Abstract
Under lean conditions, the adipose-derived hormone leptin maintains energy balance in part through CNS-mediated increases in sympathetic outflow that enhance fat burning 1,2. Triggering of beta adrenergic receptors in adipocytes stimulates energy expenditure via cAMP-dependent increases in lipolysis and fatty acid oxidation 3. Although the underlying mechanism is unclear, catecholamine signaling in fat cells is thought to be disrupted in obesity 4, where it may contribute to the ectopic accumulation of lipid in liver and to the development of insulin resistance 5,6. Here we show that the cAMP responsive CREB coactivator CRTC3 promotes obesity by attenuating beta adrenergic receptor signaling in adipose; mice with a knockout of the CRTC3 gene have increased energy expenditure, are resistant to diet induced obesity, and are protected from the development of hepatic steatosis under high fat diet feeding conditions. CRTC3 was activated in response to catecholamine signals, when it reduced adenyl cyclase activity by upregulating the expression of RGS2 7–9, a metabolic syndrome susceptibility gene 10, which we show here is also a direct target of CREB and CRTC3. RGS2 expression was down-regulated in adipocytes from CRTC3−/− mice, leading to increases in insulin and catecholamine signaling that enhanced glucose and fatty acid oxidation. As a common human CRTC3 variant (Ser72Asn), with increased transcriptional activity, is associated with several anthropometric indices of adiposity in two distinct Mexican-American cohorts, our results suggest that adipocyte CRTC3 may play a role in the development of obesity in this population.
The second messenger cAMP stimulates the expression of cellular genes via the PKA-mediated phosphorylation of CREB family members (CREB1, ATF1, CREM) and via the dephosphorylation of the TORC/CRTC coactivators 11. CREB phosphorylation promotes its association with the histone acetyl transferase (HAT) paralogs CBP and P300, while CRTC de-phosphorylation increases its nuclear entry and binding to CREB. After prolonged stimulation with cAMP agonist, CREB target gene transcription undergoes attenuation, reflecting in part the ubiquitin-mediated degradation of CRTCs 12.
Similar to other CRTC family members, CRTC3 contains CREB binding (CBD; aa. 1–50), regulatory (RD; aa. 51–549), and trans-activation domains (TAD; aa. 550–619), that are also present in CRTC1 and CRTC2 (fig. 1a). In the basal state, CRTC3 is phosphorylated at Ser162 by Salt Inducible Kinases (SIKs) and other members of the stress and energy sensing AMPK family of Ser/Thr kinases 11,13,14. Short term (0.5–1 hour) exposure to cAMP agonist promotes the de-phosphorylation and nuclear entry of CRTC3 (fig. 1a); similar to CRTC2 12, prolonged cAMP stimulation triggers CRTC3 degradation.
Figure 1.
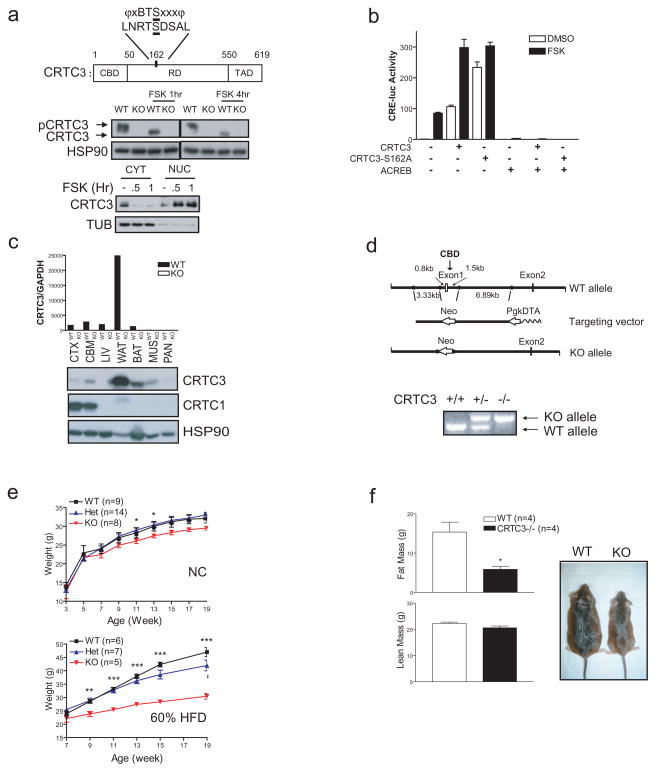
CRTC3−/− mice are resistant to diet induced obesity. A. Top, schematic of CRTC3 protein showing CREB Binding (CBD: aa. 1–50), regulatory (RD; aa. 51–549), and transactivation (TAD: aa. 550–619) domains as well as regulatory Ser162 site, which corresponds to a consensus phosphorylation site for AMPK family members. Middle, immunoblot showing effect of FSK on de-phosphorylation of CRTC3 in wild-type and CRTC3−/− mouse embryo fibroblasts (MEFs) exposed to FSK for 1 or 4 hours. Bottom, immunoblot showing nuclear and cytoplasmic amounts of CRTC3 in NIH-3T3L1 pre-adipocytes under basal conditions and following exposure to FSK. B. Effect of FSK on CRE-luciferase reporter activity in HEK293T cells following over-expression of wild-type or phosphorylation-defective S162A CRTC3. Co-transfection of dominant negative A-CREB expression vector indicated. C. Q-PCR (top) and immunoblot (bottom) analysis of CRTC3 mRNA and protein amounts in different tissues in wild-type and CRTC3 −/− (KO) mice. CRTC1 protein amounts shown for comparison. (CTX; cortex. CBM; cerebellum. LIV; liver. WAT, BAT; white and brown adipose. MUS; muscle. PAN; pancreas.) D. Top, schematic showing structure of the CRTC3 targeting vector containing Neo selection marker in place of Exon 1, which encodes the CREB binding domain (CBD). Structure of the mutant CRTC3 allele after homologous recombination indicated. Bottom, PCR analysis showing wild-type and mutant CRTC3 alleles in genomic DNA from wild-type, heterozygous, and homozygous CRTC3 knockout mice. E. Relative weight gain in wild-type and CRTC3 mutant (Het, KO) littermates under normal chow (12% of calories from fat; top) or high fat diet feeding (HFD; 60% of calories from fat; bottom) conditions. Age (in weeks) indicated. (*; P<0.05. **; P<0.01. ***; P <0.001.) F. Left, relative fat and lean mass in HFD-fed wild-type and CRTC3−/− mice by MRI analysis. Right, representative photo of HFD-fed wild-type and CRTC3−/− mice. (*; P<0.05)
CRTC3 over-expression augments the activity of a cAMP responsive (CRE-luc) reporter in cells exposed to forskolin (FSK; fig. 1b); and mutation of the regulatory Ser162 phosphorylation site to alanine further enhances CRTC3 activity under basal conditions. In keeping with the proposed role of CREB in recruiting CRTC3 to relevant promoters, expression of a dominant negative CREB inhibitor, called ACREB 15, blocks CRTC3 effects on reporter activity in cells exposed to FSK. By contrast with CRTC1, which is expressed primarily in brain, CRTC3 protein and mRNA amounts are particularly abundant in white adipose and to a lesser extent in brown adipose (sup. fig. 1, fig. 1c).
