Abstract
Background and Aims
The genus Fritillaria (Liliaceae) comprises species with extremely large genomes (1C = 30 000–127 000 Mb) and a bicontinental distribution. Most North American species (subgenus Liliorhiza) differ from Eurasian Fritillaria species by their distinct phylogenetic position and increased amounts of heterochromatin. This study examined the contribution of major repetitive elements to the genome obesity found in Fritillaria and identified repeats contributing to the heterochromatin arrays in Liliorhiza species.
Methods
Two Fritillaria species of similar genome size were selected for detailed analysis, one from each phylogeographical clade: F. affinis (1C = 45·6 pg, North America) and F. imperialis (1C = 43·0 pg, Eurasia). Fosmid libraries were constructed from their genomic DNAs and used for identification, sequence characterization, quantification and chromosome localization of clones containing highly repeated sequences.
Key Results and Conclusions
Repeats corresponding to 6·7 and 4·7 % of the F. affinis and F. imperialis genome, respectively, were identified. Chromoviruses and the Tat lineage of Ty3/gypsy group long terminal repeat retrotransposons were identified as the predominant components of the highly repeated fractions in the F. affinis and F. imperialis genomes, respectively. In addition, a heterogeneous, extremely AT-rich satellite repeat was isolated from F. affinis. The FriSAT1 repeat localized in heterochromatic bands makes up approx. 26 % of the F. affinis genome and substantial genomic fractions in several other Liliorhiza species. However, no evidence of a relationship between heterochromatin content and genome size variation was observed. Also, this study was unable to reveal any predominant repeats which tracked the increasing/decreasing trends of genome size evolution in Fritillaria. Instead, the giant Fritillaria genomes seem to be composed of many diversified families of transposable elements. We hypothesize that the genome obesity may be partly determined by the failure of removal mechanisms to counterbalance effectively the retrotransposon amplification.
Keywords: Fritillaria, Liliaceae, repetitive DNA, transposable elements, retrotransposon, heterochromatin, satellite repeats, chromosomes, genome size variation
INTRODUCTION
The genus Fritillaria (Liliaceae) has long held a special place in plant science, first as a pioneering cytogenetic model for the study of meiosis, mitosis and chromosome structure due to its exceptionally large chromosomes (Darlington, 1935, 1937; Frankel, 1940; Darlington and La Cour, 1941) and, second, as the genus containing some of the largest plant genomes so far reported (i.e. tetraploid Fritillaria assyriaca, 1C = 127·4 pg; Bennett and Smith, 1976). Recently, also other Liliales genera (Paris, Trillium) have been shown to harbour species with giant nuclear genomes (Pellicer et al., 2010; Zonneveld, 2010).
The genus Fritillaria comprises approx. 100 geophytic species found in the temperate zones of the northern hemisphere. It is considered to have diverged from the sister genus Lilium 2–14 Mya (Patterson and Givnish, 2002) and subsequently diversified to give rise to its current, bicontinental distribution. Around 20 species are found in North America (centred in California) while the remainder are distributed across Eurasia (from the Iberian Peninsula to Japan). A single species (F. camschatcensis) links both groups by its distribution in North America and eastern Asia (Japan and Siberia).
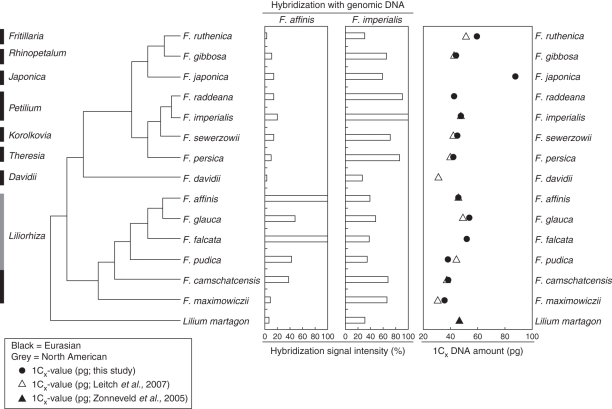
Recently, the bicontinental distribution of the genus has been shown to reflect the evolutionary relationships between species. Sequence analysis of three gene regions across 34 Fritillaria species (Rønsted et al., 2005) revealed the genus to be divided into two clades: (1) a predominantly North American clade comprising species belonging to subgenus Liliorhiza (sensu Rix, 2001) and (2) a Eurasian clade containing the remaining species (Fig. 1).
