Abstract
Background and Aims
Improving phosphorus (P) nutrient efficiency in Lolium perenne (perennial ryegrass) is likely to result in considerable economic and ecological benefits. To date, research into the molecular and biochemical response of perennial ryegrass to P deficiency has been limited, particularly in relation to the early response mechanisms. This study aimed to identify molecular mechanisms activated in response to the initial stages of P deficiency.
Methods
A barley microarray was successfully used to study gene expression in perennial ryegrass and this was complemented with gas chromatography-mass spectrometry metabolic profiling to obtain an overview of the plant response to early stages of P deficiency.
Key Results
After 24 h of P deficiency, internal phosphate concentrations were reduced and significant alterations were detected in the metabolome and transcriptome of two perennial ryegrass genotypes. Results indicated a replacement of phospholipids with sulfolipids and the utilization of glycolytic bypasses in response to P deficiency in perennial ryegrass.
Conclusions
The transcriptome and metabolome of perennial ryegrass undergo changes in response to reductions in P supply after 24 h.
Keywords: Lolium perenne, perennial ryegrass, phosphorus deficiency, metabolic profiling, transcript profiling, cross species hybridization
INTRODUCTION
Depleting phosphorus (P) reserves has been described as one of the ‘most significant sustainability issues of our time’ (Vaccari, 2009). Continually increasing demand will deplete global supplies by the end of the century (Vaccari, 2009), leading to price volatility that threatens the economic sustainability of agriculture. Overuse of fertilizers has increasingly been shown to exert a detrimental impact on the environment largely through run-off (Ferrier and Edwards, 2002) and associated environmental degradation (Tilman, 1998; Gyaneshwar et al., 2002). However, predictions suggest that worldwide fertilizer usage is projected to increase as the population increases (by 46 % to 8·9 billion people by 2050; Anon, 2004) and exerts an associated requirement for food. In the past, significant attention has been focused on the negative impacts of nitrogen but more recently there has been an increased focus on the environmental impacts of P. It was revealed in one Canadian study that decreasing inputs of P was more critical in reducing eutrophication than controlling nitrogen inputs (Schindler et al., 2008). Furthermore, there is also evidence that excess P is likely to result in a greater loss of plant species than that associated with enhanced nitrogen, therefore having a greater impact on biodiversity (Wassen et al., 2005).
Perennial ryegrass, Lolium perenne, is the principal forage used in temperate regions (Humphreys, 2005). Improving P nutrient efficiency in this species is likely to result in considerable economic and ecological benefits. To date, no studies have investigated the (bio)molecular response of perennial ryegrass to P deficiency. Publicly available genomic resources, including available microarrays, for the study of gene expression are limiting in perennial ryegrass (Ciannamea et al., 2006). To overcome this limitation, microarrays developed for related species can be employed to study gene expression in the target organism (Hammond et al., 2005; Bar-Or et al., 2007). In other species, P deficiency was reported to result in a variety of responses both at the gene expression (Hammond et al., 2003, 2005; Misson et al., 2005; Wasaki et al., 2006; Guo et al., 2008) and metabolite (Hernandez et al., 2007; Huang et al., 2008) levels.
Improving a plant's ability to acquire and efficiently utilize P is one approach to both alleviating the scarcity of the resources and reducing environmental impacts. Efforts towards this have been accelerated by legislative measures to reduce fertilizer usage and pollution (Anon, 2000). The adaptation of plants to limited P can be optimized by allocating a greater proportion of their biomass to the root system (Hermans et al., 2006), remobilization of internal P and modifications in the carbon metabolism (Raghothama, 1999; Ramirez-Rodriguez et al., 2005). Plants generally adjust their metabolism to optimize internal P utilization and improve P acquisition (Theodorou and Plaxton, 1993). In barley, carbohydrate metabolism is initially readjusted to reduce P consumption, and when stress becomes more severe, P is salvaged from small P-containing metabolites such as glucose 6-P, fructose 6-P, inositol 1-P and glycerol 3-P (Huang et al., 2008).
P uptake can be enhanced by secretion of phosphatases and organic acids, modified root growth, and enhanced expression of P transporters (Ramirez-Rodriguez et al., 2005). Organic P generally constitutes the majority of P in soils; however, it is unavailable for plant uptake (Wang et al., 2004; Richardson et al., 2009) but can be converted to a more available form with the excretion of phosphatases (Plaxton, 2004). Transgenic studies aimed at improving P use efficiency have generally focused on developing plants with better ability to make use of organic P in the soil. The expression of phytase from Aspergillus in Arabidopsis resulted in increase root phytase activity and subsequently enabled plants to meet their P requirements from phytate (Richardson et al., 2001). An alternative route to increased P uptake was taken by Wang et al. (2004), who over- expressed an Arabidopsis thaliana purple phosphatase gene (AtPAP15) containing a carrot (Daucus carota) extracellular targeting peptide in soybean to obtain inorganic P from organic P sources. This resulted in a 60–120 % increase in plant weight and an associated 60–90 % increase on plant P content (Wang et al., 2004).
The increase in the expression of genes related to organic acid metabolism and their secretion have also been reported under P deficiency, which suggests that organic acids play an important role in the adaptation to P-limiting conditions (Uhde-Stone et al., 2003a, b). A citrate synthase gene over-expressed in Arabidopsis was shown to improve growth on P-limited soils due to enhanced citrate excretion from roots (Koyama et al., 2000). The majority of the inorganic P present in soil is in precipitated form or adsorbed to soil constituents such as organic matter and clays (Richardson et al., 2009). It is thought that an increase in the levels of organic acids in the rhizosphere will chelate ions which are usually associated with phosphate groups, such as Ca2+, Fe2+ or Al3+, thus allowing the inorganic P to be released for plant uptake (Vance et al., 2003).
The objective of the present study was to determine the early response mechanisms of perennial ryegrass to P deficiency. We used a 44k barley microarray to study changes in gene expression whilst a complementary gas chromatography-mass spectrometry (GC-MS) approach was taken to study changes in primary metabolism.
MATERIALS AND METHODS
Selection of genotypes
A screen was carried out on five seedlings from each of 34 ecotypes and two cultivars of Lolium perenne (Supplementary Data 1, available online) with the aim of identifying material with a high capacity to remove P from solution. After 5 d, seedlings were weighed, transferred to 50-mL glass test tubes containing 40 mL nutrient solution [CaCl2.2H2O, 0·75 mm; MgSO4.7H2O, 0·38 mm; MS Micro Salts (Duchefa, Haarlem, The Netherlands), 0·146 g L−1; NH4NO3, 5 mm; Ca(NO3)2.4H2O, 2·33 mm; KH2PO4, 0·31 mm] and placed in a growth room maintained at 23 °C with a 16-h daylight regime [photosynthetically active radiation (PAR) = 360 µmol m−2 s−1]. After 3 d, samples were taken from each solution, the P content was determined using the molybdenum blue assay as described by He and Honeycutt (2005), and a standard curve was generated (R2 = 0·99) using dilutions of initial nutrient solution. Results were analysed in GenStat V10 (VSNi, Hemel Hempstead, UK) using a one-way ANOVA (without blocking). The fresh weights of seedlings at the start of the experiment were used as covariates in the analysis. Two genotypes (IRL-OP-02538-P and Cashel-P) were selected from this screen and propagated to provide adequate clonal replicates for the experiments described below.
Experimental conditions
Seedlings of IRL-OP-02538-P and Cashel-P were cleaned of soil and transferred to perlite medium held inside plastic plug trays (P84 Plug Trays, Carley's Bridge Potteries, Enniscorthy, Ireland) floated on the plant nutrient solution (see above). Solutions were placed in two 25-L tanks and aerated with an aquarium pump (Rena Air 300, RENA, Chalfont, PA, USA). Experiments were performed in a controlled glasshouse with a mean daily temperature of 22 °C and supplemented with lighting (PAR = 650 µmol m−2 s−1) for 16 h. The plants were allowed to acclimate for 1 week to the hydroponics growth conditions before applying treatments. At the start of the treatment the solutions in both tanks were replaced with either solution A, identical to that above, or solution B, identical to that above except that the KH2PO4 content was reduced to 0·016 mm. After 24 h (noon) root and leaf tissues were separated and flash frozen in liquid nitrogen. Two separate repeats of the experiment were performed to obtain samples for array hybridizations and metabolite profiling, respectively, and independent biological replicates (four in total) were sampled for each.