Based on the importance of the CREB Binding Domain (CBD) for CRTC-mediated induction of cAMP responsive genes 16,17, we generated CRTC3 −/− mice with a deletion of exon 1, which encodes the CBD (fig. 1d). CRTC3 −/− mice are born at the expected Mendelian frequency; they appear comparable to wild-type littermates at birth, despite the absence of detectable CRTC3 mRNA and protein amounts in all tissues (fig. 1c).
When maintained on a normal chow diet, CRTC3−/− mice appear more insulin sensitive than controls by insulin tolerance testing (sup. fig. 1, right). CRTC3−/− animals also have 50% lower adipose tissue mass, despite comparable food intake and physical activity to control mice (sup. fig. 2).
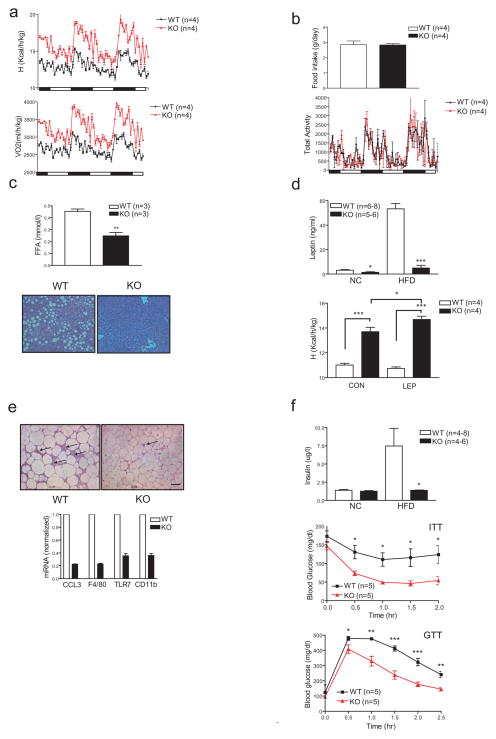
When transferred to a high fat diet (HFD; 60% of calories from fat), CRTC3 −/− mice gained 35% less weight relative to controls reflecting primarily differences in fat accumulation (fig. 1e,f). The effect of CRTC3 on adiposity appeared to be gene dosage dependent as CRTC3+/− mice show intermediate weight gains relative to wild-type and CRTC3−/− mice. Although physical activity and food intake were nearly identical, energy expenditure and oxygen consumption were substantially elevated in HFD-fed CRTC3−/− mice relative to wild-type littermates (fig. 2a,b). Pointing to parallel increases in glucose and lipid oxidation, respiratory quotients were comparable in wild-type and CRTC3−/− mice (sup. fig. 3).
Figure 2.
Increased energy expenditure and insulin sensitivity in CRTC3−/− mice. A. Relative energy expenditure and oxygen consumption in wild-type and CRTC3−/− mice under high fat diet feeding conditions. B. Metabolic cage analysis of food intake and physical activity in wild-type and CRTC3−/− mice maintained on a HFD for 12 weeks. C. Top, circulating concentrations of free fatty acids (FFAs) in wild-type and CRTC3−/− mice under ad libitum feeding conditions. Bottom, hematoxylin-eosin staining of hepatic sections showing relative amounts of lipid in HFD fed wild-type and CRTC3−/− mice. (**; P<0.01) D. Top, Circulating leptin concentrations in NC and HFD-fed wild-type and CRTC3−/− mice. Bottom, effect of leptin (3mg/kg) or vehicle injection (IP) on energy expenditure in wild-type and CRTC3−/− mice. (*; P<0.05. ***; P<0.001.) E. Top, immunohistochemical analysis of WAT sections from wild-type and CRTC3−/− mice using F4/80 antiserum to visualize resident adipose tissue macrophages. Scale bar, 50μm. Bottom, Q-PCR analysis of mRNA amounts for macrophage-specific genes in WAT from HFD-fed wild-type and CRTC3−/− mice. F. Circulating concentrations of insulin (top), insulin tolerance testing (middle), and glucose tolerance testing (bottom) of wild-type and CRTC3−/− mice maintained a high fat diet (HFD) for 10 weeks. Insulin levels on NC diet (top) shown for comparison. (*; P<0.05. **; P<0.01. ***; P<0.001.)
Circulating concentrations of FFAs were decreased in CRTC3−/− mice, and they were protected from the effects of HFD feeding on hepatic steatosis (fig. 2c). Consistent with their reduced fat mass, CRTC3−/− mice had decreased circulating leptin concentrations than wild-type littermates, although the reduction in leptin levels (10-fold) appeared disproportionately low relative to the difference in fat mass (3-fold) (fig. 2d; sup. fig. 4). Indeed, intraperitoneal (IP) administration of leptin stimulated energy expenditure to a greater extent in CRTC3 mutant mice relative to wild-type. Taken together, these results indicate that disruption of CRTC3 activity leads to increases in energy expenditure, which maintain leptin sensitivity and protect against ectopic lipid accumulation.
Under obese conditions, increases in inflammatory infiltrates in adipose contribute to the development of systemic insulin resistance 18. Although they were readily observed in wild-type mice, adipose-tissue macrophages were less abundant in CRTC3−/− tissue (fig. 2e; sup. fig. 5). Arguing against an effect of the CRTC3 knockout on macrophage function per se, Tumor Necrosis Factor alpha (TNFα) release from peritoneal macrophages in response to lipopolysaccharide (LPS) appeared comparable between CRTC3 mutant and control cells (sup. fig. 5). In line with these differences, circulating insulin concentrations were lower in HFD-fed CRTC3−/− mice relative to wild-type, and whole body insulin sensitivity was correspondingly improved by insulin and glucose tolerance testing (fig. 2f). As a result, glucose uptake into muscle was increased in CRTC3−/− mice compared to control littermates (sup. fig. 6).