Fig. 1.
Phylogenetic relationships of selected species according to Rønsted et al. (2005) with their genome size and dot-blot hybridizations of total DNAs from these Fritillaria species using F. affinis and F. imperialis genomic DNAs as a probe. Genome size data for Lilium martagon taken from Zonneveld et al. (2005), otherwise C-values are estimated from this study or correspond to data published by Leitch et al. (2007), as indicated. Cross-hybridization signals of genomic DNAs were related to hybridization intensity of the species used as a probe (100 %).
In addition, a recent analysis of genome size variation and evolution in 23 Fritillaria species has extended our knowledge of genome size data for this genus, revealing a four-fold infrageneric variation in genome size (mean 1C = 50·9 pg). Superimposing these data onto the phylogenetic tree of Rønsted et al. (2005) suggests that there have been two independent increases in genome size, one in each clade (Leitch et al., 2007; Fig. 1). The largest monoploid genomes were identified in species of the subgenus Japonica (two species, Leitch et al., 2007; present study) endemic to Japan.
Despite the significant genome size variation reported in Leitch et al. (2007), Fritillaria karyotypes are surprisingly stable. Most species are diploids (2n = 24) with a uniform karyotype (Darlington, 1937). Only a few species possess different chromosome numbers, i.e. 2n = 26 (two species), 2n = 22 (two) and 2n = 18 (three) (Darlington, 1937; Noda, 1964; Li and Shang, 1989), and these are considered to be derived from the presumably ancestral 12 chromosome pairs via fusion or fission events (Darlington, 1935, 1937). Polyploidy within the genus is virtually non-existent and only triploid plants are repeatedly found (Darlington, 1937; Marchant and Macfarlane, 1980; Peruzzi et al., 2009). Heterochromatin content has been shown to be variable between the species. Early studies using Feulgen staining revealed conspicuous differences in the amount of heterochromatic bands in the North American (subgenus Liliorhiza) versus the Eurasian species (La Cour, 1951). Although the heterochromatin content of North American species was variable (range 15–50 %), its proportion was always greater than that found in Eurasian species where typically the heterochromatin content was less than 5 % (La Cour, 1951).
Ever since the genome size record for F. assyriaca was published by Bennett and Smith (1976), its genome size has been widely quoted in textbooks. However, insights into the composition, organization and evolution of the DNA sequences which comprise the enormous Fritillaria genomes remain largely unknown. To address the gap in our knowledge, this paper aims to provide the first comparative study of the repetitive DNA composition of Fritillaria species. We selected two Fritillaria species of similar genome size and chromosome number for the analysis, one from each phylogeographical clade: F. affinis (1C = 45·6 pg, 2n = 24, from subgenus Liliorhiza, North America) and F. imperialis (1C = 43·0 pg, 2n = 24, from subgenus Petillium, Eurasia). Screening of partial genomic DNA libraries of the two species and restriction digestion-based analysis of genomic DNAs were employed to identify the most abundant dispersed and tandem repeats, respectively. The occurrence, distribution and copy numbers of highly repeated sequences isolated from both species were also investigated in a further nine species, selected to cover the phylogenetic diversity of Fritillaria (Fig. 1 and Table 1). Together, these data are used to address the following questions. (1) What is the nature of genome obesity in Fritillaria? Are these genomes composed of a (high) number of different repeat families, or are there only a few repeats amplified to extremely high copy numbers? (2) What are the main components of heterochromatin chromosome bands in Fritillaria species?
Table 1.
