Abstract
Background and Aims
The main assemblage of the grass subfamily Chloridoideae is the largest known clade of C4 plant species, with the notable exception of Eragrostis walteri Pilg., whose leaf anatomy has been described as typical of C3 plants. Eragrostis walteri is therefore classically hypothesized to represent an exceptional example of evolutionary reversion from C4 to C3 photosynthesis. Here this hypothesis is tested by verifying the photosynthetic type of E. walteri and its classification.
Methods
Carbon isotope analyses were used to determine the photosynthetic pathway of several E. walteri accessions, and phylogenetic analyses of plastid rbcL and ndhF and nuclear internal transcribed spacer DNA sequences were used to establish the phylogenetic position of the species.
Results
Carbon isotope analyses confirmed that E. walteri is a C3 plant. However, phylogenetic analyses demonstrate that this species has been misclassified, showing that E. walteri is positioned outside Chloridoideae in Arundinoideae, a subfamily comprised entirely of C3 species.
Conclusions
The long-standing hypothesis of C4 to C3 reversion in E. walteri is rejected, and the classification of this species needs to be re-evaluated.
Keywords: C4 photosynthesis, evolution, reversion, Eragrostis, Chloridoideae, Arundinoideae, Poaceae, Africa, Namibia
INTRODUCTION
Complex traits have received a great deal of attention by evolutionary biologists, but key questions remain regarding the directionality of transitions between their states and, in particular, about their reversibility (e.g. Collin and Miglietta, 2008; Tripp and Manos, 2008; Lynch and Wagner, 2009). C4 photosynthesis is a prime example of a complex trait due to the numerous morphological, anatomical and biochemical adaptations relative to ancestral C3 photosynthesis that are required for proper function. These adaptations are thought to involve hundreds of genetic changes (Bräutigam et al., 2010), but nonetheless have been demonstrated to be evolutionarily labile. C4 photosynthesis has evolved numerous times independently in distantly related plant families during the past 30 million years, with >50 independent origins inferred in the angiosperms (Giussani et al., 2001; Kadereit et al., 2003; Sage, 2004; Muhaidat et al., 2007; Christin et al., 2008; Besnard et al., 2009). The majority of C4 plant species belong to the grass family, in which C4 taxa form a minimum of 17 different lineages separated in the phylogeny by C3 taxa within the PACMAD clade (Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae and Danthonioideae; Sage, 2004; Christin et al., 2008). Comparisons of the phenotype and genotype of the C4 traits used by these different C4 phylogenetic groups in grasses suggest that most derive from independent C4 origins (Christin et al., 2010), ranking C4 photosynthesis amongst the most convergent of complex traits (Conway-Morris, 2006). Surprisingly, very few putative losses of the C4 pathway and recovery of the ancestral C3 trait have been identified. Two exceptions are Alloteropsis semialata (R.Br.) Hitchc. subsp. eckloniana (Nees) Gibbs Russ., a C3 subspecies nested in a C4 clade of Panicoideae (Ibrahim et al., 2009), and Eragrostis walteri Pilg.
Eragrostis walteri is a grass endemic to Namibia that was described in 1941 (Pilger, 1941) and that possesses many morphological features typical of Eragrostis species, including multifloreted spikelets, paniculate inflorescences and ciliate ligules. This placement within Eragrostis was confirmed by phylogenetic analyses of morphological data (van den Borre and Watson, 1994). Eragrostis contains about 400 species distributed worldwide and is placed in the main assemblage of Chloridoideae, which contains >1400 species. Numerous Chloridoideae species have been studied for leaf anatomy and/or carbon isotope composition, and all have been found to be C4, making this group the largest wholly C4 clade in plants. This fact made the report of non-C4 leaf anatomy in E. walteri by Ellis (1984) particularly striking. The photosynthetic pathway employed by Ellis' E. walteri specimens was subsequently confirmed by δ13C analysis (Schulze et al., 1996). The presence of a C3 plant in an otherwise C4 clade was strongly suggestive of an evolutionary loss of C4 photosynthesis. Consequently, E. walteri has been repeatedly cited as the best candidate for C4 to C3 reversion for the last 25 years (e.g. Renvoize, 1987; Morrone and Zuloaga, 1991; Kellogg, 1999; Kubien et al., 2008; Ibrahim et al., 2009; Edwards and Smith, 2010; Roalson, 2011).