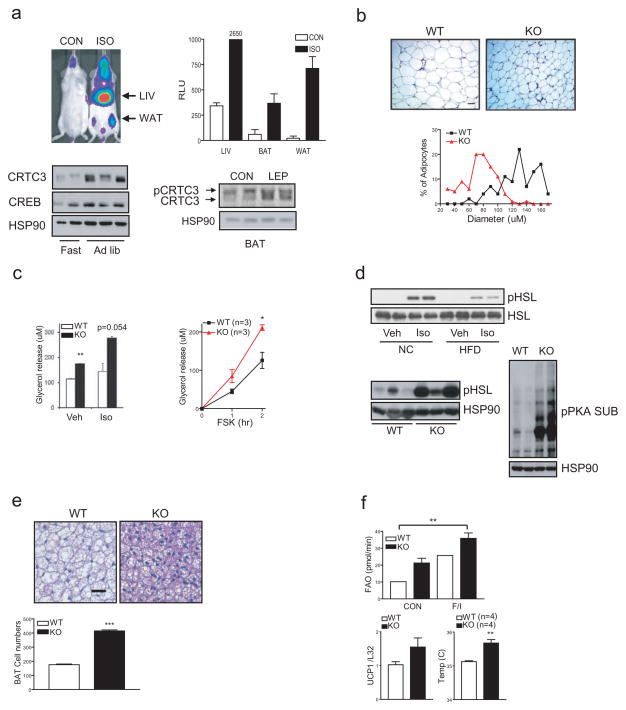
We considered that CRTC3 activity in adipose may be modulated by hormonal signals. In line with its effects on CRTC3 dephosphorylation in cell cultures (sup. fig. 7), IP administration of β adrenergic agonist isoproterenol (ISO) increased the activity of a CRE-luc reporter transgene in WAT and BAT by live imaging analysis (fig. 3a). Leptin administration (IP) also promoted CRTC3 de-phosphorylation. CRTC3 protein amounts in WAT are elevated under ad libitum conditions; they decreased after 6 hour fasting, when CRTC3 appeared to undergo degradation (fig. 3a). Consistent with an increase in protein stability under obese conditions, CREB and CRTC3 protein amounts were upregulated in WAT from HFD-fed compared to NC-fed mice (sup. fig. 8).
Figure 3.
Increased catecholamine signaling in CRTC3−/− adipose. A. Top, In vivo imaging analysis showing CRE-luc reporter activity in tissues from transgenic reporter mice following IP administration of isoproterenol (10mg/kg). Bar graph shows reporter activity in extracts from BAT, WAT, and liver. Bottom left, immunoblot of CRTC3 protein amounts in WAT from wild-type mice under fasting or fed conditions. Bottom right, immunoblot showing effect of leptin injection IP (3mg/kg; s.i.d. for 3d) on CRTC3 de-phosphorylation in BAT. B. Top, H&E sections of WAT from wild-type and CRTC3−/− mice. Scale bar, 50μm. Bottom, relative size distribution of adipocytes from wild-type and CRTC3 mutant mice. C. Left, in vitro lipolysis rates in cultured wild-type or CRTC3−/− adipocytes exposed to isoproterenol (ISO; 2μM). Right, relative lipolysis rates in wild-type and CRTC3−/− adipocytes exposed to FSK for 1 or 2 hours. Lipolysis rates determined by measuring glycerol release into the medium. (*; P<0.05. **; P<0.01). D. Top, immunoblot analysis of phospho (Ser660) HSL amounts in WAT from NC or HFD-fed mice following IP ISO administration as indicated. Bottom left, immunoblot of P-HSL amounts in WAT from WT or CRTC3−/− mice under HFD conditions. Bottom right, PKA activity, as measured with phospho- PKA substrate antiserum in epididymal WAT from HFD-fed wild-type and CRTC3−/− mice. E. Top, H&E sections of BAT from wild-type and CRTC3−/− mice. Scale bar, 50μm. Bottom, relative numbers of brown adipocytes recovered from fat pads of wild-type and CRTC3 mutant mice. (***; P<0.001). F. Top, in vitro fatty acid oxidation rates in brown adipocytes, normalized to total cell number. Exposure to FSK (10μM) plus isoproterenol (10μM; F/I) indicated. Bottom left, UCP1 mRNA amounts, determined by Q-PCR analysis of BAT mRNA from wild-type and CRTC3−/− mice (**; P<0.01). Bottom right, core body temperatures in wild-type and CRTC3−/− mice..
Under HFD feeding conditions, increases in catecholamine signaling maintain energy balance by mobilizing triglyceride stores in WAT 19. Although the total number of adipocytes in WAT fat pads was nearly identical in both groups, adipocytes from CRTC3−/− mice were substantially smaller than from wild-type mice (fig. 3b). Arguing against a disruption in triglyceride synthesis, mRNA amounts for lipogenic genes (ACC, LPL, SCD) appeared comparable between CRTC3 mutant and wild-type adipocytes (sup. fig. 9). Rather, basal and ISO-induced lipolysis rates were increased in CRTC3−/− compared to control adipocytes (fig. 3c). Exposure to FSK also increased lipolysis to a greater extent in CRTC3−/− adipocytes (fig. 3c), pointing to the potential upregulation of the cAMP signaling pathway in these cells.
Triggering of β-adrenergic receptors has been found to promote lipolysis via the cAMP dependent protein kinase A (PKA)-mediated phosphorylation of hormone sensitive lipase (HSL) 20. In keeping with the proposed down-regulation of β adrenergic receptor signaling in obesity, administration of ISO had only modest effects on HSL phosphorylation in HFD-fed relative to NC-fed animals (fig. 3d). Indeed, amounts of phospho (Ser 660) HSL were substantially elevated in CRTC3−/− WAT compared to wild-type, even though circulating concentrations of norepinephrine and epinephrine were similar between the two groups (fig 3d, sup. fig. 10). PKA activity in WAT was also increased in CRTC3−/− mice by immunoblot assay using a phospho-specific PKA substrate antiserum. Consistent with the predominant expression of CRTC3 in adipose, PKA activity in other tissues appeared similar between wild-type and CRTC3−/− mice (sup. fig. 11).