List of Fritillaria and Lilium species used in the present study, including infrageneric classification, origin, chromosome number and genome size; species used for dot-blot quantification of dispersed repetitive elements are included
| Species | Geographical origin* | Origin/source | Chromosome number (2n)‡ | 1C-value (pg; ± s.d.) (this study) | 1C-value (pg) (Leitch et al., 2007) |
|---|---|---|---|---|---|
| Subgenus Fritillaria | |||||
| F. ruthenica Wikst. | EA | RBG, Kew (2004-3479) | 18 | 59·0 ± 0·4 | 51·1 |
| Subgenus Liliorhiza | |||||
| F. affinis (Schult.) Sealy | NW | RBG, Kew (2010-905)†¶ | 24 | 45·7 ± 0·5¶ | 45·6 |
| RBG, Kew (2000-1744) | |||||
| F. camschatcensis (L.) Ker Gawler subsp. camschatcensis | NW/EA | RBG, Kew (2007-2037) | 36 | 57·6 (Cx = 38·4) ± 0·8 | 56·1 (Cx = 37·4) |
| F. falcata (Jepson) D. E. Beetle¶ | NW | L. Hill (112) | 24 (lit.) | 51·9 ± 0·7 | |
| F. glauca Greene | NW | RBG, Kew (1967-59504) | 24 | 53·6 ± 0·7 | 48·9 |
| F. maximowiczii Freyn¶ | EA | RBG, Kew (2005-2043) | 24 (lit.) | 35·6 ± 0·1 | 30·8 |
| F. pudica (Pursh) Sprengel | NW | RBG, Kew (2x: 1986-6110; 3x: 2004-3477†) | 2x: 24, 3x: 36 | 2x: 38·3 ± 0·9 (=Cx) | 3x: 66·4 (Cx = 44·3) |
| Subgenus Petilium | |||||
| F. imperialis L. ‘Maxima Lutea’ | EA | RBG, Kew | 24 + 4 B | 47·2 ± 0·1 | 47·4 |
| F. raddeana Regel¶ | EA | RBG, Kew (1969-6107) | 24 | 42·7 ± 1·1 | |
| Subgenus Korolkowia | |||||
| F. sewerzowii Regel | EA | RBG, Kew (2010-916) RBG, Kew (1995-4397)† | approx. 24 | 44·8 ± 0·4 | 42·3 |
| Subgenus Theresia | |||||
| F. persica L. ‘Adiyaman’ | EA | RBG, Kew (2005-2033; 2003-81†) | 24 | 42·0 ± 0·4 | 39·9 |
| Subgenus Japonica | |||||
| F. japonica Miq. | EA | L. Hill (323); RBG, Kew (1981-5579)† | 22 (lit.) | 87·3 ± 0·0 | |
| Subgenus Rhinopetalum | |||||
| F. gibbosa Boiss. | EA | RBG, Kew (2004-3469) | 24 | 43·9 ± 0·3 | 42·4 |
| Subgenus Davidii | |||||
| F. davidii Franch. | EA | RBG, Kew (2004-3461) | approx. 24 | 31·1 | |
| Genus Lilium | |||||
| L. martagon | EA | RBG, Kew (1978-2452) | 24 (lit.) | 46·6§ | |
* EA, Eurasia; NW, New World.
†Accession used for genomic work, otherwise the same accessions were used for genomic work and genome size estimation (RBG, Kew = Royal Botanic Gardens, Kew; L. Hill = accessions acquired from Laurence Hill).
‡Chromosome numbers counted by us or taken from the literature (lit.).
§Genome size taken from Zonneveld et al. (2005).
¶Species that were not used for copy number estimation of repetitive elements by dot-blot hybridization.
MATERIALS AND METHODS
Plant material and genome size estimation
The origin of Fritillaria L. species analysed in the present study is given in Table 1. Genome size was estimated for all but one (F. davidii) Fritillaria species using a CyFlow SL flow cytometer (Partec, Görlitz, Germany). Small amounts of fresh leaf tissue of sample and internal standard (onion Allium cepa ‘Ailsa Craig’, 2C = 33·5 pg; Van't Hof, 1965) were chopped in 1 mL of Tris/MgCl2buffer (Pfosser et al., 1995) to which 100 µL of propidium iodide (1 mg mL−1) was added. A further 1 mL of buffer was added and the suspension was then filtered through a 50-μm nylon mesh and analysed. Due to the large genome sizes of Fritillaria, the G2 peak of onion was used to calculate the fluorescence intensity ratio between sample and standard. When sample and onion peaks overlapped, a different Fritillaria species was used as a secondary internal standard. In this case, results were consequently recalculated using ratios between C-values of the secondary standard and onion. Each species was measured three times on different days and the results were averaged. The average coefficient of variance of all peaks used for measurements was 3·97 %.