Microarray processing: genomic DNA hybridizations
Genomic DNA (gDNA) was isolated according to the method of Doyle and Doyle (1987) with minor modifications. Throughout this study, a custom microarray SCRI-Hv35-44k-v1 (Agilent design 020599) representing approx. 42 000 barley unigene sequences from the public HarvEST database (assembly 35; http://www.harvest-web.org/) was used (ArrayExpress accession A-MEXP-1728). This microarray along with a smaller barley array design (Chen et al., 2010) have been successfully used for gene expression analysis in our laboratory. Array procedures followed MIAME guidelines (Brazma et al., 2001). The design of the microarray experiment and the data derived from it are detailed in the public database ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/; accession E-TABM-950). For gDNA hybridizations, fluorescent labelling and purification was carried out according to the protocol detailed by Ducreux et al. (2008).
Microarray processing: cDNA hybridizations
Total RNA was isolated independently from four biological replicates of leaf and root tissues using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) with on-column DNase I digestion according to the manufacturer's protocol. Samples were run on Bioanalyzer RNA 6000 Nano Chips (Agilent Technologies, Santa Clara, CA, USA) to assess quality and integrity.
The design of the microarray experiment and the data derived from it are detailed in the public database ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/; accession E-TABM-943). In total, four biological replicates of each sample were processed (two genotypes, two treatments, two tissues, four replicates; 32 samples; 16 arrays). The experimental design was balanced with respect to fluorescent dyes, with two replicates of each sample labelled with either Cy3 or Cy5 to minimize dye bias. RNA samples were labelled as cDNA and purified using the Quick Amp Labelling Kit (Agilent Technologies) as recommended using 1 µg total RNA per sample.
Microarray processing: data extraction and analysis
For data extraction, microarray images were imported into Agilent Feature Extraction (FE) (v·10·5·1·1) software and aligned with the array grid template (020599-D-F-20080612). Intensity data for each spot were extracted using a defined FE protocol (GE2-v5-95-Feb07) and data from each array were normalized using the LOWESS (locally weighted polynomial regression) algorithm (Yang et al., 2002). Normalized datasets for each array were subsequently loaded into GeneSpring software (v.7·3·1; Agilent Technologies) for analysis.
For the RNA-based microarray study, specific samples were subjected to dye-swap according to the experimental design, and data with consistently low probe intensity level – flagged as absent in all replicate samples – were discarded. Comparisons were made between the two P treatments for each tissue and genotype independently using volcano plots (Cui and Churchill, 2003), with thresholds of >two-fold change and a Student's t-test P value <0·05 applied, to identify significantly regulated genes. Unique and overlapping genes between the lists were selected using Venn diagrams.
Quantitative RT-PCR
Total RNA was isolated as described above and 200 ng was converted to cDNA using SuperScript III and Oligo(dt)20 primer (Invitrogen, Carlsbad, CA, USA) as recommended by the manufacturer. Real-time assays and melting curve analysis were performed with SYBR Green I Master (Roche) on the LightCycler 480 according to the pack insert (http://www.roche-applied-science.com/pack-insert/4707516a.pdf). Analysis was carried out on three biological replicates (each in technical triplicates). Reaction efficiency values were calculated by running each primer set on serial dilutions of a cDNA mixture comprising leaf and root material. Normalization was performed using the housekeeping gene LpGAPDH (Petersen et al., 2004). Primers were designed from the barley sequences used for microarray probe design (Supplementary Data 2, available online). Statistical analyses were performed in REST 2008 (Pfaffl et al., 2002).
Sample preparation for metabolite profiling
Frozen tissues of roots and leaves were freeze dried and homogenized using a mortar and pestle. Frozen tissue powder (approx. 50 mg) was extracted as described by Foito et al. (2009). In summary, samples were extracted sequentially with methanol (3 mL), water (0·75 mL), chloroform (6 mL) and an additional volume of water (1·5 mL) resulting in a biphasic extract in order. Aliquots from the resulting polar and non-polar fractions (750 µL and 4 mL, respectively) were derivatized as described by Foito et al. (2009).
Analysis of metabolites by GC-MS
The polar and non-polar samples were analysed similarly using a GC-DSQ-MS system (Thermo Finnigan, Manchester, UK). Samples (1 µL) were injected into a programmable temperature vaporizing (PTV) injector with a split of 40 : 1. The PTV and chromatography conditions used are described by Foito et al. (2009). Mass spectra were acquired under electron impact ionization conditions at 70 eV and 100 mA of emission current over the mass range 35–900 a.m.u at 6 scans s−1 with a source temperature of 200 °C and a solvent delay of 1·3 min. Acquisition rates were set to give approximately ten data points across a chromatographic peak. Data were acquired using the XcaliburTM software package v. 1·4. Acquired total ion chromatograms and mass spectra were analysed using XcaliburTM software package v2·0·7 (Thermo Finnigan) as described by Foito et al. (2009). Analysis of variance was used to compare across P treatments and identify significantly regulated metabolites (P < 0·05). The complete profiles were initially compared by principal components analysis (PCA) and the results for metabolite analysis with respect to data acquisition controls are described in Supplementary Data 3 (Figs S1 and S2, available online). All statistical analysis was performed using GenStat version 12·1·0·3338 (VSNi).
RESULTS
Genotype selection
The purpose of the pre-screen was to identify seedlings with increased rates of Pi removal from solution. The results showed that ecotype had an effect on P removal (F35,142 = 3·07, P < 0·001). The ecotype with the highest removal of P from solution was IRL-OP-02538 (Supplementary Data 1). The seedling with the highest P removal (63·56) was propagated together with a seedling from the cultivar ‘Cashel’ (41·51), which displayed an average removal of P from solution. We chose to test two genotypes as opposed to a single genotype in order to identify more general response mechanisms of perennial ryegrass to 24 h of P deficiency and avoid making assumptions based on the response of a single genotype.
Array hybridization across species
Initial experimentation used genomic DNA to determine the overall level of hybridization of perennial ryegrass genome to probes on the barley microarray. This took advantage of the extreme sensitivity of the Agilent microarray platform. Approximately 40 % of probes (16000 genes) hybridized with measurable signal to perennial ryegrass genomic DNA. This level of hybridization reflects the levels of sequence diversity between the two species, although the 60-mer probes utilized in the Agilent microarray allow for a certain degree of mismatches, as reported previously for analysis of diverse potato genotypes (Ducreux et al., 2008). After hybridization with cDNA derived from leaf and root RNA, approx. 6800 and approx. 6300 probes, respectively, produced an acceptable signal amongst replicates.
The validity of the array results was verified by real-time RT-PCR of seven selected genes (Table 1) identified as significantly regulated by using the barley arrays. The results were in general agreement with the array results with the exception of U35-44k-v1-8347, a putative MAP3K-like protein kinase, which was not identified as being significantly up-regulated by real-time RT-PCR (Table 1). Indeed, the strong agreement confirms the efficacy of the barley arrays for analysing gene expression in perennial ryegrass.
Table 1.
Validation of microarray leaf and root data by real-time RT-PCR of seven genes identified as significantly regulated from leaf or root hybridizations
| Description | Rice match | Putative function | Array |
RT-PCR |
||
|---|---|---|---|---|---|---|
| Cashel-P | IRL-OP-02538-P | Cashel-P | IRL-OP-02538-P | |||
| Leaf | ||||||
| U35-44k-v1-12109 | LOC-Os02g33710·1 | Histidine decarboxylase | 5·9 (0·001) | 5·8 (0·000) | 8·2 (0·000) | 7·4 (0·000) |
| U35-44k-v1-37470 | LOC-Os12g37600·1 | Glycerol-3-phosphate acyltransferase 1 | 0·9 (0·951) | 5·8 (0·005) | 0·9 (0·904) | 2·7 (0·029) |
| U35-44k-v1-8347 | LOC-Os03g30130·2 | Phospholipase C | 2·8 (0·007) | 4·3 (0·006) | 1·4 (0·374) | 1·6 (0·178) |
| U35-44k-v1-7335 | LOC-Os01g54620·1 | CESA4 – cellulose synthase | 1·4 (0·702) | 7·0 (0·038) | 1·2 (0·626) | 4·0 (0·013) |
| Root | ||||||
| U35-44k-v1-22715 | LOC-Os01g67126·1 | 60S ribosomal protein L5-2 | 3·3 (0·585) | 3·6 (0·032) | 0·6 (0·714) | 8·2 (0·000) |
| U35-44k-v1-49610 | LOC-Os08g43870·1 | Hypothetical protein | 1·0 (0·939) | 2·6 (0·024) | 0·9 (0·543) | 2·8 (0·003) |
| U35-44k-v1-43490 | LOC-Os08g39300·1 | Serine-glyoxylate aminotransferase | 1·2 (0·718) | 0·3 (0·021) | 0·7 (0·487) | 0·3 (0·000) |
The fold change for both array and RT-PCR data are shown with associated significance values in parentheses; significant differences in bold. Array data were analysed in GeneSpring and RT-PCR data in REST.