Having seen that lipolysis rates are increased in WAT, and realizing that circulating free fatty acid concentrations are reduced in CRTC3 mutant mice, we considered that fatty acid oxidation should also be upregulated in this setting. Under high fat diet conditions, leptin has been proposed to trigger catecholamine-mediated increases in fat burning in BAT, a process known as diet-induced thermogenesis 21,22. In keeping with the ability for catecholamines to stimulate BAT expansion, brown adipocyte numbers were increased 2-fold in intra-scapular fat pads from CRTC3−/− mice compared to controls (fig. 3e). Suggesting a parallel increase in fat burning, CRTC3 −/− brown adipocytes also had smaller intracellular lipid vacuoles than wild-type cells. Moreover, fatty acid oxidation rates were increased in primary brown adipocytes from CRTC3−/− mice relative to controls, and uncoupling protein 1 (UCP1) mRNA amounts were also higher (fig. 3f). Correspondingly, core body temperatures were elevated in CRTC3−/− mice compared to control animals. Taken together, these results indicate that loss of CRTC3 increases fat burning in part through increases in brown adipocyte numbers in BAT.
We reasoned that the loss of CRTC3 expression could increase cellular PKA activity by altering the subunit composition of the PKA holoenzyme. Over-expression of the forkhead protein FOXC2 in WAT, for example, has been found to promote energy expenditure by up-regulating mRNA amounts for Regulatory Subunit I, which has a higher affinity for cAMP compared to RII 23. Arguing against this possibility, however, mRNA amounts for regulatory subunits I and II in WAT were comparable between wild-type and CRTC3 mutants (sup. fig. 12, left).
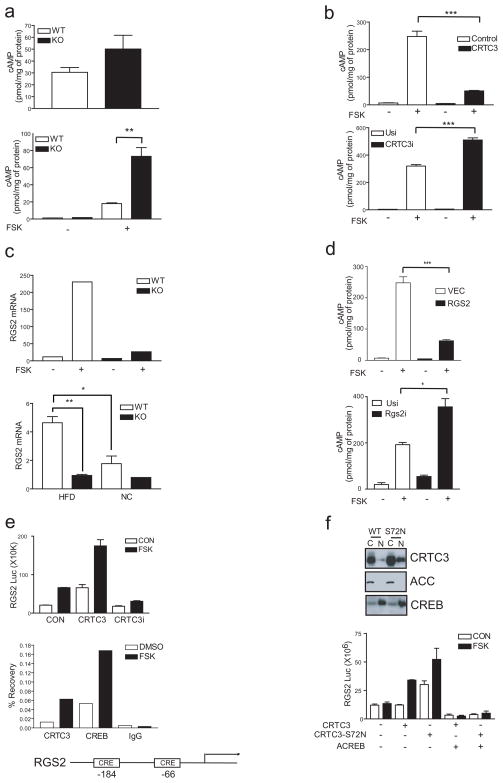
Alternatively, disruption of the CRTC3 gene may enhance PKA activity by increasing cellular cAMP accumulation in response to hormonal signals. Supporting this idea, cAMP concentrations were elevated in WAT from CRTC3 mutant mice relative to controls (fig. 4a). Exposure to FSK also triggered cAMP accumulation to a greater extent in CRTC3−/− MEFs and in CRTC3−/− brown adipose stromal-vascular cells compared to wild-type cells (fig. 4a, sup. fig. 12, right). Moreover, CRTC3 over-expression in wild-type cells reduced cAMP production in response to FSK, while acute RNAi-mediated depletion of CRTC3 increased it (fig. 4b; sup. fig. 13).
Figure 4.
CRTC3 attenuates cAMP signaling in adipose. A. cAMP content in WAT (top) and cultured MEFs (bottom) from wild-type and CRTC3−/− mice. Exposure to FSK indicated. (**; P<0.01) B. Effect of CRTC3 over-expression (top) or RNAi mediated knockdown (bottom) on cAMP accumulation in wild-type MEFs exposed to FSK as indicated. (***; P<0.001) C. Top, Q-PCR analysis showing relative mRNA amounts for RGS2 in primary cultured adipocytes (WT, CRTC3−/−) exposed to FSK as indicated. Bottom, Q-PCR analysis of RGS2 mRNA amounts in WAT from NC or HFD-fed mice (WT, CRTC3−/−). (*; P<0.05. **; P<0.01) D. Effect of RGS2 over-expression (top) or RNAi mediated knockdown (bottom) on cAMP accumulation in wild-type MEFs exposed to FSK as shown. (*; P<0.05. ***; P<0.001) E. Top, transient assay of RGS2-luc reporter activity in HEK293T cells exposed to FSK. Effect of CRTC3 over-expression or RNAi-mediated depletion indicated. Bottom, chromatin immunoprecipitation (ChIP) assay showing occupancy of CRTC3 over the RGS2 promoter in MEFs exposed to FSK as indicated. F. Top, immunoblot showing relative amounts of wild-type and S72N variant epitope-tagged CRTC3 polypeptides in cytoplasmic and nuclear fractions from transfected HEK293T cells. Fractionation of control cytoplasmic (ACC) and nuclear (CREB) proteins shown. Bottom, transient transfection assay of HEK293T cells showing relative activities of wild-type and S72N CRTC3 expression vectors co-transfected with RGS2-luc reporter plasmid. Exposure to FSK indicated.
In principle, the enhanced accumulation of cAMP in CRTC3-deficient cells could reflect a decrease in cellular phosphodiesterase (PDE) activity. In that event, treatment with non-selective PDE inhibitor should lead to comparable increases in cAMP concentrations between wild-type and mutant cells exposed to β adrenergic agonist. However, intracellular cAMP concentrations remained higher in CRTC3−/− compared to wild-type cells following co-stimulation with ISO plus Isobutyl-methyl xanthine (IBMX; sup. fig. 14). Based on these results, we reasoned that CRTC3 likely inhibits cAMP signaling by modulating cellular adenyl cyclase activity.
In gene profiling studies to identify cellular genes that mediate inhibitory effects of CRTC3 on cAMP signaling, we identified the Regulator of G protein signaling 2 (RGS2) as the most highly upregulated gene of 15 that are induced 3-fold or better in adipocytes exposed to FSK 24. We confirmed these effects in cultured primary adipocytes, where exposure to FSK increased the expression of RGS2 and other targets in wild-type cells, but to a much lesser extent in CRTC3−/− cells (fig. 4c, top; sup. fig. 15). By contrast with its reduction in adipose from CRTC3−/− mice, CREB target gene expression in skeletal muscle appeared comparable between wild-type and CRTC3 mutant animals, likely reflecting regulatory contributions from CRTC2, which has been shown to promote the expression of PGC1α and mitochondrial genes in muscle cells 25.