Assessing global repeat similarity between Fritillaria species
Serial dilutions (10, 1, 0·1 ng) of genomic DNA from 14 Fritillaria species (see Table 1 and Fig. 1) were quantitatively dot-blotted onto Hybond-N+ membranes (Amersham Biosciences, Uppsala, Sweden). The membranes were probed with genomic DNA from either F. affinis or F. imperialis which was radioactively labelled with [α-32P]ATP (MP Biomedicals) using the DecaLabel™ DNA Labeling Kit (Fermentas, St. Leon-Rot, Germany). Hybridization in buffer containing 0·25 m Na phosphate, 7 % SDS and 10 mm EDTA and washing in 2× SSC (1× SSC = 0·15 m NaCl, 0·015 m Na 3-citrate) and 0·1 % SDS were performed at 50 °C. Probe signals were detected using a Fujifilm FLA-7000 phosphoimager (Fuji Photo Film Co., Tokyo, Japan).
Construction of fosmid libraries and sequencing
The libraries were constructed for F. affinis and F. imperialis using a Copy Control Fosmid Library Production Kit (Epicentre, Madison, WI, USA). High-molecular-weight genomic DNA was prepared from isolated nuclei as described by Macas et al. (2007), mechanically sheared and approx. 40-kb fragments were purified from agarose gel and cloned into a pCC2FOS vector. Resulting colonies were picked into 384-well plates (36 plates for each library) and stored at –80 °C. The libraries were gridded onto hybridization membranes using a GeneTAC G3 robot (Genomic Solutions, Huntington, UK) and sequentially screened by colony hybridization using sonicated genomic DNAs of both Fritillaria species as probes to reveal clones containing shared or species-specific abundant repetitive sequences. Colony hybridization screening was performed with the AlkPhos Direct™ hybridization kit (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer's protocol. The temperature of hybridization and washing was 55 °C and probe hybridization was detected using CDP-Star substrate and signals were captured on X-ray film. Four clones giving strong signal from each library were selected, subcloned into randomly fragmented shotgun libraries using dephosphorylated Sma I-cut pBluescript II SK+ as a vector and sequenced.
Sequence analysis
Sequences were processed and assembled using the Staden software package (Staden, 1996). Dot-plot comparisons and multiple sequence comparisons were performed with Dotter (Sonnhammer and Durbin, 1995) and ClustalW software (Thompson et al., 1994), respectively. Classification of long terminal repeat (LTR) retrotransposons into distinct lineages was done based on phylogenetic analyses of the reverse transcriptase (RT) protein sequences. Alignments of RT sequences were carried out with Muscle software (Edgar, 2004) and phylogenetic trees were constructed via the neighbour-joining method. The trees were drawn and edited using the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/). Similarity searches were performed using BLAST (Altschul et al., 1997), and similarities to Repbase (Jurka et al., 2005) were found using Censor (Kohany et al., 2006; http://www.girinst.org/censor/index.php). Conserved protein domains were found by RPS-BLAST (Marchler-Bauer et al., 2003; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Sequences of tRNA used for identification of primer binding sites were obtained from the Arabidopsis thaliana tRNA database (Lowe and Eddy, 1997).
