM methodology. S/T investigated the full rebound mechanism for four different snapshots of a molecular dynamics (MD) trajectory,2,5 while G/F addressed the initial H-abstraction step in the quartet state starting from a geometry close to the X-ray structure (pdb code: 1DZ9). The S/T system includes a water layer of 16 Å and has 24394 atoms with a net charge of −10e, whereas the G/F system is neutral and has 7448 atoms (no solvent layer, polar surface residues neutralized). The heme side chains were incorporated into the QM region by G/F, but not by S/T in their standard setup. The QM/MM boundary was handled by frozen orbitals (G/F) and by link atoms with charge shift model (S/T) as implemented in QSite6 and ChemShell,7 respectively.
According to the unrestricted open-shell B3LYP (UB3LYP)/CHARMM calculations2 by S/T on the snapshots considered, TSH and HYD are ca. 21 (22) and 15 (11) kcal/mol higher in energy than RC (Scheme 1), on the quartet surface at the R1/B1 (R2/B2) level (definitions see below). Lower values of 11.7 and 4.5 kcal/mol are found in the restricted open-shell B3LYP (ROB3LYP)/OPLS-AA study of G/F at the R2s/Bg* level;3,4 the A-propionate (A-prop) heme side chain carries significant spin density which decreases during the H-abstraction reaction, with concomitant increase of negative charge at the A-prop carboxylate that is proposed to provide a differential stabilization of TSH and HYD through electrostatic interactions with Arg299.3,4 By contrast, single-point UB3LYP/CHARMM calculations by S/T on their snapshots (with extended QM regions) and UB3LYP calculations on corresponding model systems do not give such spin density, neither in the enzyme nor in the gas phase, as long as the propionates are screened by neighboring Arg residues.2,8 In view of these significant differences in relative energies, spin densities, and role of the propionates, both groups decided to pinpoint the source of these discrepancies, by comparing the species common to their previous studies, namely, the lowest-energy electromers on the quartet surface, RC(IV), TSH(III), and HYD(III), during H-abstraction in the enzyme environment.2–4
For this purpose new UB3LYP/CHARMM calculations were carried out at Mülheim to treat the G/F and S/T systems on equal footing. The MM region was represented by the CHARMM22 force field.9 QM basis sets (B1, B2 and Bg*) and QM regions (R1, R2, R1p, R2s, R1’ and R1’p; 51–120 QM atoms) were taken from our previous work2–5 (see Supporting Information for details). QM/MM protocols were as described previously.2,5
UB3LYP/CHARMM single-point calculations on the G/F geometries3,4 yield barriers (endothermicities) in the range between 7.7–14.2 (0.3–6.4) kcal/mol (depending on the choice of QM region and basis set, see Supporting Information), consistent with the published R2s/Bg* values of 11.5 (4.7) kcal/mol3,4 which cannot be reproduced exactly because of the different treatment of the QM/MM boundary (see above). Focusing on the closest analogue to G/F,3,4 the present single-point calculations for R2s/B1, R2s/Bg* and R2s/B2 give relative energies that are within 2 kcal/mol of the published data, and non-vanishing spin densities at the A-prop carboxylate for RC/TSH/HYD (B1: 0.17/0.09/0.12; Bg*: 0.20/0.12/0.11; B2: 0.07/0.01/0.01) that are in accord with the published values of G/F3,4 (Bg*: 0.20/0.10/0.12) at least in case of the smaller basis sets (B1, Bg*). This confirms that the QM/MM implementations in Qsite and ChemShell yield similar results at a given geometry.
When UB3LYP/CHARMM geometry optimizations are performed along reaction paths starting from any of the G/F structures (RC, TSH, and HYD), considerably higher R1/B1 barriers (endothermicities) of ca. 16 (10) kcal/mol are obtained in each case (see Supporting Information). Reoptimization of the G/F system along pathways with a consistent protein environment increases the computed relative energies by ca. 5 kcal/mol, which accounts for about half of the reported discrepancies.2–4
As mentioned before, different types of starting geometries were used for QM/MM minimization by S/T (MD snapshots) and by G/F (X-ray structure). Going back to the S/T setup procedure,2,5 we have now adopted the protonated and solvated X-ray structure (unmodified backbone) as starting geometry (labeled snapX), which was relaxed by CHARMM followed by a QM/MM energy scan along the O–H5exo distance. The resulting R1/B1 energies of 16.7 (8.3) kcal/mol (Table 1) are reasonably close to those obtained after rescanning the reaction path for the G/F structures (see above), but significantly lower than those obtained previously by starting from the MD snapshots.2 The choice of the snapX setup thus affects the relative energies by about 5 kcal/mol (further analysis see below) and is responsible roughly for the other half of the reported discrepancies.2-4
Table 1.