First identified as a GTPase activating protein that blocks Gq signaling, RGS2 has also been shown to inhibit the cAMP pathway by binding directly to a subset of adenyl cyclases (types III, V and, VI) 7–9, isoforms that are enriched in BAT and WAT 26,27. Moreover, mutations that enhance RGS2 expression have been associated with increased risk of metabolic syndrome in humans 10. In keeping with the upregulation of CRTC3 in obesity, HFD feeding stimulated RGS2 mRNA amounts in wild-type WAT, but RGS2 expression remained low in WAT from HFD-fed CRTC3−/− mice (fig. 4c, bottom). Consistent with its proposed role as an adenyl cyclase inhibitor, RGS2 over-expression reduced cAMP production in cells exposed to FSK, while RNAi-mediated knockdown of RGS2 increased it (fig. 4d; sup. fig. 16, 17).
We examined whether the RGS2 gene is a direct target of CREB and CRTC3. In line with the presence of conserved CREB binding sites at −184 and −66 on the RGS2 promoter, exposure to FSK upregulated RGS2-luciferase reporter activity in transient transfection assays (fig. 4e). CRTC3 over-expression further enhanced RGS2 promoter activity, whereas RNAi-mediated depletion of CRTC3 reduced it. Consistent with a direct effect of these activators, exposure to FSK increased CREB and CRTC3 occupancy over the RGS2 promoter in wild-type cells (fig. 4e).
Having seen the effects of CRTC3 on energy expenditure, we wondered whether this coactivator also contributes to obesity in humans. Within the human database (dbSNP) of single nucleotide polymorphisms (SNPs), we noticed a common CRTC3 variant allele, which encodes a missense variant (S72N) near a predicted nuclear export sequence 28. Supporting this idea, nuclear amounts of 72N CRTC3 were elevated relative to 72S CRTC3 under basal conditions (fig. 4f). Correspondingly, 72N variant CRTC3 was more potent than 72S CRTC3 in stimulating RGS2 promoter activity, particularly under basal conditions (fig. 4f).
We examined the potential association between the S72N variant CRTC3 and adiposity in a Mexican-American cohort of 779 individuals (Table 1). The allele frequency of the 72N variant in this population was 34%. In keeping with its increased activity relative to wild-type CRTC3, the 72N allele was also associated with several anthropometric indices of adiposity including weight, BMI, and hip circumference. Similar to the gene dosage effect of CRTC3 on weight gain in mice, Mexican-Americans with two 72N alleles had increased adiposity compared to those with only one variant allele; and 72S/72N heterozygous individuals had intermediate adiposity relative to individuals homozygous for the wild-type and variant CRTC3 alleles.
Table 1.
Association of S72N with anthropometric indices in the MACAD cohort
| S/S (n=346) | S/N (n=338) | N/N (n=95) | P value | |
|---|---|---|---|---|
| Weight (kg) | 73.3 (19.6) | 75.1 (20.6) | 76.0 (22.2) | 0.033 |
| BMI (kg/m2) | 28.1 (6.1) | 28.3 (6.1) | 29.1 (5.6) | 0.038 |
| Hip circumference (cm) | 103.0 (12.3) | 104.5 (13.0) | 104.5 (12.8) | 0.033 |
| Waist (cm) | 90.5 (15) | 93.0 (16) | 93.3 (17.3) | 0.15 |
| BSA (m2) | 1.77 (0.28) | 1.81 (0.30) | 1.84 (0.30) | 0.075 |
| Waist-hip-ratio | 0.88 (0.10) | 0.89 (0.11) | 0.88 (0.12) | 0.43 |
Values are median (interquartile range). Significant P values are in bold.
S/S: Individuals homozygous for wild-type CRTC3 (Ser72); S/N: Individuals heterozygous for variant CRTC3 (Asn72); N/N: Individuals homozygous for variant (Asn72) CRTC3.
We then sought to confirm the association of 72N with increases in adiposity indices, by assessing the association of a perfect proxy SNP rs3862434 (r2=1 with S72N) with adiposity measures in 987 Mexican Americans from the Multi-Ethnic Study of Atherosclerosis (MESA). The minor allele of rs3862434 (G, frequency 34%), which corresponds to 72N, was associated with increased body surface area (BSA) and a trend to increased weight (Table 2). In an analysis combining the two cohorts, 72N exhibited associations with weight, hip circumference, BMI, and BSA (Table 3). Because these traits are interrelated, we did not employ multiple testing correction, which would be overly conservative in light of the fact that our other data suggested 72N altered the biological function of CRTC3. Taken together, these results indicate that CRTC3 is associated with increased obesity risk in Mexican Americans.
Table 2.
Association of rs3862434 with anthropometric indices in MESA Mexican Americans
| A/A (n=423) | A/G (n=449) | G/G (n=115) | P value | |
|---|---|---|---|---|
| Weight (kg) | 76.9 (19.6) | 76.4 (20.3) | 79.1 (18.8) | 0.069 |
| BMI (kg/m2) | 29.5 (5.7) | 28.7 (6.2) | 28.6 (6.2) | 0.75 |
| Hip circumference (cm) | 103.6 (12.0) | 103.1 (12.6) | 103.9 (13.0) | 0.50 |
| Waist (cm) | 100.5 (14.4) | 99.5 (15.2) | 100.6 (14.8) | 0.79 |
| BSA (m2) | 1.81 (0.27) | 1.81 (0.28) | 1.88 (0.27) | 0.0073 |
| Waist-hip-ratio | 0.97 (0.08) | 0.97 (0.09) | 0.97 (0.07) | 0.53 |
Values are median (interquartile range). Significant P values are in bold.
A/A: Individuals homozygous for the major allele of rs3862434; A/G: Individuals heterozygous at the SNP; G/G: Individuals homozygous for the minor allele. The minor allele of rs3862434 corresponds to the minor allele of rs8033595 (Asn72) in Mexican Americans in CRTC3.
Table 3.
Association of S72N with anthropometric indices in the MACAD and MESA Mexican-American cohorts combined
| S/S or A/A (n=769) | S/N or A/G (n=787) | N/N or G/G (n=210) | P value | |
|---|---|---|---|---|
| Weight (kg) | 74.8 (20.2) | 75.9 (20.6) | 78.3 (20.2) | 0.0039 |
| BMI (kg/m2) | 28.8 (5.7) | 28.6 (6.2) | 29.0 (6.0) | 0.39 |
| Hip circumference (cm) | 103.5 (12.0) | 103.5 (13.0) | 104.0 (13.2) | 0.037 |
| Waist (cm) | 97.0 (16.6) | 97.2 (17.0) | 97.9 (16.5) | 0.22 |
| BSA (m2) | 1.79 (0.28) | 1.81 (0.29) | 1.85 (0.28) | 0.0065 |
| Waist-hip-ratio | 0.94 (0.12) | 0.94 (0.11) | 0.95 (0.11) | 0.049 |
Values are median (interquartile range). Significant P values are in bold.