Isolation and characterization of satellite repeat FriSAT1
A search for satellite repeats was performed by digesting genomic DNA of ten Fritillaria species (F. affinis, F. camschatcensis, F. davidii, F. gibbosa, F. gussichiae, F. imperialis, F. persica, F. pudica, F. sewerzowii and F. thunbergii) with the following restriction endonucleases: AfaI, AluI, HaeIII, MboI, MseI, MspI, NlaIII and TaqI (4-cutters); BamHI, BclI, BglII, DraI, EcoRI, EcoRV, HindIII, KpnI, PstI, SacI, SacII, XhoI and XbaI (6-cutters). The high-molecular-weight ‘relic’ band in MboI-digested F. affinis DNA was purified from the gel using Wizard SV Gel and PCR Clean-Up kit (Promega, Madison, WI, USA). Degenerate oligonucleotide primed (DOP)-PCR amplification of the DNA isolated from the band was performed with primer Frit-W6-Mse (5′GGCACCTGCAGAATACWWWWWWTTAA3′) for ten cycles of 94 °C for 1 min, 30 °C for 1·5 min, transition to 72 °C at 0·2 °C s−1, 72 °C for 3 min, preceeded by denaturation (92 °C for 2 min) and followed by incubation at 72 °C for 10 min. The reaction product was then re-amplified using the nested primer (5′TCGACGGCACCTGCAGAATAC3′) for 30 cycles with an annealing temperature of 50 °C. Purified PCR products were cut with PstI (recognition site was present in primer sequences), cloned into plasmid vectors and screened by colony hybridization with F. affinis genomic DNA. Selected clones giving strong hybridization signals were sequenced.
A DOP-PCR clone c1063 (GenBank accession number GU182252) carrying the satellite sequence (FriSAT1) was used as a probe for screening F. affinis short-insert plasmid libraries to retrieve genuine genomic sequences of this satellite. The libraries were constructed from fragments of 600–800 bp and 900–1200 bp, which were gel-purified from DraI digests of genomic DNA. Four positive clones were identified and sequenced. The selected size-ranges were previously found to be enriched for FriSAT1 repeats based on Southern hybridization of F. affinis genomic DNA digested with DraI.
Copy number estimations
To estimate copy number of selected repetitive elements in 11 Fritillaria species and Lilium martagon (see Table 1), serial dilutions of their genomic DNA (5 × 102 to 1·2 × 104copies of the monoploid genome given as Cx-value) were dot blotted onto Hybond-N+ membrane together with dilutions of the particular probe itself as a hybridization standard (1 × 106 to 3 × 109copies). For the probes, radioactively [α-32P] ATP-labelled restriction fragments of retrotransposon RT-domains and DNA-transposon transposase were used. Quantification of the repetitive elements was performed at the same stringency, whereby the hybridization temperature and salt concentration were calculated according to the G + C content of the probe, allowing up to 15 % of mismatches. Prehybridization and hybridization steps were carried out in hybridization buffer (0·25 m Na phosphate, 7 % SDS, 10 mm EDTA) followed by washing in 0·2× or 2× SSC with 0·1 % SDS. In all cases, probes were labelled using the DecaLabelTM DNA Labeling Kit following the manufacturer's instructions. Results were visualized using a Fujifilm FLA-7000 phosphoimager and quantified using the MultiGauge software (Fuji Photo Film Co.).
Quantification of the FriSAT1 satellite in various Fritillaria species (Table 1) was carried out using serial dilutions of genomic DNA (500, 100, 50, 5 and 1 ng) and c1063 PCR clone (2·5, 1, 0·5, 0·25, 0·1 and 0·05 ng). Based on these hybridization standards the approximate percentage of the satellite in each genome was estimated using the same buffers as above, allowing up to 20 % of mispairing. The radioactively [α-32P] ATP-labelled restriction fragment from the PstI-cut c1063 PCR clone was used as a probe.
Alternatively, estimations of repeat copy numbers in F. affinis and F. imperialis genomes were based on the number of positive clones observed after screening the fosmid libraries using the same probes and conditions as in dot-blot hybridizations. Copy numbers were calculated using a formula described in Neumann et al. (2006).