UB3LYP/CHARMM barriers (endothermicities) in kcal/mol at snapX-based optimized geometries (R1/B1 first column, R1/B2 otherwise).
| Asp297 | R1/B1 | R1/B2 | R1p/B2 | R1’/B2 | R1’p/B2 | R2/B2 | R2s/B2 |
|---|---|---|---|---|---|---|---|
| deprotonated | 16.7 (8.3) | 16.3 (4.1) | 16.9 (4.4) | 19.0 (4.6) | 19.9 (4.9) | 18.8 (4.6) | 19.7 (5.0) |
| protonated | 16.9 (10.1) | 16.7 (6.1) | 16.9 (6.1) | 19.3 (6.8) | 19.7 (6.9) | 18.7 (6.5) | 19.1 (6.4) |
The G/F3,4 and S/T2,5 structures have several different protonation state assignments for titratable residues. We address only Asp297 and His355 here. Both groups have assigned Asp297 to be deprotonated,2–5 contrary to Poisson-Boltzmann results10 for the pentacoordinated ferric P450cam complex and the preferred assignment in a recent QM/MM study of the resting state.11 QM/MM optimizations for several P450cam intermediates with protonated Asp297 afford geometries close to the corresponding X-ray structures12 (see Supporting Information) whereas this is not true for deprotonated Asp297 (treated as part of the MM region, see ref 13): here the plane of the A-prop carboxylate group rotates by ca. 45° and Arg299 aligns such that it screens Asp297 rather than the O2A atom of A-prop. The computed barriers are, however, almost the same for both protonation states of Asp297 while the endothermicities differ by 2 kcal/mol (Table 1).
His355 is the only residue near the active center that has been assigned a different protonation state by G/F3,4 (deprotonated) and S/T2 (protonated10). To check the implications of these choices, we have performed UB3LYP/CHARMM single-point calculations after removing the H atom at the ε nitrogen of His355, in the snapX-based R1/B2-optimized geometries. The R2s/B2 barriers (endothermicities) are almost independent of the protonation state of His355: 20.9 (5.8) / 19.7 (5.0) kcal/mol for the deprotonated and 19.5 (6.3) / 19.1 (6.4) kcal/mol for the protonated Asp297 systems with deprotonated / protonated His355. By contrast, the spin densities at the A-prop carboxylate of RC/TSH/HYD are affected: in the deprotonated Asp297 systems they are 0.19/0.16/0.16 (R2s/B2) for deprotonated His355 and zero for protonated His355. In the protonated Asp297 systems, the spin densities at the A-prop carboxylate are always zero. These and other computational experiments show that, when the carboxylates are only partially screened, spin density at the A-prop carboxylate can indeed appear. However, even in these cases, the barrier is not affected much (typically by less than 2 kcal/mol) showing that there is no clear correlation between the computed A-prop spin density and barrier height.