S/S or A/A: Individuals homozygous for wild-type Ser72 at rs8033595 or of A/A genotype at rs3862434; S/N or A/G: Individuals heterozygous at rs8033595 or at rs3862434; N/N or G/G: Individuals homozygous for variant rs8033595 (Asn72) or of G/G genotype at rs3862434. rs8033595 (S72N) was genotyped in MACAD subjects; the perfect proxy rs3862434 was genotyped in MESA.
This association did not extend to 2,527 non-Hispanic whites from MESA, in whom we observed no association of rs3862434 with any of the six available obesity traits (Supplementary Table 1). Of note, the G allele of rs3862434 (proxy for the adiposity-associated 72N allele) is the minor allele in Mexican Americans and the major allele in non-Hispanic whites. We then conducted a z-score meta-analysis in over 63,000 subjects from the CHARGE and GIANT consortia, examining the association of S72N with BMI. In this meta-analysis, the 72N allele was associated with increased BMI; however, this did not reach statistical significance (z-score 0.74, P=0.45). This suggests that the effect of 72N to promote obesity, while substantial in Mexican Americans, is very weak or nonexistent in non-Hispanic whites. As the 72N allele is more frequent in non-Hispanic whites, our results are consistent with the observation that obesity is less frequent in non-Hispanic whites than Mexican Americans 29. The weaker effect of 72N in non-Hispanic whites may be due to environmental/lifestyle factors and/or differences in genetic background.
High fat diet feeding has been shown to promote obesity and insulin resistance through increases in energy intake that lead to the ectopic deposition of lipid in liver. Our results suggest that CRTC3 contributes to these changes in part by attenuating catecholamine signaling in adipose (sup. fig. 18).
Although the proposed role of CRTC3 as a negative feedback regulator of adipocyte cAMP signaling was unexpected, we note that intracellular signaling pathways often self-attenuate as part of a homeostatic mechanism to limit cellular responses to hormonal stimuli 30. Thus the chronic upregulation of sympathetic nerve activity under high fat diet conditions 19 may attenuate the intracellular cAMP pathway through the CRTC3-mediated induction of RGS2. By limiting catecholamine-dependent increases in lipolysis and fatty acid oxidation, CRTC3 may also function as a so-called “thrifty” gene that enhances survival under starvation conditions. Future studies may reveal the extent to which CRTC3 also contributes to the development of insulin resistance and type II diabetes.
Materials and Methods
Animal Studies
Mice were housed in a temperature-controlled environment under a 12 hour light/dark cycle with free access to water and standard rodent chow diet (Lab diet 5001). For high fat diet studies, 6–8 week old mice were transferred to a 60% high fat diet (Research Diets, D12492). Magnetic Resonance Imaging (MRI) scans for fat and lean mass were performed using an Echo MRI-100 instrument according to the manufacturer’s instructions. All animal procedures were performed with an approved protocol from the Salk institutional Animal Care and Use Committee.
CRTC3−/− mice
The targeting vector was constructed by replacing exon 1 of the CRTC3 gene, which encodes the CREB Binding Domain, with a phosphoglycerol kinase (pgk)-neomycin selection cassette. The vector also contained a pgk-diphtheria toxin-A cassette for negative selection. The targeting vector was linearized and electroporated into R1 embryonic stem cells. G418-resistant clones were screened for homologous recombination by southern blot analysis. CRTC3−/− mice were backcrossed to C57/BL6 for up to 3 generations for metabolic studies.
CRE-luciferase transgenic mice
A cassette of eight tandem full CREB binding sites (CRE) in the context of a minimal CFTR promoter (−126 bp of 5′ flanking sequence from the human CFTR gene) was amplified by PCR from lysed recombinant CRE-luc adenovirus 31. The CRE-luciferase transgene with flanking H19 insulator sequences was microinjected into 129/Sv oocytes for implantation into pseudopregnant female mice.
Transgenic founders were identified by PCR analysis of genomic DNA. Founder lines were back-crossed onto albino C57/BL6 mice (C57BL/6-Tyrc-2J, Jackson) for three generations. To maintain consistent copy number, hemizygous transgenic mice were bred to wild-type mice; the transgene segregated to 50% of offspring in each litter and was stable for at least three generations. In vivo luciferase activity was measured by intra-vital imaging. Mice were anesthetized using isofluorane gas (2% to effect), injected with D-luciferin (150 mg/kg, i.p.) and imaged in an IVIS-100 instrument (Caliper Life Sciences) for 1–5 min.
Genotyping
Genomic DNA was prepared from tail biopsies and genotyped using the following primer sets: CRTC3 WT allele, CCTGAGTTATTGGCGGATGT and CACTCAGGCTGTAGCAAGCA; CRTC3 KO allele, ATGGAAGGATTGGAGCTACG and CACTCAGGCTGTAGCAAGCA; CRE-luciferase transgene, GCTGGGCGTTAATCAGAGAG and TTTTCCGTCATCGTCTTTCC.
Histology
Mouse tissues were fixed in zinc-buffered formalin (Anatech) and paraffin embedded. 5μm sections were used for hematoxylin and eosin staining or immunohistochemistry. For immunohistochemical staining of adipose tissue macrophages, rehydrated antigen retrieved sections were incubated with F4/80 (Serotec) antiserum and visualized by the avidin-biotin-complex method using the chromogen, diamino-benzidine (Vector Labs).
Immunoblot and Chromatin Immunoprecipitation
Antibodies for CRTC3, phospho-HSL, and phospho-PKA substrate were obtained from Cell Signaling. Immunoblots were performed as previously described 32. Chromatin immunoprecipitation studies were performed as previously described 11.
Cell counting
Images of hematoxylin and eosin stained sections (5μm) were taken at 200× magnification (1300×1030 pixels/picture). The NIH image J program was used to perform cell counts on brown adipose tissue sections.
GTT, ITT
For glucose tolerance testing, mice were fasted for 16hrs and then injected with glucose (2g/kg; i.p.). For insulin tolerance testing, mice were fasted 2–3 hours and injected with insulin (Humulin; 1U/kg, i.p.).
Metabolites
Mouse blood was collected from the tail vein and glucose levels were measured with a One Touch Ultra Glucometer (Johnson & Johnson). Circulating insulin (Cayman), leptin (Milipore), epinephrine/norepinephrine (LDN GmbH&Co. KG) and free fatty acids (WAKO Chemicals) levels were determined by ELISA assay.
Core body Temperature
Core body temperature was measured with a Thermistor Thermometer (Model 8402-10, Cole Parmer Thermometers).
cAMP
Tissue and cellular cAMP concentrations were determined using a cAMP ELISA kit according to the manufacturer’s instruction (R&D Systems or Cayman Chemical Company). Cells were exposed to Forskolin (10μM) for times indicated.