Fluorescence in situ hybridization (FISH)
Actively growing, young roots were pretreated with a saturated solution of alpha-bromo naphthalene for 24 h at 4 °C and stored in 70 % ethanol at –20 °C until use. Selected root tips were rinsed in distilled water (twice for 5 min) and citrate buffer (10 mm sodium citrate, pH 4·8; twice for 5 min), and digested in 0·3 % (w/v) cellulase, cytohelicase and pectolyase (all Sigma-Aldrich, St Louis, MO, USA) in citrate buffer at 37 °C for 90 min. After digestion, individual root tips were dissected on a microscope slide in approx. 10 µL acetic acid and covered with a cover slip. The cell material was then evenly spread using tapping, thumb pressing and gentle flame-heating. Finally, the slide was quick frozen in liquid nitrogen and the cover slip flicked off with a razor blade. Slides were fixed in ethanol/acetic acid (3 : 1) and air-dried. Prior to FISH, slides were incubated in RNase (AppliChem, Darmstadt, Germany; 100 µg mL−1in distilled water) at 37 °C for 1 h, and washed in 2× SSC twice for 5 min. To remove cytoplasm, slides were treated with pepsin (Sigma-Aldrich; 0·1 g L−1in 0·01 m HCl) for 5 min and washed in 2× SSC twice for 5 min. Subsequently, slides were post-fixed in 4 % formaldehyde in 2× SSC (v/v) for 10 min, washed in 2× SSC twice for 5 min, dehydrated in an ethanol series (70, 80 and 96 %) and air-dried.
Probes were labelled with either biotin-, digoxigenin- or Cy3-dUTP by nick translation as described previously (Mandáková and Lysak, 2008). Labelled probes (approx. 200–500 bp) were ethanol-precipitated and dissolved in 20 µL hybridization mix of 50 or 20 % (FISH with the clone c1063) formamide and 10 % dextran sulfate in 2× SSC per slide. The probe and chromosome preparations were denatured together on a hot plate at 80 °C for 2 min and incubated in a moist chamber at 37 °C overnight. Post-hybridization washing was performed in 20 % formamide in 2× SSC at 42 °C. Detection of hybridization signals was carried out as described previously (Mandáková and Lysak, 2008). Chromosomes were counterstained with 0·5 µg mL−1DAPI in Vectashield (Vector Laboratories, Peterborough, UK). Preparations were analysed using an Olympus BX-61 epifluorescence microscope and AxioCam CCD camera (Zeiss, Jena, Germany). The monochromatic images were pseudocoloured and merged using Adobe Photoshop CS2 software (Adobe Systems).
RESULTS
Genome size and global repeat similarity of Fritillaria species
As reliable genome size estimates were unavailable for three Fritillaria species analysed, we estimated their DNA amount by flow cytometry and combined these data with C-values published by Leitch et al. (2007) (Table 1). For ten species genome sizes were also estimated by us and compared with C-values previously estimated using Feulgen microdensitometry in Leitch et al. (2007). Although there was generally good agreement between the estimates obtained using both methods, in a few cases (e.g. F. ruthenica and F. maximowiczii) the values differed more substantially. Overall, monoploid genome size varied 2·5-fold across the 14 Fritillaria species studied (Cx = 31·1–87·3 pg) (Table 1 and Fig. 1).
To cover the phylogenetic diversity of the genus Fritillaria, 14 species belonging to eight recognized subgenera (Table 1 and Fig. 1) were analysed to determine the composition of highly repeated genomic components isolated from F. affinis and F. imperialis. To get an overview of global repeat similarity among the Fritillaria species analysed, dot-blot hybridizations of labelled F. affinis and F. imperialis genomic DNAs to genomic DNAs of all investigated species were carried out (Fig. 1). Using F. affinis as a probe gave a strong hybridization signal for all species of subgenus Liliorhiza analysed with the exception of F. maximowczii which showed only weak hybridization. F. affinis and F. falcata had the strongest hybridization signals, whereas those of F. camschatcensis, F. glauca and F. pudica were weaker. All Eurasian species tested showed only weak hybridization signal, similar in strength to Lilium martagon, which was used as an outgroup. The same experiment but using F. imperialis genomic DNA as a probe gave a rather different pattern. Whereas the probe hybridized most strongly to the majority of Eurasian species (except F. ruthenica and F. davidii), significant signal was also observed on some species in subgenus Liliorhiza, especially F. camschatcensis and F. maximowiczii, which are the only species in subgenus Liliorhiza analysed which occur in Eurasia.