Transcriptomic changes in leaf tissue under P deficiency
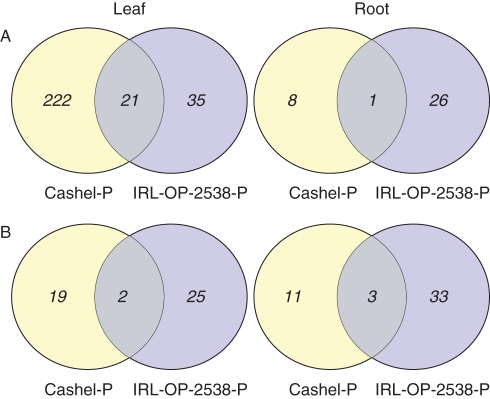
Using the barley arrays the expression profiles of two perennial ryegrass genotypes changed in response to 24 h of P deficiency (Fig. 1; Supplementary Data 4 and 5, available online). The genes significantly regulated (≥two-fold and P < 0·05) in each genotype are shown in Supplementary Data 4. The greatest number of significantly regulated genes (222) was discovered in the leaf tissue of Cashel-P under P deficiency, with 95 % of these genes being up-regulated. In contrast, a much smaller number of genes (35) were significantly regulated in the leaf tissue of IRL-OP-02538-P (Fig. 1A).
Fig. 1.
(A) Number of genes from barley array hybridizations with ≥ two-fold change in expression (P < 0·05) under limited phosphorus for each genotype. (B) Number of metabolites with significant fold change (P < 0·05) under limited phosphorus for each genotype. Leaf tissue on left and root tissue on right.
Only 21 genes were significantly regulated in a similar fashion in both genotypes, some of which have an unknown function (Table 2). No down-regulated genes were common between genotypes. The gene with the greatest induction in both genotypes encodes a putative glycerol 3-phosphate permease (G3PP).
Table 2.
Genes significantly (P < 0·05) induced ≥ two-fold after 24 h of P deficiency in the leaf tissue of both Cashel-P and IRL-OP-02538-P
| IRL-OP-02538-P |
Cashel-P |
|||||
|---|---|---|---|---|---|---|
| Array ID | Best hit rice pp5 | E value | Fold change | P | Fold change | P |
| U35-44k-v1-27526 | Glycerol 3-phosphate permease | 1 × 10−45 | 7·6 | <0·01 | 17·1 | <0·01 |
| U35-44k-v1-31414 | Glycerol 3-phosphate permease | 7 × 10−70 | 7·0 | <0·01 | 12·3 | <0·01 |
| U35-44k-v1-12109 | Histidine decarboxylase | 1 × 10−121 | 5·9 | <0·01 | 5·8 | <0·01 |
| U35-44k-v1-7536 | Purple acid phosphatase precursor | 1 × 10−166 | 5·5 | <0·01 | 5·3 | <0·01 |
| U35-44k-v1-26073 | Nucleotide pyrophosphatase/phosphodiesterase | 1 × 10−130 | 4·5 | <0·01 | 5·4 | <0·01 |
| U35-44k-v1-31178 | No hits found | 3·7 | <0·01 | 2·9 | 0·01 | |
| U35-44k-v1-7801 | UDP-sulfoquinovose synthase chloroplast precursor | 1 × 10−68 | 3·4 | 0·01 | 4·2 | <0·01 |
| U35-44k-v1-27915 | ids4-like protein | 4 × 10−18 | 3·4 | <0·01 | 5·3 | <0·01 |
| U35-44k-v1-48069 | No hits found | 3·1 | <0·01 | 2·4 | 0·02 | |
| U35-44k-v1-24642 | Expressed protein | 2 × 10−15 | 2·9 | 0·04 | 5·7 | <0·01 |
| U35-44k-v1-8347 | Phospholipase C | 1 × 10−159 | 2·8 | 0·01 | 4·3 | 0·01 |
| U35-44k-v1-30444 | Expressed protein | 4 × 10−43 | 2·8 | <0·01 | 2·0 | <0·01 |
| U35-44k-v1-36750 | Expressed protein | 4 × 10−85 | 2·7 | <0·01 | 2·4 | 0·04 |
| U35-44k-v1-28139 | Phosphate transporter 1 | 1 × 10−82 | 2·7 | 0·04 | 10·5 | 0·01 |
| U35-44k-v1-27385 | Acid phosphatase/vanadium-dependent haloperoxidase-related | 4 × 10−5 | 2·5 | 0·01 | 2·4 | <0·01 |
| U35-44k-v1-28600 | Expressed protein | 4 × 10−65 | 2·4 | 0·01 | 2·5 | <0·01 |
| U35-44k-v1-2801 | Expressed protein | 3 × 10−21 | 2·3 | 0·05 | 2·6 | <0·01 |
| U35-44k-v1-9242 | Diacylglycerol O-acyltransferase 1 putative expressed | 1 × 10−97 | 2·1 | 0·05 | 6·3 | 0·01 |
| U35-44k-v1-11282 | Xyloglucan endotransglucosylase/hydrolase protein 30 precursor | 8 × 10−47 | 2·1 | 0·05 | 2·3 | 0·04 |
| U35-44k-v1-1867 | Pyrophosphate–fructose 6-phosphate 1-phosphotransferase alpha subunit | 0·0 | 2·1 | 0·01 | 2·6 | 0·01 |
| U35-44k-v1-6244 | Glycerophosphoryl diester phosphodiesterase precursor | 3 × 10−93 | 2·1 | 0·03 | 2·4 | <0·01 |
In all cases there were a number (three) of genes encoding phosphatases up-regulated under P deficiency in leaf tissue (Supplementary Data 4). In addition, one gene encoding a phosphate transporter was also up-regulated but to a lesser extent. One group of genes was notably up-regulated, particularly in the Cashel-P genotype, and these were those involved in cell-wall synthesis with five cellulose synthase genes up-regulated under P deficiency (CESA4, CESA9, CESA1, CESA2 and CESA8). Other genes involved in cell-wall modification were also identified with the gene encoding ENDO-1, 4-beta-xylanase, which catalyses the hydrolysis of the major plant hemicellulose, xylan, showing the greatest increase in expression in Cashel-P under P deficiency.
Metabolite changes in leaf tissue under P deficiency
Metabolite profiling was carried out on genotypes from IRL-OP-02538 and ‘Cashel’ when grown under normal and restricted P supply. The profiles included the levels of 206 metabolites (140 identified), which included 116 polar metabolites and 90 non-polar metabolites. PCA revealed distinguishing differences between the profiles of leaves from different genotypes (Supplementary Data 3, Fig. S3, available online) and, to a lesser extent, between treatments. An ANOVA identified a number of significantly different (P < 0·05) metabolites in the different tissues and genotypes (Fig. 1B). In contrast to the transcriptomic data, no major difference in the number of up-regulated metabolites between genotypes was found, although the majority of the significantly regulated metabolites were different. In the leaf tissue of IRL-OP-02538-P there were 27 regulated metabolites (including nine unidentified). One unidentified metabolite experienced an increase under P deficiency, while levels of the remaining 26 (Table 3) declined. This is in contrast to the genotype Cashel-P in which levels of 11 metabolites increased and levels of ten decreased (Table 4). From the 21 significantly regulated metabolites in Cashel-P in leaf tissue, four remain unidentified. These differences in trends between genotypes contributed significantly to the segregation seen in the PCA of the leaf tissue. In Cashel-P, there was a significant decrease in amino acids (asparagine, glutamine, homoserine and histidine) while the levels of putative phytol-derived metabolites appeared to be up-regulated (phytol A-C and phytil methyl ether). Despite the overall differences in the metabolic response to P limitation, both genotypes experienced a significant decrease in levels of Pi.