Indirect Calorimetry
Mice were individually housed for at least 3 days prior to calorimetry experiments. Food intake, locomotor activity, oxygen consumption and carbon dioxide production were simultaneously measured for individually-housed mice with a LabMaster system (TSE Systems). Data were collected for 2–3 days and analyzed. For leptin studies, mice were treated with saline or leptin (3μg/g, i.p.) 90 min prior to the onset of the dark cycle.
Metabolic Studies
Rates of fatty acid oxidation and lipolysis were measured as previously described 24,33. Basal rates of glucose uptake were determined by measuring the rate of [U-14C] 2-deoxyglucose uptake by isolated soleus muscle. Pairs of soleus muscle were rapidly dissected from WT or CRTC3 −/− mice. Isolated soleus strips were incubated twice for 15 min at 37°C in continuously gassed (95% O2, 5% CO2) Krebs-Henseleit bicarbonate buffer containing 5.5mM glucose and 0.5mM oleate complexed to 2% fatty-acid free bovine serum albumin (Millipore). After the 30 min pre-incubation period, muscle strips were transferred into fresh identical buffer with [U-14C] 2-deoxyglucose (2μCi/mL). Soleus strips were continuously gassed and incubated at 37°C for 30min. At the end of the experiment, individual soleus strips were washed three times with ice-cold Hank’s balanced salt solution. Muscle strips were weighed and incubated for 1h at 55°C in digestion buffer (50mM Tris, pH 8; 100mM NaCl, 100mM EDTA, 1% SDS, and 500 μg/mL proteinase K). All of the digested tissue was transferred into scintillant (EcoLite) and the radioactivity was measured by liquid scintillation counting.
Mouse Embryonic Fibroblasts (MEFs)
Mouse embryos were obtained from gravid female mice at embryonic days E13-14. Embryos were minced, trypsinized, and washed with PBS. MEFs were plated in DMEM with 10% FBS, and 1% penicillin-streptomycin.
Primary Adipocyte and Stromal Vascular Fraction (SVF) Cultures
Primary adipocytes and the stromal vascular fraction were isolated from epididymal white adipose and brown adipose, as described previously 24,33. Primary adipocytes fractions were plated in DMEM containing 5.5mM Glucose, 2% fatty acid free BSA and 1% penicillin-streptomycin. For SVF cells, pellets from adipocyte isolations were washed 3 times with HDB, and cultured in DMEM with 10% fetal bovine serum and 1% penicillin-streptomycin.
RNA studies
Total RNA was isolated by Trizol (Invitrogen) and RNeasy mini kit (Qiagen). 1–2ug of total RNA was used for cDNA synthesis with Superscript II according to the manufacturer’s instruction (Invitrogen). Relative mRNA amount was determined by real time qPCR on LightCycler 480 instrument (Roche).
Statistics
Data are presented as means ± s.e.m. Statistical analysis was performed using an unpaired t test with Graph Pad Prism software. Statistical significance is indicated as * for P<0.05, ** for P<0.01, and *** for P<0.001. All transient luciferase assays were performed on at least three independent occasions.
Human Subjects
Associations with adiposity parameters (weight, body mass index (BMI), waist circumference, hip circumference, waist-hip ratio) were first assessed in participants in the Cedars-Sinai/UCLA Mexican-American Coronary Artery Disease (MACAD) study, a study of Mexican Americans families from Los Angeles. 34,35 In the present report, 206 two-generation Mexican-American families were included, comprising 779 subjects (adult offspring of probands with CAD and the spouses of those offspring) who underwent anthropometric measurements and genotyping. By design, the offspring were free of diabetes and clinically manifest cardiovascular disease, thus avoiding secondary changes in phenotype caused by overt disease. All studies were approved by Human Subjects Protection Institutional Review Boards at UCLA and Cedars-Sinai Medical Center.
Confirmatory studies were undertaken in Mexican-American subjects from the Multi-Ethnic Study of Atherosclerosis (MESA). A detailed description of the MESA study design and methods has been published previously 36. Briefly, 6,814 participants 45 to 84 years of age who identified themselves as white (2,748), black (1,930), Hispanic/Latino (1,496), or Chinese (806) were recruited from six US communities between 2000 and 2002. To obtain a replication cohort most similar to that of MACAD, we studied MESA Hispanics with exclusion of those recruited from the New York site, as the latter are mainly from the Caribbean and may thus have genetic differences from the Mexican-Americans of MACAD 37. This resulted in a cohort of 987 MESA Mexican Americans.
To determine whether the genetic associations observed in Mexican Americans would also be seen in other ethnic groups, we also examined 2,527 non-Hispanic white subjects from MESA who had available anthropometric data. We then also accessed data from two large consortia of non-Hispanic whites, the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium (n=31,373) 38 and the GIANT (Genetic Investigation of ANthropometric Traits) Consortium (n=32,504) 39, both of which had conducted genome-wide association studies of BMI. These datasets did not overlap in subjects.
Genotyping of Human Samples
Genotyping of SNP rs8033595 (S72N) in MACAD was performed using TaqMan MGB technology as previously described 35,40. The genotyping success rate was 98.3%. In the MESA Mexican-American and white cohorts, S72N was represented by a proxy SNP rs3862434 (A/G, r2=1 with S72N in the Mexican-American cohort of the phase III HapMap data, and r2=1 in the Caucasian European cohort of the phase II HapMap) that was directly genotyped (not imputed) in the genome wide association study (GWAS) conducted in MESA.. In CHARGE and GIANT, rs8033595 was either directly genotyped or imputed, depending on the GWAS arrays used in the individual cohorts comprising each consortium.
Human Genetic Association Analysis
The MACAD cohort is composed of small families and marrying-in spouses, therefore the generalized estimating equation (GEE141) approach was selected to utilize data from all phenotyped subjects. We evaluated association using this robust variance estimation approach to test hypothesized associations between phenotypes and genotypes while accounting for familial correlations present in the data. The PROC GENMOD procedure in SAS (version 9.0, SAS Institute, Cary NC) was used for the association analysis in which a sandwich estimator was used assuming exchangeable correlation. Family was taken as the cluster factor, i.e., members from the same family were assumed to be correlated. The adiposity traits were log transformed to better approximate conditional normality and homogeneity of variance. An additive genetic model was assumed in all the association analyses. Analyses were conducted using age and sex as covariates, unless otherwise specified. The same analytic techniques were used to assess association of rs3862434 with adiposity traits within MESA Mexican Americans and non-Hispanic whites. We also conducted an analysis combining the MACAD and MESA Mexican-Americans, in which 72N and the minor allele of rs3862434 were considered the same. In CHARGE and GIANT, we conducted weighted z-score-based meta-analysis of association of rs8033595 with BMI, combining P values obtained from similar z-score-based meta-analysis conducted in each consortium. The meta-analysis was carried out using the program METAL (http://www.sph.umich.edu/csg/abecasis/metal/).