Given that the leaf anatomy of E. walteri shows no evidence of partially C4 characters, confirmation of this putative C4 to C3 reversion in Eragrostis would demonstrate that C4 evolution is reversible. This case is particularly remarkable because the reversion would probably have occurred tens of millions of years after the initial C4 origin in Chloridoideae, which is estimated to have occurred between 25 and 32 million years ago (Christin et al., 2008). Furthermore, the Chloridoideae encompasses numerous species with well-optimized C4 characters that confer ecological success in many of the world's biomes. If the hypothesis of reversion from C4 to C3 photosynthesis was proven, E. walteri would represent an outstanding system in which to investigate the genetic mechanisms and ecological pressures involved (Christin et al., 2010). However, it is necessary first to confirm that E. walteri does in fact use C3 photosynthesis with multiple independent collections of the species. Additionally, the assumption that E. walteri is nested in a C4 clade relies solely on morphological evidence that has not been confirmed with genetic markers. This is crucial, since polyphyly has been demonstrated for several grass genera upon phylogenetic analysis of genetic data (e.g. Aliscioni et al., 2003; Peterson et al., 2010).
In this study, we investigated the likelihood of an evolutionary reversion from C4 to C3 photosynthesis in E. walteri to gain insights into the reversibility of C4 evolution. To test the hypothesis of reversion, our work aimed to: (a) verify the photosynthetic type of several E. walteri specimens using unambiguous methods; and (b) determine the phylogenetic position of E. walteri using genetic markers from the plastid and nuclear genomes.
MATERIALS AND METHODS
Carbon isotope ratio
The C4 pathway is defined by the fixation of atmospheric CO2 through a coupling of carbonic anhydrase and phosphoenolpyruvate carboxylase, whereas in C3 plants this fixation is performed by ribulose-1,5-bisphosphate carboxylase. These enzymes differentially discriminate between the carbon isotopes naturally present in the atmosphere, resulting in different ratios of carbon isotopes in the plants that can be determined by mass spectrometry. Values of δ13C between –21 ‰ and –32 ‰ are indicative of C3 photosynthesis, while C4 plants have δ13C between –9 ‰ and –16 ‰. Some well-developed C3–C4 intermediates can have δ13C values between –16 ‰ and –19 ‰, but there is no overlap between the δ13C values of wholly C3 and C4 species (von Caemmerer, 1992).
Foliar δ13C values were determined on six herbarium samples whose identification as Eragrostis walteri had been verified by T. A. Cope and M. Vorontsova by reference to the specimen collection held at RBG Kew. A 20 mg sub-sample from each was analysed using an ANCA GSL preparation module coupled to a 20–20 stable isotope analyser (PDZ Europa, Cheshire, UK). Measurements on the same sample had a reproducibility of 0·5 ‰, and the isotopic composition of each (δ13C) was calculated as the sample 13C/12C ratio relative to the PDB standard (‰).
Molecular phylogenies
Two herbarium specimens of E. walteri (K:Kolberg & Tholkes 695 and PRE:Hines 262) were selected for genetic analyses. The Kolberg & Tholkes 695 sample was analysed in Lausanne, while the Hines 262 sample was analysed at Wabash College. No laboratory products or PCR primers were shared between these laboratories, excluding the possibility of contamination. At Lausanne, two plastid markers (rbcL and ndhF) were amplified and sequenced using a previously published methodology (Christin et al., 2008). At Wabash College, rbcL was amplified and sequenced with new primers designed specifically for grasses: 7F (5′-GGGACTTATGTCACCACAAAC-3′) and 1433R (5′-ACTTAATCGATGGTATCTACCG-3′). Amplifications were carried out as described in Ingram and Doyle (2003) with an annealing temperature of 55 °C. DNA sequencing was completed by the Cornell BioResource Center. All plastid sequences were added to a previously published data set with representatives of all Poaceae subfamilies (Christin et al., 2008). Phylogenetic methods are as previously described (Christin et al., 2008). An independent estimate of phylogeny was obtained with sequences from the nuclear ribosomal DNA internal transcribed spacer (ITS). New grass-specific ITS primers were used for amplification and sequencing [ITS 18S-F-grass (5′-ATTGAATGGTCCGGTGAAG-3′) and ITS 26S-R-grass (5′-GACGCCTCTCCAGACTACAA-3′)]. PCR was as described in Ingram and Doyle (2003) with annealing temperatures of 56 °C. ITS PCR products were sequenced directly. An ITS data set was assembled from sequences deposited in GenBank, which contains several thousand ITS sequences for grasses. The selected sequences included numerous representatives of all PACMAD families, with Pooideae outgroups. Details on the species and GenBank accession numbers can be found in the Supplementary Data (available online). The sequences were aligned with ClustalX (Larkin et al., 2007), and a phylogenetic tree was inferred as described for the plastid markers (Christin et al., 2008). All E. walteri sequences were deposited in GenBank (accession numbers HQ329788–HQ329791).