Table 3.
Comparison of identified significantly different (P < 0·05) metabolites in the leaves and roots of IRL-OP-2538-P under P-sufficient and P-deficient phosphorous
| Average response ratio of IRL-OP-2538-P under sufficient P supply | Average response ratio of IRL-OP-2538-P under limited P supply | Low/high | P | Log(ratio) | |
|---|---|---|---|---|---|
| Leaf | |||||
| Unknown | 1·83 × 10−4 | 3·10 × 10−4 | 1·69 | 0·007 | 0·23 |
| Phytil methyl ether 2nd peak | 4·47 × 10−1 | 4·01 × 10−1 | 0·90 | 0·028 | –0·05 |
| Unknown | 5·17 × 10−1 | 4·49 × 10−1 | 0·87 | 0·045 | –0·06 |
| Unknown | 1·10 × 10−2 | 9·22 × 10−3 | 0·84 | 0·037 | –0·08 |
| Phenylalanine | 5·64 × 10−4 | 4·71 × 10−4 | 0·84 | 0·015 | –0·08 |
| n-Tetracosanol | 1·11 × 10−2 | 9·01 × 10−3 | 0·81 | 0·010 | –0·09 |
| Octadecanol | 5·25 × 10−3 | 4·02 × 10−3 | 0·76 | 0·044 | –0·12 |
| Isoleucine | 3·09 × 10−4 | 2·13 × 10−4 | 0·69 | 0·002 | –0·16 |
| Unknown | 3·71 × 10−4 | 2·53 × 10−4 | 0·68 | 0·001 | –0·17 |
| Unknown | 5·13 × 10−4 | 3·48 × 10−4 | 0·68 | 0·007 | –0·17 |
| Citric acid | 5·48 × 10−3 | 3·50 × 10−3 | 0·64 | 0·032 | –0·19 |
| Pentadecenoic acid | 5·43 × 10−3 | 3·37 × 10−3 | 0·62 | 0·002 | –0·21 |
| β-Alanine | 4·94 × 10−5 | 3·01 × 10−5 | 0·61 | 0·016 | –0·22 |
| Leucine | 4·13 × 10−4 | 2·49 × 10−4 | 0·60 | 0·030 | –0·22 |
| Fructose* | 1·19 × 10−2 | 7·15 × 10−3 | 0·60 | 0·005 | –0·22 |
| Tyrosine | 2·40 × 10−4 | 1·37 × 10−4 | 0·57 | 0·013 | –0·24 |
| Unknown | 4·19 × 10−4 | 2·39 × 10−4 | 0·57 | 0·049 | –0·24 |
| Malic acid | 1·22 × 10−2 | 6·73 × 10−3 | 0·55 | <0·001 | –0·26 |
| Unknown | 5·80 × 10−5 | 2·58 × 10−5 | 0·44 | <0·001 | –0·35 |
| n-Pentadecanoic acid | 8·70 × 10−4 | 3·82 × 10−4 | 0·44 | 0·029 | –0·36 |
| Lysine | 3·61 × 10−5 | 1·56 × 10−5 | 0·43 | 0·017 | –0·36 |
| Fucosterol | 1·84 × 10−3 | 7·44 × 10−4 | 0·41 | 0·020 | –0·39 |
| Phosphate | 3·06 × 10−3 | 1·20 × 10−3 | 0·39 | 0·004 | –0·41 |
| 2-Piperidinecarboxylic acid | 2·27 × 10−5 | 8·58 × 10−6 | 0·38 | 0·015 | –0·42 |
| Unknown | 2·26 × 10−3 | 8·37 × 10−4 | 0·37 | 0·023 | –0·43 |
| Unknown | 2·12 × 10−3 | 7·73 × 10−4 | 0·36 | <0·001 | –0·44 |
| Proline | 2·20 × 10−3 | 5·26 × 10−4 | 0·24 | 0·043 | –0·62 |
| Root | |||||
| Unknown | 5·63 × 10−4 | 0·00 | * | <0·001 | * |
| Stigmastadienol | 6·68 × 10−5 | 6·03 × 10−4 | 9·03 | 0·032 | 0·96 |
| Diamino-1,3-propane | 3·25 × 10−6 | 1·93 × 10−5 | 5·94 | 0·009 | 0·77 |
| Unknown | 8·96 × 10−4 | 3·65 × 10−3 | 4·08 | 0·006 | 0·61 |
| Unknown | 1·72 × 10−3 | 5·29 × 10−3 | 3·07 | 0·003 | 0·49 |
| Unknown | 4·07 × 10−5 | 9·54 × 10−5 | 2·34 | 0·002 | 0·37 |
| d-5-Avenasterol | 1·63 × 10−3 | 3·63 × 10−3 | 2·23 | 0·010 | 0·35 |
| Putrescine | 7·52 × 10−5 | 1·62 × 10−4 | 2·15 | <0·001 | 0·33 |
| Galactose | 2·89 × 10−5 | 5·78 × 10−5 | 2·00 | 0·046 | 0·30 |
| Fructose* | 1·22 × 10−3 | 2·35 × 10−3 | 1·93 | <0·001 | 0·29 |
| Glucose* | 1·23 × 10−3 | 2·27 × 10−3 | 1·85 | <0·001 | 0·27 |
| Unknown | 5·51 × 10−5 | 9·49 × 10−5 | 1·72 | 0·008 | 0·24 |
| Shikimic acid | 2·66 × 10−5 | 4·00 × 10−5 | 1·50 | 0·001 | 0·18 |
| Unknown | 2·07 × 10−4 | 3·09 × 10−4 | 1·49 | 0·011 | 0·17 |
| Sucrose | 7·85 × 10−2 | 1·16 × 10−1 | 1·48 | 0·027 | 0·17 |
| n-Octadecanoic acid | 7·44 × 10−3 | 9·96 × 10−3 | 1·34 | 0·020 | 0·13 |
| Linoleic acid | 2·06 × 10−1 | 2·62 × 10−1 | 1·27 | 0·001 | 0·10 |
| α-Linolenic acid | 5·63 × 10−2 | 6·82 × 10−2 | 1·21 | 0·016 | 0·08 |
| β-Sitosterol | 1·55 × 10−1 | 1·86 × 10−1 | 1·20 | 0·017 | 0·08 |
| n-Hexadecanoic acid | 2·05 × 10−1 | 2·47 × 10−1 | 1·20 | 0·017 | 0·08 |
| Valine | 1·61 × 10−3 | 1·27 × 10−3 | 0·79 | 0·031 | –0·10 |
| Eicosanol | 1·08 × 10−2 | 7·60 × 10−3 | 0·70 | 0·026 | –0·15 |
| Leucine | 9·10 × 10−4 | 6·26 × 10−4 | 0·69 | 0·015 | –0·16 |
| Unknown | 4·80 × 10−4 | 3·02 × 10−4 | 0·63 | 0·041 | –0·20 |
| Lysine | 2·29 × 10−4 | 1·42 × 10−4 | 0·62 | 0·006 | –0·21 |
| Phosphate | 1·24 × 10−2 | 7·33 × 10−3 | 0·59 | <0·001 | –0·23 |
| Cinnamic acid | 4·50 × 10−2 | 2·60 × 10−2 | 0·58 | 0·045 | –0·24 |
| 4- or 3-Hydroxycinnamic acid | 2·23 × 10−2 | 1·23 × 10−2 | 0·55 | <0·001 | –0·26 |
| Alanine | 1·83 × 10−3 | 9·89 × 10−4 | 0·54 | 0·001 | –0·27 |
| Unknown | 5·29 × 10−4 | 2·75 × 10−4 | 0·52 | 0·006 | –0·28 |
| Allantoin | 8·70 × 10−4 | 4·16 × 10−4 | 0·48 | <0·001 | –0·32 |
| Unknown | 1·25 × 10−3 | 5·05 × 10−4 | 0·40 | <0·001 | –0·39 |
| Unknown | 1·36 × 10−4 | 4·74 × 10−5 | 0·35 | <0·001 | –0·46 |
| n-Docosanol | 8·50 × 10−4 | 2·97 × 10−4 | 0·35 | 0·007 | –0·46 |
| Heneicosanol | 9·84 × 10−4 | 3·15 × 10−4 | 0·32 | 0·002 | –0·49 |
| Mannitol | 3·14 × 10−4 | 3·64 × 10−5 | 0·12 | <0·001 | –0·94 |
* Both glucose and fructose result in two peaks corresponding to their isomers. The levels of both isomers were combined to calculate the total value of glucose and fructose.