What feature of snapX is responsible for lowering the barrier from ca. 20 to 16 kcal/mol? A crystallographic water molecule (w903) close to the oxo ligand of Cpd I is present in snapX and G/F, but not in the S/T snapshots where it has moved away from the oxo ligand to some accessible space around camphor during the dynamics runs of the setup.2 The associated energetic effects can easily be tested by removing w903 from the snapX-based optimized structures and reoptimizing them at the UB3LYP/CHARMM level: the R1/B1 barrier (endothermicity) increases to 20.8 (13.9) kcal/mol, very close to the published R1/B1 values of 20.6 (14.9) kcal/mol.2 Hence, the presence of w903 lowers the barrier (endothermicity) by ca. 4 (6) kcal/mol. This reduction is found regardless whether w903 is included in the QM region or not (changes of less than 1 kcal/mol), indicating that the stabilization due to w903 has mainly electrostatic origin. Single-point gas-phase B3LYP calculations of QM region R1’ with and without w903 at the snapX-based QM/MM geometries yield analogous differential stabilizations of TSH (HYD) by 3.0 (5.2) kcal/mol, which can be traced to the different interaction energies between w903 and R1’ for RC/TSH/HYD (−8.9/−11.9/−14.2 kcal/mol at the B3LYP/B1 level without counterpoise corrections). These values are well reproduced by a simple point-charge model for the electrostatic interactions in the H-bond between w903 and R1’: in the series RC/TSH/HYD, the Mulliken charges remain almost constant for w903, but increase for the oxo atom and for H5exo, which upon application of Coulomb’s law translates into an increasing electrostatic stabilization (−8.6/−10.9/−14.5 kcal/mol, see Supporting Information for details). The computed charge shifts in the Fe=O…H…C moiety are in line with qualitative expectations for H-abstraction by the electronegative oxo atom (see Scheme 1). The concomitant spin density transfer from Fe=O to C is energetically less relevant for the H-bond with w903.
In summary, the stabilization by w903 arises from favorable electrostatic interactions in a hydrogen bond14 that is stronger in TSH and HYD than in RC mostly due to an increasing negative charge at the oxo atom (charge transfer from camphor). The w903 molecule thus acts as a catalyst for H-abstraction, lowering the barrier by about 4 kcal/mol. Since one water molecule is liberated during the conversion of Cpd 0 to Cpd I, the catalyst for the consumption of Cpd I is generated during the formation of Cpd I.
Supplementary Material
Acknowledgment
This work was supported by NIH (grant GM40526 to RAF).
Footnotes
Supporting Information Available: Detailed numerical results; complete refs 7 and 9. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd Ed. New York: Kluwer Academic/Plenum Publishers; 2004. [Google Scholar]
- 2.Schöneboom JC, Cohen S, Lin H, Shaik S, Thiel W. J. Am. Chem. Soc. 2004;126:4017. doi: 10.1021/ja039847w. [DOI] [PubMed] [Google Scholar]
- 3.Guallar V, Baik M, Lippard SJ, Friesner RA. Proc. Natl. Acad. Sci. USA. 2003;100:6998. doi: 10.1073/pnas.0732000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guallar V, Friesner RA. J. Am. Chem. Soc. 2004;126:8501. doi: 10.1021/ja036123b. [DOI] [PubMed] [Google Scholar]
- 5.Schöneboom JC, Lin H, Reuter N, Thiel W, Cohen S, Ogliaro F, Shaik S. J. Am. Chem. Soc. 2002;124:8142. doi: 10.1021/ja026279w. [DOI] [PubMed] [Google Scholar]
- 6.QSite. Portland, OR: Schrödinger, Inc.; 2001. [Google Scholar]
- 7.Sherwood P, et al. J. Mol. Struct. (Theochem) 2003;632:1. [Google Scholar]
- 8.Bathelt CM, Zurek JL, Mulholland AJ, Harvey JN. J. Am. Chem. Soc. 2005;127:12900. doi: 10.1021/ja0520924. [DOI] [PubMed] [Google Scholar]
- 9.MacKerell J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 10.Lounnas V, Wade RC. Biochemistry. 1997;36:5402. doi: 10.1021/bi9622940. [DOI] [PubMed] [Google Scholar]
- 11.Schöneboom JC, Thiel WJ. Phys. Chem. B. 2004;108:7468. [Google Scholar]
- 12.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DA, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 13.When Asp297 is included in the QM region and both Asp297 and His355 are deprotonated, QM/MM optimizations yield an X-ray-like RC structure with a short O-O distance between the A-prop and Asp297 carboxylates (see Supporting Information) which will be discussed elsewhere Guallar V, Olsen B. J. Inorg. Biochem. doi: 10.1016/j.jinorgbio.2006.01.019. accepted.
- 14.Ogliaro F, Cohen S, de Visser SP, Shaik S. J. Am. Chem. Soc. 2000;122:12892. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.