RESULTS AND DISCUSSION
The δ13C values (Table 1) ranged between –24·3 ‰ and –29·1 ‰ for the six E. walteri accessions, which unambiguously indicates that these plants assimilated carbon via C3 photosynthesis. This confirms previous conclusions (Ellis, 1984; Schulze et al., 1996) with independent samples of E. walteri.
Table 1.
Stable carbon isotope ratio (δ13C) for leaf material of Eragrostis walteri (Pilg.)
| Collector and collection number | δ13C |
|---|---|
| Giess, W. 8977 | –27·3 |
| Giess, W. 8104A | –25·8 |
| Kolberg, H. and Tholkes, T. 695 | –28·2 |
| Giess, W. 10413 | –28·1 |
| Giess, W. and Müller, M. 14316 | –29·1 |
| Range, P. 14831 | –24·3 |
All specimens were collected in southern Africa and were identified and archived in the herbarium of the Royal Botanic Gardens, Kew.
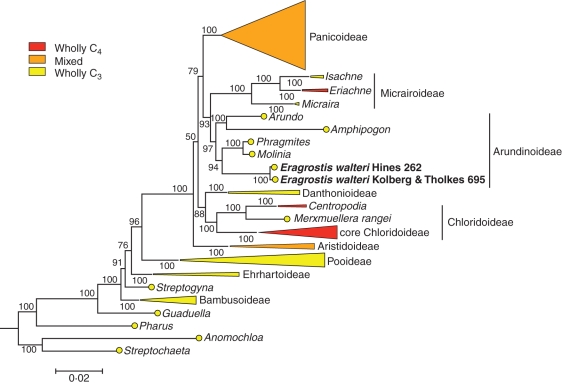
The rbcL sequences for E. walteri obtained independently in the two different laboratories were identical. In the phylogeny, however, they did not group with Chloridoideae as expected from morphology (van den Borre and Watson, 1994). Instead, E. walteri was placed within the Arundinoideae, sister to a clade composed of Molinia and Phragmites (Fig. 1). This position within Arundinoideae was strongly supported (Bayesian support values >0·95). Blasting the E. walteri ITS sequence showed the highest similarity with Molinia caerulea (94 %) followed by Phragmites spp. (90–91 %). The Bayesian inference confirmed this close relationship between E. walteri ITS and those of Molinia and Phragmites (Fig. 2), congruent with the results from the plastid markers. This relationship was also highly supported (Bayesian support value of 1·0). Therefore, both nuclear and plastid markers show that E. walteri does not belong to Eragrostis, nor to Chloridoideae, but is unambiguously a member of Arundinoideae. The positioning of E. walteri outside Eragrostis is not entirely surprising when morphological features are more carefully examined. Eragrostis is a highly heterogeneous group, but E. walteri is an outlier in some otherwise invariable traits. For example, Eragrostis lemmas are consistently three-nerved, but Pilger (1941) noted in his original description of E. walteri that this species has three prominent and two inconspicuous nerves on the lemmas. In addition, E. walteri lemma apices have been described as ‘nearly awned’ (Watson and Dallwitz, 1992 onwards), in contrast to the acute lemma apices found in most other species in the genus. The discrepancy between morphological classification and molecular phylogenies mirrors the numerous cases of polyphyletic genera in grasses (e.g. Giussani et al., 2001; Aliscioni et al., 2003; Peterson et al., 2010).
Fig. 1.
Phylogenetic position of E. walteri inferred from plastid markers. This tree was obtained through Bayesian inference based on ndhF and rbcL sequences. Bayesian support values are indicated near nodes. The main groups are compressed. Clades containing only C3 taxa are in yellow, those containing only C4 taxa are in red, and those containing both C3 and C4 taxa are in orange. For further details on the data set see Christin et al. (2008).