Table 4.
Comparison of identified significantly different (P < 0·05) metabolites in the leaves and roots of Cashel-P under P-sufficient and P-deficient conditions
| Average response ratio of Cashel-P under sufficient P supply | Average response ratio of Cashel-P under limited P supply | Low/high | P | Log(ratio) | |
|---|---|---|---|---|---|
| Leaf | |||||
| γ-Aminobutyric acid | 3·41 × 10−4 | 6·86 × 10−4 | 2·01 | 0·006 | 0·30 |
| Unknown | 3·56 × 10−5 | 5·65 × 10−5 | 1·59 | 0·029 | 0·20 |
| Galactose/glycerol conjugate | 8·19 × 10−5 | 1·25 × 10−4 | 1·53 | 0·042 | 0·18 |
| Threonic acid | 8·27 × 10−4 | 1·21 × 10−3 | 1·46 | 0·034 | 0·16 |
| Unknown | 2·60 × 10−1 | 3·62 × 10−1 | 1·4 | <0·001 | 0·14 |
| Phytol B | 2·12 × 10−2 | 2·85 × 10−2 | 1·34 | 0·002 | 0·13 |
| Phytol A | 1·27 × 10−2 | 1·66 × 10−2 | 1·31 | <0·001 | 0·12 |
| Sucrose | 4·01 × 10−1 | 5·04 × 10−1 | 1·26 | 0·038 | 0·10 |
| Phytol C | 6·33 × 10−3 | 7·88 × 10−3 | 1·25 | 0·025 | 0·10 |
| n-Hexadecenoic acid | 4·43 × 10−2 | 5·54 × 10−2 | 1·25 | 0·014 | 0·10 |
| Phytil methyl ether | 1·49 × 10−1 | 1·79 × 10−1 | 1·2 | 0·004 | 0·08 |
| Homoserine | 2·16 × 10−4 | 1·17 × 10−4 | 0·54 | 0·047 | –0·27 |
| 2-Piperidinecarboxylic acid | 3·95 × 10−5 | 2·03 × 10−5 | 0·51 | 0·048 | –0·29 |
| Unknown | 7·45 × 10−5 | 3·18 × 10−5 | 0·43 | 0·010 | –0·37 |
| 5-Oxoproline | 5·29 × 10−2 | 2·27 × 10−2 | 0·43 | 0·049 | –0·37 |
| Histidine | 7·89 × 10−6 | 2·04 × 10−6 | 0·26 | 0·024 | –0·59 |
| Spermidine | 5·38 × 10−6 | 1·18 × 10−6 | 0·22 | 0·048 | –0·66 |
| Glutamine* | 2·66 × 10−3 | 5·3 × 10−4 | 0·20 | 0·026 | –0·70 |
| Phosphate | 7·64 × 10−3 | 1·17 × 10−3 | 0·15 | 0·002 | –0·82 |
| Asparagine* | 1·11 × 10−3 | 1·5 × 10−4 | 0·14 | 0·009 | –0·87 |
| Unknown | 3·48 × 10−4 | 3·10 × 10−5 | 0·09 | 0·046 | –1·05 |
| Root | |||||
| Unknown | 2·40 × 10−4 | 5·71 × 10−4 | 2·38 | 0·045 | 0·38 |
| Unknown | 1·08 × 10−4 | 2·28 × 10−4 | 2·11 | 0·019 | 0·32 |
| Unknown | 3·69 × 10−4 | 7·56 × 10−4 | 2·05 | 0·046 | 0·31 |
| Methionine | 3·62 × 10−5 | 7·12 × 10−5 | 1·97 | 0·027 | 0·29 |
| Unknown | 5·11 × 10−5 | 9·36 × 10−5 | 1·83 | 0·046 | 0·26 |
| Sucrose | 5·50 × 10−2 | 9·70 × 10−2 | 1·76 | 0·022 | 0·25 |
| Aspartic acid | 5·14 × 10−3 | 8·28 × 10−3 | 1·61 | 0·002 | 0·21 |
| Glutamic acid | 6·61 × 10−3 | 9·84 × 10−3 | 1·49 | 0·034 | 0·17 |
| n-Tetracosanoic | 1·35 × 10−2 | 9·20 × 10−3 | 0·68 | 0·043 | –0·17 |
| Unknown | 1·21 × 10−4 | 7·60 × 10−5 | 0·63 | 0·028 | –0·20 |
| Octadecenoic acid | 4·61 × 10−3 | 2·53 × 10−3 | 0·55 | 0·005 | –0·26 |
| Unknown | 2·09 × 10−3 | 1·05 × 10−3 | 0·50 | 0·044 | –0·30 |
| Unknown | 6·10 × 10−4 | 2·69 × 10−4 | 0·44 | 0·003 | –0·36 |
| Unknown | 4·32 × 10−5 | 1·39 × 10−5 | 0·32 | 0·035 | –0·49 |
* Both glutamine and asparagine generate two trimethylsilyl (TMS) derivatives which were combined to generate the total levels of asparagine and glutamine. For glutamine the TMS4 derivative was multiplied by a factor of 2 before being combined with its other derivative.
Trancriptomic changes in root tissue under P deficiency
There were a much lower number of significantly regulated genes under P deficiency in the root tissue in comparison with leaf tissue. Furthermore, the majority of the significantly regulated genes were down-regulated in both genotypes (Supplementary Data 5). In Cashel-P the down-regulated genes included those which encode a pair of ribosomal proteins, an asparagine synthetase (EC 6·3·5·4), a serine-glyoxylate aminotransferase (EC _2·6·1·45), a fructose-1, 6-bisphosphatase (EC 3·1·3·11) and a sedoheptulose-1, 7-bisphosphatase (EC 3·1·3·37; full list in Supplementary Data 5). Only one gene was commonly regulated in both genotypes and this encoded a 1,4-beta-xylanase (EC 3·2·1·8), which was up-regulated.
Metabolite changes in root tissue under P deficiency
In the PCA of the metabolite profiles from root tissue (Supplementary Data 3, Fig. S4) there was no segregation between genotypes as observed in the leaf tissue. However, the ANOVA highlighted differences which could not be visualized using PCA of the root tissue. A greater difference in the number of significantly regulated metabolites was found between genotypes in the root tissue: 14 metabolites (eight unidentified) were regulated in Cashel-P (Table 4) whereas 36 (ten unidentified) were regulated in IRL-OP-02538-P under P deficiency (Table 3). Furthermore, the latter genotype experienced a general increase in sugar levels (glucose, fructose, galactose and sucrose) and fatty acids (C16:0, C18:0, C18:2 and C18:3) whilst levels of some of the pyruvate-derived amino acids (alanine, leucine and valine) decreased. Among the metabolites significantly regulated in Cashel-P (14), only six were putatively identified. For Cashel-P, the imposition of limiting P conditions resulted in an increase in the levels of sucrose and some amino acids (methionine, aspartic acid and glutamic acid) whilst levels of some fatty acid (C24:0 and C18:1) decreased.
DISCUSSION
This study aimed to uncover changes in the transcriptome and metabolome of two perennial ryegrass genotypes during the initial stages of P deficiency. It is the first study looking at the global responses to low P availability in perennial ryegrass and provides an insight into the early-stage response mechanisms of the plant to P deficiency. Metabolic profiling demonstrated that P deficiency resulted in a Pi decrease in the leaves of both genotypes (Tables 3 and 4), thus confirming that the experimental conditions have successfully produced an effect on the levels of Pi and that the time frame (24 h) was sufficient to elicit this response. However, no significant differences in growth were observed (data not shown). Hammond et al. (2003) reported that in A. thaliana the concentration of P in the shoots decreases without causing an effect of plant growth between 24 and 72 h after P withdrawal, suggesting that 24 h of exposure to P withdrawal is a plausible time frame to expect a decrease in shoot P in perennial ryegrass. Interestingly, Hammond et al. (2003) used the promoter of the SDQ1 gene (UDP-sulfoquinovose synthase), which specifically responds to P limitation, to control the expression of the GUS marker gene in an attempt to develop smart plants allowing the detection of P-limitation before it affected plant growth. In this study we observed an increase in the expression of SDQ1 after 24 h (Table 2), which appears to confirm that the experimental conditions have resulted in a specific response from the plant to P limitation. Furthermore, an increase in the expression of SDQ1 in A. thaliana pho1 mutants (with low internal Pi concentration) and in wild-type plants exposed to P imitation has been reported (Essigmann et al., 1998). This led the authors to suggest that this effect is caused by low shoot P (Essigmann et al., 1998), which appears to be consistent with our results.