Supplementary Material
Acknowledgments
This study was supported in part by NIH grants R01-DK049777, R01-DK083834, P30-DK063491, R01-HL088457, R01-DK79888, R01-HL071205, N01-HC95159, N02-HL64278and M01-RR00425 (General Clinical Research Center Grant from the NCRR), The Keickhefer Foundation, The Clayton Foundation for Medical Research, The Helmsley Foundation, the Cedars-Sinai Winnick Clinical Scholars Award (to M.O.G) and the Cedars-Sinai Board of Governor’s Chair in Medical Genetics (J.I.R.). We thank Bruce Beutler and Owen Siggs (The Scripps Research Institute) for help with macrophage studies.
References
- 1.Bartness TJ, Song CK. Thematic review series: adipocyte biology. Sympathetic and sensory innervation of white adipose tissue. J Lipid Res. 2007;48:1655–72. doi: 10.1194/jlr.R700006-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Lafontan M, Langin D. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res. 2009;48:275–97. doi: 10.1016/j.plipres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Bachman ES, et al. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–5. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 4.van Baak MA. The peripheral sympathetic nervous system in human obesity. Obes Rev. 2001;2:3–14. doi: 10.1046/j.1467-789x.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 5.Denzer C, et al. Gender-specific prevalences of fatty liver in obese children and adolescents: roles of body fat distribution, sex steroids, and insulin resistance. J Clin Endocrinol Metab. 2009;94:3872–81. doi: 10.1210/jc.2009-1125. [DOI] [PubMed] [Google Scholar]
- 6.Vega GL, Chandalia M, Szczepaniak LS, Grundy SM. Metabolic correlates of nonalcoholic fatty liver in women and men. Hepatology. 2007;46:716–22. doi: 10.1002/hep.21727. [DOI] [PubMed] [Google Scholar]
- 7.Roy AA, et al. RGS2 interacts with Gs and adenylyl cyclase in living cells. Cell Signal. 2006;18:336–48. doi: 10.1016/j.cellsig.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Salim S, Sinnarajah S, Kehrl JH, Dessauer CW. Identification of RGS2 and type V adenylyl cyclase interaction sites. J Biol Chem. 2003;278:15842–9. doi: 10.1074/jbc.M210663200. [DOI] [PubMed] [Google Scholar]
- 9.Sinnarajah S, et al. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–5. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- 10.Freson K, et al. −391 C to G substitution in the regulator of G-protein signalling-2 promoter increases susceptibility to the metabolic syndrome in white European men: consistency between molecular and epidemiological studies. J Hypertens. 2007;25:117–25. doi: 10.1097/HJH.0b013e3280109c6c. [DOI] [PubMed] [Google Scholar]
- 11.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–73. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh Y, et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 2006;273:2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 14.Fu A, Screaton RA. Using kinomics to delineate signaling pathways: control of CRTC2/TORC2 by the AMPK family. Cell Cycle. 2008;7:3823–8. doi: 10.4161/cc.7.24.7241. [DOI] [PubMed] [Google Scholar]
- 15.Ahn S, et al. A dominant negative inhibitor of CREB reveals that it is a general mediator stimulus-dependent transcription of c-fos. Molec Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iourgenko V, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci U S A. 2003;100:12147–52. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–23. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56:16–23. doi: 10.2337/db06-1076. [DOI] [PubMed] [Google Scholar]
- 19.Landsberg L. Feast or famine: the sympathetic nervous system response to nutrient intake. Cell Mol Neurobiol. 2006;26:497–508. doi: 10.1007/s10571-006-9010-7. [DOI] [PubMed] [Google Scholar]
- 20.Carmen GY, Victor SM. Signalling mechanisms regulating lipolysis. Cell Signal. 2006;18:401–8. doi: 10.1016/j.cellsig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 22.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function--of mice and men. Genes Dev. 2009;23:788–97. doi: 10.1101/gad.1779209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cederberg A, et al. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–73. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 24.Qi L, et al. Adipocyte CREB promotes Insulin Resistance in Obesity. Cell Metab. 2009 doi: 10.1016/j.cmet.2009.01.006. In the Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci U S A. 2006;103:14379–84. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohlfs EM, Daniel KW, Premont RT, Kozak LP, Collins S. Regulation of the uncoupling protein gene (Ucp) by beta 1, beta 2, and beta 3-adrenergic receptor subtypes in immortalized brown adipose cell lines. J Biol Chem. 1995;270:10723–32. doi: 10.1074/jbc.270.18.10723. [DOI] [PubMed] [Google Scholar]
- 27.Granneman JG. Expression of adenylyl cyclase subtypes in brown adipose tissue: neural regulation of type III. Endocrinology. 1995;136:2007–12. doi: 10.1210/endo.136.5.7720648. [DOI] [PubMed] [Google Scholar]
- 28.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–5. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 29.Ford ES, Zhao G, Li C, Pearson WS, Mokdad AH. Trends in obesity and abdominal obesity among hypertensive and nonhypertensive adults in the United States. Am J Hypertens. 2008;21:1124–8. doi: 10.1038/ajh.2008.246. [DOI] [PubMed] [Google Scholar]
- 30.Hagiwara M, et al. Transcriptional Attenuation Following cAMP Induction Requires PP-1-Mediated Dephosphorylation of CREB. Cell. 1992;70:105–113. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- 31.Koo SH, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–11. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 32.Altarejos JY, et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat Med. 2008;14:1112–7. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi L, et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–6. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- 34.Goodarzi MO, et al. Lipoprotein lipase is a gene for insulin resistance in Mexican Americans. Diabetes. 2004;53:214–20. doi: 10.2337/diabetes.53.1.214. [DOI] [PubMed] [Google Scholar]
- 35.Goodarzi MO, et al. Determination and use of haplotypes: ethnic comparison and association of the lipoprotein lipase gene and coronary artery disease in Mexican-Americans. Genet Med. 2003;5:322–7. doi: 10.1097/01.GIM.0000076971.55421.AD. [DOI] [PubMed] [Google Scholar]
- 36.Bild DE, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–81. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 37.Choudhry S, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118:652–64. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 38.Heard-Costa NL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–9. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 41.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–30. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.