Fig. 2.
Phylogenetic position of E. walteri inferred from nuclear markers. This tree was obtained through Bayesian inference based on nuclear ribosomal DNA internal transcribed spacer sequence. Bayesian support values are indicated near nodes. The main groups are compressed, and their delimitation follows Christin et al. (2008) and Fig. 1. Clades are coloured as in Fig. 1.
Because molecular data indicate that E. walteri belongs to Arundinoideae, its C3 type is no longer surprising, as all other species of Arundinoideae are also C3. Therefore, the hypothesis that this species is a C4 to C3 revertant should be abandoned. With our current understanding of grass phylogenetics and photosynthetic pathways, Alloteropsis semialata subsp. eckloniana is the only plausible C4 revertant in grasses and should now be more closely investigated to detect traces of C4 loss (Christin et al., 2010). However, this new discovery regarding the misclassification of E. walteri clearly demonstrates the dominance of C3 to C4 transitions over reversions, suggesting that C4 evolution is almost always a one-way event. The resolution of the enigma created by the peculiar foliar anatomy of E. walteri also highlights the importance of working with species or even accessions as evolutionary units, and the risks of extrapolating phylogenetic positions from congenerics. While grass phylogeny is far from being resolved at the species level, efforts should be put into incorporating as many of the evolutionarily interesting taxa as possible, until an exhaustive phylogeny is obtained. This could reveal other surprises, including the non-monophyly of numerous morphological taxonomic units, even at the subfamily level, as for E. walteri.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank David Simpson, Rosa Cerros and other herbarium staff at Royal Botanic Gardens, Kew (K) and the South African National Biodiversity Institute (PRE) for assistance in acquiring E. walteri material. We also thank J. Travis Columbus for designing the rbcL and ITS primers. This work was supported by the US National Science Foundation [grant number DEB-0921203 to A.L.I.]; the Swiss National Science Foundation [grant number PBLAP3-129423 to P.A.C.]; and a Royal Society University Research Fellowship to C.P.O.
LITERATURE CITED
- Aliscioni SS, Giussani LM, Zuloaga FO, Kellogg EA. A molecular phylogeny of Panicum (Poaceae: Paniceae): tests of monophyly and phylogenetic placement within the Panicoideae. American Journal of Botany. 2003;90:796–821. doi: 10.3732/ajb.90.5.796. [DOI] [PubMed] [Google Scholar]
- Besnard G, Muasya AM, Russier F, Roalson EH, Salamin N, Christin PA. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Molecular Biology and Evolution. 2009;26:1909–1919. doi: 10.1093/molbev/msp103. [DOI] [PubMed] [Google Scholar]
- Brautigam A, Kajala K, Wullenweber J, et al. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology. 2010 doi: 10.1104/pp.110.159442. in press. doi:10.1104/pp.110.159442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Besnard G, Samaritani E, et al. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Current Biology. 2008;18:37–43. doi: 10.1016/j.cub.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Christin PA, Freckleton RP, Osborne CP. Can phylogenetics identify C4 origins and reversals? Trends in Ecology and Evolution. 2010;25:403–409. doi: 10.1016/j.tree.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Collin R, Miglietta MP. Reversing opinions on Dollo's Law. Trends in Ecology and Evolution. 2008;23:602–609. doi: 10.1016/j.tree.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Conway-Morris S. Evolutionary convergence. Current Biology. 2006;16:R826–R827. doi: 10.1016/j.cub.2006.08.077. [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Smith SA. Phylogenetic analyses reveal the shady history of C4 grasses. Proceedings of the National Academy of Sciences, USA. 2010;107:2532–2537. doi: 10.1073/pnas.0909672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RP. Eragrostis walteri – a 1st record of non-Kranz anatomy in the subfamily Chloridoideae (Poaceae) South African Journal of Botany. 1984;3:380–386. [Google Scholar]