Lipid membrane remodelling
One mechanism of adaptation to P-deficient growth conditions is the remodelling of plant lipid membranes in order to liberate Pi bound in membrane phospholipids (Plaxton, 2004). The increase reported in the expression levels of genes encoding phospholipase A1 (EC 3·1·1·32) together with increases in glycerophosphoryl diester phosphodiesterase (EC 3·1·4·46, hydrolysis of deacylated phospholipids to glyceraldehyde 3-phosphate), acid phosphatase/vanadium-dependent hydrolase and diacylglycerol O-acyltransferase 1 (EC 2·3·1·20) also appears to suggest a certain degree of phospholipid remodelling in the response to P deficiency in perennial ryegrass. Photosynthetic membranes are characterized by a substantial fraction of non-phosphorus lipids, such as galactolipids or sulfolipids (e.g. sulfoquinovosyldiacylglycerol), which reflects the need to conserve P by plants (Benning, 2009). Here we observed a significant increase in expression of genes encoding UDP-sulfoquinovose synthase (EC 3·13·1·1), a key enzyme in the sulfoquinovosyldiacylglycerol (SQDG) synthesis pathway, during P deficiency in the leaf tissue of both genotypes. Hammond et al. (2003) demonstrated that, in Arabidopsis, the expression of a gene involved in sulfolipid biosynthesis (SQD1) is specifically increased in response to P limitation. Furthermore, Misson et al. (2005) demonstrated in Arabidopsis that under Pi deficiency SQDG increased, whereas phosphatidylglycerol (PG) decreased. These results are in agreement with elevated expression of genes involved in SQDG synthesis and phospholipid degradation.
We also observed an increase in gene expression levels of a sulphate transporter in both genotypes, although the increase was only significant in Cashel-P (Cashel-P: 5·2-fold, P < 0·001; IRL-OP-02538-P: 2·8-fold, P = 0·054). This has previously been observed in Arabidopsis and was proposed to support an increased demand for sulphur during P deficiency to meet the needs for increased sulfolipid synthesis (Misson et al., 2005). An Arabidopsis mutant, sqd2, disrupted in a putative identified sulfolipid synthase (SQD2) gene, displayed no measurable sulfolipid content and reduced growth under P-deficient conditions (Yu et al., 2002). The authors of that study hypothesized that sulfolipids may substitute for anionic phospholipids under P-deficient conditions and therefore allow photosynthesis to continue. Further support for lipid membrane remodelling in perennial ryegrass can be seen from the strong induction of a gene encoding G3PP under P deficiency (7·5- and 17·1-fold, P < 0·001 in IRL-OP-02538-P and Cashel-P, respectively). This suggests the enhanced mobilization of glycerol 3-phosphate from the cytosol into the chloroplast or endoplasmic reticulum where it could be utilized in lipid biosynthesis (Fig. 2).
Fig. 2.
Diagram depicting the primary metabolism in Lolium perenne, adapted from Hammond et al. (2004). Bold arrows represent a downstream set of reactions, not single reactions. Dashed arrows highlight alternative pathways for glycolysis to conserve Pi. Microarray results indicate an increase in expression of transcripts encoding genes involved in reactions 1, 6, 7, 8 and 10. Abbreviations: Suc, sucrose; Glc, glucose; Fru, fructose; UDPG, UDP-glucose; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F2,6P, fructose 2,6-bisphosphate; F1,6P, fructose 1,6-bisphosphate; G3P, glycerol 3-phosphate; DHAP, dihidroxyacetone phosphate; OAA, oxalo acetic acid; Ga3P, glyceraldehyde 3-phosphate; 1,3-DPGA, 1,3- diphosphoglyceraldehyde; 3-PGA, 3-phosphoglycerate; PEP, phosphoenol pyruvate; Phe, phenylalanine; Ala, alanine; Val, valine; Ile, isoleucine; Leu, leucine. Reactions: (1) Pyrophosphate fructose 6-phosphate 1-phosphotransferase, EC 2·7·1·90; (2) ATP : fructose 6-phosphate 1-phosphotransferase, EC 2·7·1·11; (3) phosphoenolpyruvate carboxykinase (ATP), EC 4·1·1·49; (4) pyruvate kinase, EC 2·7·1·40; (5) malate dehydrogenase (oxaloacetate-decarboxylating), EC 1·1·1·38; (6) involvement of anthranilate phosphoribosyltransferase, EC 2·4·2·18 (7) involvement of flavonol synthase/flavanone 3-hydroxylase; (8) glycerol 3-phosphate dehydrogenase, EC 1·1·5·3; (9) glycerol 3-phosphate dehydrogenase, EC 1·1·5·3; (10) glycerol 3-phosphate permease; (11) UDP-glucose pyrophosphorylase, EC 2·7·7·9.
Our results indicate that the replacement of phospholipids with sulfolipids in the membranes of photosynthetic tissue is also occurring in perennial ryegrass under P deficiency and that this is a rapid response to changes in P status.
Carbon partitioning
At the metabolome level in the leaves of IRL-OP-02538-P, P deficiency was accompanied by an overall decrease in metabolite levels whilst there were no putatively identified up-regulated metabolites. The significantly regulated metabolites included a variety of metabolite classes such as sugars, amino acids, fatty acids and organic acids. The overall decrease in metabolite levels may be symptomatic of either a decrease of source activity due to decreases in photosynthetic activity, an increase in sink activity or both. P limitation has been reported to impact upon photosynthetic activity (Foyer and Spencer, 1986; Lauer et al., 1989; Rodriguez et al., 1998; Hammond and White, 2008) although increases in the root/shoot ratio are also considered typical effects of P limitation (Freedan et al., 1989; Hermans et al., 2006; Hammond and White, 2008) and may account for increased sink activity as carbohydrates are exported from the leaves to the roots. A study by Wissuwa et al. (2005) focused on determining whether root growth in rice is affected by either source or sink limitations when grown under P-deficient conditions and they did this by comparing rice genotypes under four P levels and two light treatments. The authors reported that net photosynthesis was 70 % greater in plants grown under high, compared with low, light but it was found that this did not translate into greater root growth under P limitation, leading them to suggest that assimilate supply from source leaves was not a limiting factor under P limitation. Conversely, low P supply appears to limit root growth directly. It was proposed that the most tolerant genotype used in the experiment of Wissuwa et al. (2005) preferably mobilized P to the roots, which would increase P concentrations and consequently promote growth. The levels of sugars in the roots of IRL-OP-02538-P increase in response to P limitation, which seem to suggest a decrease in sink activity, resulting from a reduction in root growth. Root growth inhibition in response to P limitation has been suggested to be mediated by putrescine in rice (Shih and Kao, 1996), and although no root/shoot ratios were measured, the significant up-regulation of putrescine levels found in this study suggests a limitation of root growth in IRL-OP-02538-P plants as an early response to limited P availability.
Glycolytic bypasses
The results showed evidence of glycolytic bypasses being utilized and these have previously been reported in other species as being valuable mechanisms to enable the efficient use of P during periods of deficiency (Theodorou and Plaxton, 1993). We observed a significant increase in the expression of a gene encoding an α-subunit of pyrophosphate : fructose 6-phosphate 1-phosphotransferase (PFP, EC 2·7·1·90) during P deficiency in the leaf tissue of both genotypes. The phosphorylation of fructose 6-phosphate to fructose 1-6 bisphosphate during glycolysis is efficiently catalysed by ATP : fructose 6-phosphate 1-phosphotransferase (PFK, EC 2·7·1·11), using ATP (Stryer, 1995). PFP can also catalyse this reaction but employing PPi rather than ATP (Fig. 2). It has already been demonstrated in black mustard (Brassica nigra) suspension cells that PFP activity is increased under P deficiency and its activity falls to below the level of PFK as solute P levels increase (Duff et al., 1989). Furthermore, it was shown that the α-subunit of PFP has a regulatory role over the β-subunit and that its activity is tightly regulated by intracellular Pi status (Theodorou et al., 1992). Our results demonstrate that a switch to alternative pathways during glycolysis occurs at an early stage of P deficiency in perennial ryegrass.
Cell-wall metabolism
One class of genes stands out as being abundant within the list of transcripts significantly up-regulated in leaf tissue under P deficiency, particularly in the Cashel-P genotype. These are genes involved in cell-wall synthesis and remodelling. Previous reports have also reported the up-regulation of genes involved in cell-wall synthesis under P-limiting conditions (Wasaki et al., 2003). We identified the up-regulation of five genes encoding the catalytic subunit of cellulose synthase (CESA) between both genotypes: four in Cashel-P and one in IRL-OP-02538-P. Ten CESA genes are reportedly present in the Arabidopsis genome and the role of these in cell-wall synthesis has previously been described by Scheible and Pauly (2004). The significant up-regulation of this gene family points to an increased production of cellulose during the early stages of P deficiency. However, the gene with the highest increase in expression under P deficiency in Cashel-P leaves encodes an ENDO-1, 4-beta-xylanase (EC 3·2·1·8), a xylan hydrolase. Together, these results may suggest a remodelling of cell walls, with cellulose content increasing and xylan content decreasing, although direct evidence is still needed to corroborate this. The exact role, if any, that this modification could have under P deficiency remains unknown. Interestingly, it has been recently demonstrated that the over-expression of a wall-bound purple acid phosphatase from tobacco (NtPAP12) resulted in an increased deposition of cellulose in transgenic cells (Kaida et al., 2009). Transcripts coding for acid phosphatases (purple acid phosphatase precursor and acid phosphatase 1) are up-regulated in both genotypes in our study. The induction of acid phosphatases is regarded as a universal symptom/response of P limitation (Duff et al., 1994) and their role in hydrolysing Pi from mono-esters makes them important in intracellular and extracellular Pi salvage systems (Plaxton, 2004).
Signalling mechanisms
The microarray results indicated changes in the expression levels of genes involved in signal transduction that may also be involved in the leaf response to P deprivation. A gene encoding phospholipase C (PLC, EC 3·1·4·3) was significantly up-regulated in the leaf tissue of both genotypes. PLC catalyses the hydrolysis of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] into diacylglycerol (DAG) and inositol triphosphate (IP3), the latter diffusing through the cytosol releasing calcium from intracellular stores or being converted to inositol hexakisphosphate (IP6), both of which may play roles as signalling molecules (Meijer and Munnik, 2003).
We also identified the up-regulation of an ids4-like gene in both genotypes, which has been identified in previous transcriptomic studies as being enhanced under P deficiency (Misson et al., 2005; Guo et al., 2008). The ids4-like gene up-regulated here shares most significant homology with At2g45130 from Arabidopsis that contains an SPX domain, a region in PHO1 genes identified as being important for inorganic phosphate homeostasis (Wang et al, 2004). This gene, AtSPX3, has recently been proposed to play a role in a phosphate-signalling network (Duan et al., 2008). It was shown that when AtSPX3 was repressed by RNAi it resulted in augmentation of P-deficient symptoms and altered allocation of internal P, leading to the conclusion that AtSPX3 plays an important role in plant adaptation to P deficiency (Duan et al., 2008). The high induction of an AtSPX3 homologue in the early stages of P deficiency in both genotypes of our study indicates it may have a similar role to play in adaptation to P deficiency in perennial ryegrass.
Aromatic secondary metabolites
A common physiological response in a wide variety of plants to long-term P limitation is the increase in anthocyanin content in leaf tissue (Hammond et al., 2004) and a number of studies have demonstrated the up-regulation of genes involved in anthocyanin biosynthesis during P deficiency (Hammond et al., 2003; Misson et al., 2005). It is thought that anthocyanin helps protect the photosynthetic machinery during leaf senescence (Hoch et al., 2001). The biosynthesis of anthocyanins involves the shikimate pathway to produce phenylalanine, which eventually leads to flavonoid biosynthesis (Fig. 2). The accumulation of anthocyanin is generally a feature of long-term P deficiency and the majority of the genes are specifically induced during long-term deficiency (Misson et al., 2005). In IRL-OP-02538-P leaves after 24 h of P deficiency, two transcripts involved in phenylalanine and flavonoid biosynthesis (anthranilate phosphoribosyltransferase, EC 2·4·2·18; and flavonol synthase/flavanone 3-hydroxylase, EC 1·14·11·9) were up-regulated, which may suggest the early onset of a secondary metabolite response in this genotype.
Summary
Although most metabolic profiling reports aimed at studying the responses of plants to P deficiency have not been performed at early stages of the response, it seems that a significant degree of metabolic control is evident in the early stages of such response. This was particularly evident in the leaves, suggesting that P sensing mechanisms readily signal to the leaf tissue eliciting a metabolic response. We also observed significant changes in the transcriptome, which pointed to the utilization of glycolytic bypasses, remodelling of lipid membranes and P-scavenging mechanisms under early P deficiency. Although at an early stage, these studies are providing insights at the biochemical and genetic level that will undoubtedly facilitate hypotheses testing via transgenic methodologies and ultimately identify a way forward to improve P-use efficiency in ryegrass by employing candidate genes and marker-assisted breeding approaches.
SUPPLEMENTARY DATA
Supplementary data are available online at w.aob.oxford-journals.org and consist of the following files. (1) Mean values for Pi removal from solution by 34 ecotypes and two cultivars. (2) Primer sequences used in real-time RT-PCR to verify array results. (3) Metabolomics quality control and PCA. (4) Genes significantly regulated (>two-fold, P < 0·05) by P deficiency in leaf tissue. (5) Genes significantly regulated (>two-fold, P < 0·05) by P deficiency in root tissue.
ACKNOWLEDGEMENTS
We thank Tom Shepherd (Scottish Crop Research Institute) for expert technical assistance. This study was financed by the Irish Department of Agriculture, Fisheries and Food under the Stimulus programme (RSF 06-346; S.L.B., A.F. and S.B.). D.S., P.H. and J.M. acknowledge support from the Scottish Government Rural and Environment Research and Analysis Directorate.