- Giussani LM, Cota-Sanchez JH, Zuloaga FO, Kellogg EA. A molecular phylogeny of the grass subfamily Panicoideae (Poaceae) shows multiple origins of C4 photosynthesis. American Journal of Botany. 2001;88:1993–2012. [PubMed] [Google Scholar]
- Ibrahim DG, Burke T, Ripley BS, Osborne CP. A molecular phylogeny of the genus Alloteropsis (Panicoideae, Poaceae) suggests an evolutionary reversion from C4 to C3 photosynthesis. Annals of Botany. 2009;103:127–136. doi: 10.1093/aob/mcn204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram AL, Doyle JJ. The origin and evolution of Eragrostis tef (Poaceae) and related polyploids: evidence from nuclear waxy and plastid rps16. American Journal of Botany. 2003;90:116–122. doi: 10.3732/ajb.90.1.116. [DOI] [PubMed] [Google Scholar]
- Kadereit G, Borsch T, Weising K, Freitag H. Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. International Journal of Plant Science. 2003;164:959–986. [Google Scholar]
- Kellogg EA. Phylogenetic aspects of the evolution of C4 photosynthesis. In: Sage RF, Monson RK, editors. C4 plant biology. San Diego: Academic Press; 1999. pp. 411–444. [Google Scholar]
- Kubien DS, Whitney SM, Moore PV, Jesson LK. The biochemistry of Rubisco in Flaveria. Journal of Experimental Botany. 2008;59:1767–1777. doi: 10.1093/jxb/erm283. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2·0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. Did egg-laying boas break Dollo's law? Phylogenetic evidence for reversal to oviparity in sand boas (Eryx: Boidae) Evolution. 2010;64:207–216. doi: 10.1111/j.1558-5646.2009.00790.x. [DOI] [PubMed] [Google Scholar]
- Morrone O, Zuloaga F. Revision of the genus Streptostachys (Poaceae, Panicoideae), its systematic position in the tribe Paniceae. Annals of the Missouri Botanical Garden. 1991;78:359–376. [Google Scholar]
- Muhaidat R, Sage RF, Dengler NF. Diversity of Kranz anatomy and biochemistry in C4 eudicots. American Journal of Botany. 2007;94:362–381. doi: 10.3732/ajb.94.3.362. [DOI] [PubMed] [Google Scholar]
- Peterson PM, Romaschenko K, Johnson G. A classification of the Chloridoideae (Poaceae) based on multi-gene phylogenetic trees. Molecular Phylogenetics and Evolution. 2010;55:580–598. doi: 10.1016/j.ympev.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Pilger RKF. Eragrostis walteri. Notizblatt des Botanischen Gartens und Muzeums zu Berlin-Dahlem. 1941;15(452) [Google Scholar]
- Renvoize SA. A survey of leaf blade anatomy in grasses XI. Paniceae. Kew Bulletin. 1987;42:739–768. [Google Scholar]
- Roalson EH. C4 photosynthesis origins in the monocots: a review and reanalysis. In C4 photosynthesis and related CO2 concentrating mechanisms. In: Raghavendra AS, Sage RF, editors. C4 photosynthesis and related CO2 concentrating mechanisms. Berlin: Springer; 2011. (in press) [Google Scholar]
- Sage RF. The evolution of C4 photosynthesis. New Phytologist. 2004;161:341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- Schulze ED, Ellis R, Schulze W, Trimborn P, Ziegler H. Diversity, metabolic types and δ13C carbon isotope ratios in the grass flora of Namibia in relation to growth form, precipitation and habitat conditions. Oecologia. 1996;106:352–369. doi: 10.1007/BF00334563. [DOI] [PubMed] [Google Scholar]
- Tripp EA, Manos PS. Is floral specialization an evolutionary dead-end? Pollination system transitions in Ruellia (Acanthaceae) Evolution. 2008;62:1712–1737. doi: 10.1111/j.1558-5646.2008.00398.x. [DOI] [PubMed] [Google Scholar]
- Van den Borre A, Watson L. The infrageneric classification of Eragrostis (Poaceae) Taxon. 1994;43:383–422. [Google Scholar]
- Von Caemmerer S. Stable carbon isotope discrimination in C3–C4 intermediates. Plant, Cell and Environment. 1992;15:1063–1072. [Google Scholar]
- Watson L, Dallwitz MJ. The grass genera of the world: descriptions, illustrations, identification, and information retrieval; including synonyms, morphology, anatomy, physiology, phytochemistry, cytology, classification, pathogens, world and local distribution, and references. 1992 onwards. http://delta-intkey.com/grass/ , accessed 17 August 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.