LITERATURE CITED
- Anon. Indicators for the integration of environmental concerns into the common agricultural policy. 2000 Brussels: COM(2000) 20 final. [Google Scholar]
- Anon. The United Nations on world population in 2300. Population and Development Review. 2004;30:181–187. doi:10.1111/j.1728-4457.2004.00009.x. [Google Scholar]
- Bar-Or C, Czosnek H, Koltai H. Cross-species microarray hybridizations: a developing tool for studying species diversity. Trends in Genetics. 2007;23:200–207. doi: 10.1016/j.tig.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Benning C. Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annual Review of Cellular and Developmental Biology. 2009;25:71–91. doi: 10.1146/annurev.cellbio.042308.113414. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME) – toward standards for microarray data. Nature Genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Chen X, Hackett CA, Niks RE, et al. An eQTL analysis of partial resistance to Puccinia hordei in Barley. PLoS ONE. 2010;5:e8598. doi: 10.1371/journal.pone.0008598. doi:10.1371/journal.pone.0008598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciannamea S, Busscher-Lange J, de Folter S, Angenent GC, Immink RGH. Characterization of the vernalization response in Lolium perenne by a cDNA microarray approach. Plant and Cell Physiology. 2006;47:481–492. doi: 10.1093/pcp/pcj015. [DOI] [PubMed] [Google Scholar]
- Cui XQ, Churchill GA. Statistical tests for differential expression in cDNA microarray experiments. Genome Biology. 2003;4:210. doi: 10.1186/gb-2003-4-4-210. doi:10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant Journal. 2008;54:965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- Ducreux LJM, Morris WL, Prosser IM, et al. Expression profiling of potato germplasm differentiated in quality traits leads to the identification of candidate flavour and texture genes. Journal of Experimental Botany. 2008;59:4219–4231. doi: 10.1093/jxb/ern264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Moorhead GBG, Lefebvre DD, Plaxton WC. Phosphate starvation inducible ‘bypasses’ of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiology. 1989;90:1275–1278. doi: 10.1104/pp.90.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. The role of acid-phosphatases in plant phosphorus metabolism. Physiologica Plantarum. 1994;90:791–800. [Google Scholar]
- Essigmann B, Guler S, Narang RA, Link D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier RC, Edwards AC. Sustainability of Scottish water quality in the early 21st century. Science of the Total Environment. 2002;294:57–71. doi: 10.1016/s0048-9697(02)00052-9. [DOI] [PubMed] [Google Scholar]
- Foito A, Byrne SL, Shepherd T, Stewart D, Barth S. Transcriptional and metabolic profiles of Lolium perenne L. genotypes in response to a PEG-induced water stress. Plant Biotechnology Journal. 2009;7:719–732. doi: 10.1111/j.1467-7652.2009.00437.x. [DOI] [PubMed] [Google Scholar]
- Foyer C, Spencer C. The relationship between phosphate status and photosynthesis in leaves – effects on intracellular ortho-phosphate distribution, photosynthesis and assimilate partitioning. Planta. 1986;167:369–375. doi: 10.1007/BF00391341. [DOI] [PubMed] [Google Scholar]
- Freedan AL, Rao IM, Terry N. Influence of phosphorus nutrition on growth and carbon partitioning in Glycine max. Plant Physiology. 1989;89:225–230. doi: 10.1104/pp.89.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhang L, Zhao J, Liao H, Zhuang C, Yan X. Identification of temporally and spatially phosphate-starvation responsive genes in Glycine max. Plant Science. 2008;175:574–584. [Google Scholar]
- Gyaneshwar P, Kumar GN, Parekh LJ, Poole PS. Role of soil microorganisms in improving P nutrition of plants. Plant and Soil. 2002;245:83–93. [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, et al. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology. 2003;132:578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ. Genetic responses to phosphorus deficiency. Annals of Botany. 2004;94:323–332. doi: 10.1093/aob/mch156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, Craigon DJ, et al. Using genomic DNA-based probe-selection to improve the sensitivity of high-density oligonucleotide arrays when applied to heterologous species. Plant Methods. 2005;1:10. doi: 10.1186/1746-4811-1-10. doi:10.1186/1746-4811-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, White PJ. Sucrose transport in the phloem: integrating root responses to phosphorus starvation. Journal of Experimental Botany. 2008;59:93–109. doi: 10.1093/jxb/erm221. [DOI] [PubMed] [Google Scholar]
- He Z, Honeycutt CW. A modified molybdenum blue method for orthophosphate determination suitable for investigating enzymatic hydrolysis of organic phosphates. Communications in Soil Science and Plant Analysis. 2005;36:1373–1383. [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N. How do plants respond to nutrient shortage by biomass allocation? Trends in Plant Science. 2006;11:610–617. doi: 10.1016/j.tplants.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hernandez G, Ramirez M, Valdes-Lopez O, et al. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiology. 2007;144:752–767. doi: 10.1104/pp.107.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch WA, Zeldin EL, McCown BH. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiology. 2001;21:1–8. doi: 10.1093/treephys/21.1.1. [DOI] [PubMed] [Google Scholar]
- Huang CY, Roessner U, Eickmeier I, et al. Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.) Plant and Cell Physiology. 2008;49:691–703. doi: 10.1093/pcp/pcn044. [DOI] [PubMed] [Google Scholar]
- Humphreys MO. Genetic improvement of forage crops – past, present and future. Journal of Agricultural Science. 2005;143:441–448. [Google Scholar]
- Kaida R, Satoh Y, Bulone V, et al. Activation of beta-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiology. 2009;150:1822–1830. doi: 10.1104/pp.109.139287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D. Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant and Cell Physiology. 2000;41:1030–1037. doi: 10.1093/pcp/pcd029. [DOI] [PubMed] [Google Scholar]
- Lauer MJ, Pallardy SG, Blevins DG, Randall DD. Whole leaf carbon exchange characteristics of phosphate deficient soybeans (Glycine max L) Plant Physiology. 1989;91:848–854. doi: 10.1104/pp.91.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJG, Munnik T. Phospholipid-based signaling in plants. Annual Review of Plant Biology. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Didion T, Andersen CH, Nielsen KK. MADS-box genes from perennial ryegrass differentially expressed during transition from vegetative to reproductive growth. Journal of Plant Physiology. 2004;161:439–447. doi: 10.1078/0176-1617-01212. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research. 2002;30:e36. doi: 10.1093/nar/30.9.e36. Nucleic Acids Research 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC. Plant response to stress: biochemical adaptations to phosphate deficiency. In: Goodman R, editor. Encyclopedia of plant and crop science. New York, NY: Marcel Dekker; 2004. pp. 976–980. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annual Review of Plant Biology. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE. Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant Journal. 2001;25:641–649. doi: 10.1046/j.1365-313x.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant and Soil. 2009;321:305–339. [Google Scholar]
- Ramirez-Rodriguez V, Lopez-Bucio J, Herrera-Estrella L. Adaptive responses in plants to nonoptimal soil pH. In: Jenks MA, Hasegawa PM, editors. Plant abiotic stress. Oxford: Blackwell Publishing; 2005. pp. 145–170. [Google Scholar]
- Rodriguez D, Zubillaga MM, Ploschuk EL, Keltjens WG, Goudriaan J, Lavado RS. Leaf area expansion and assimilate production insunflower (Helianthus annuus L.) growing under low phosphorus conditions. Plant and Soil. 1998;202:133–147. [Google Scholar]
- Scheible WR, Pauly M. Glycosyltransferases and cell wall biosynthesis: novel players and insights. Current Opinion in Plant Biology. 2004;7:285–295. doi: 10.1016/j.pbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Schindler DW, Hecky RE, Findlay DL, et al. Eutrophication of lakes cannot be controlled by reducing nitrogen input: results of a 37-year whole-ecosystem experiment. Proceedings of the National Academy of Sciences USA. 2008;105:11254–11258. doi: 10.1073/pnas.0805108105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CY, Kao CH. Growth inhibition in suspension-cultured rice cells under phosphate deprivation is mediated through putrescine accumulation. Plant Physiology. 1996;111:721–724. doi: 10.1104/pp.111.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L. Biochemistry. 4th edn. New York: WH Freeman; 1995. [Google Scholar]
- Theodorou ME, Plaxton WC. Metabolic adaptations of plant respiration to nutritional phosphate deprivation. Plant Physiology. 1993;101:339–344. doi: 10.1104/pp.101.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou ME, Cornel FA, Duff SM, Plaxton WC. Phosphate starvation-inducible synthesis of the alpha-subunit of the pyrophosphate-dependent phosphofructokinase in black mustard suspension cells. Journal of Biological Chemistry. 1992;267:21901–21905. [PubMed] [Google Scholar]
- Tilman D. The greening of the green revolution. Nature. 1998;396:211–212. [Google Scholar]
- Uhde-Stone C, Gilbert G, Johnson JMF, et al. Acclimation of white lupin to phosphorus deficiency involves enhanced expression of genes related to organic acid metabolism. Plant and Soil. 2003a;248:99–116. [Google Scholar]
- Uhde-Stone C, Zinn KE, Ramirez-Yanez M, Li A, Vance CP, Allan DL. Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to phosphorus deficiency. Plant Physiology. 2003b;131:1064–1079. doi: 10.1104/pp.102.016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari DA. Phosphorus: a looming crisis. Scientific American. 2009;300:42–47. doi: 10.1038/scientificamerican0609-54. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist. 2003;157:423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiology. 2004;135:400–411. doi: 10.1104/pp.103.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki J, Yonetani R, Kuroda S, et al. Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant, Cell and Environment. 2003;26:1515–1523. [Google Scholar]
- Wasaki J, Shinano T, Onishi K, et al. Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. Journal of Experimental Botany. 2006;57:2049–2059. doi: 10.1093/jxb/erj158. [DOI] [PubMed] [Google Scholar]
- Wassen MJ, Venterink HO, Lapshina ED, Tanneberger F. Endangered plants persist under phosphorus limitation. Nature. 2005;437:547–550. doi: 10.1038/nature03950. [DOI] [PubMed] [Google Scholar]
- Wissuwa M, Gamat G, Ismail AM. Is root growth under phosphorus deficiency affected by source or sink limitations? Journal of Experimental Botany. 2005;56:1943–1950. doi: 10.1093/jxb/eri189. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30:e15. doi: 10.1093/nar/30.4.e15. e15 doi:10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proceedings of the National Academy of Sciences USA. 2002;99:5732–5737. doi: 10.1073/pnas.082696499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.