Sequence analysis of fosmid clones carrying highly repeated genomic elements
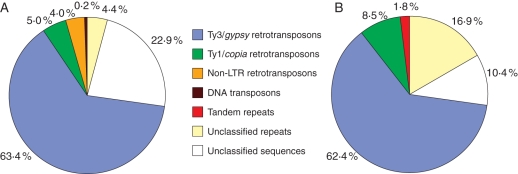
Libraries of fosmid clones carrying genomic DNA fragments of approx. 40 kb were constructed from F. affinis and F. imperialis and sequentially screened with genomic DNA of both species to identify clones containing repetitive genomic elements. Four clones were selected from each fosmid library based on their strong hybridization to the genomic DNA probes from their species of origin. Clones were completely or partially sequenced and this yielded over 140 kb of sequence data per species which was analysed for the presence of repetitive DNA. Overall, identifiable repetitive elements comprised 77·1 and 89·6 % of the F. affinis and F. imperialis clones, respectively, with LTR retrotransposons representing the dominant class of repeats (Fig. 2). The only other repeat types found in F. affinis fosmid clones were a single non-LTR retrotransposon and part of an En/Spm type of DNA transposon whereas in F. imperialis a short array of a tandem repeat was identified. Supplementary Data Fig. S1 (available online) shows detailed annotation of the sequenced fosmid clones.
Fig. 2.
Proportion of different classes of repetitive elements identified in 140 kb of DNA sequenced from four fosmid clones of F. affinis (A) and F. imperialis (B).
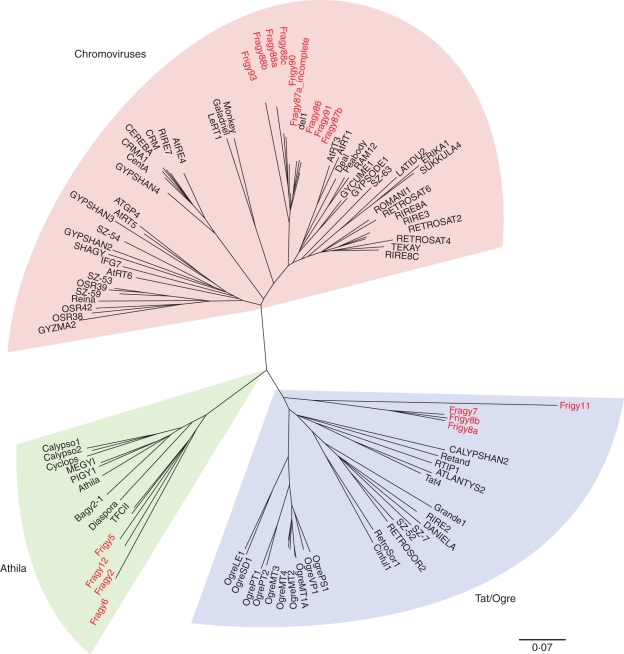
From a phylogenetic analysis of their RT domains, the identified Ty3/gypsy elements were shown to belong to three major lineages of plant Ty3/gypsy retrotransposons, namely Athila, Tat and chromoviruses (Fig. 3; Table 2). Fritillaria elements from the lineage of chromoviruses were found to be phylogenetically very close to retrotransposon del 1–46 which was isolated from the closely related Lilium henryi (Smyth et al., 1989). However, at the DNA sequence level the similarity between Fritillaria elements and del 1–46 was very low (data not shown), suggesting that they are not new members of the del family but rather represent novel retrotransposon families. Fritillaria elements from the Athila lineage revealed significant similarity to retrotransposon TFCII found in F. camschatcensis (GenBank accession numbers AF219201·1 to AF219220·1). As the similarity was significant at both the protein and DNA level (up to approx. 80 %), these sequences presumably diverged from a common ancestral element relatively recently.
Fig. 3.
Neighbour-joining tree of Ty3/gypsy-type retrotransposons inferred from alignment of reverse transcriptase protein domains. The elements identified in F. affinis and F. imperialis are highlighted. Classification of lineages is according to Llorens et al. (2008).
Table 2.
Characterization of Ty3/gypsy and Ty1/copia class LTR retrotransposons identified in this study
| Name (according to Wicker et al., 2007) | Species of origin | Parts identified* | Length (bp)* | Percentage identity of LTRs† | |
|---|---|---|---|---|---|
| Ty3/gypsy class: Athila lineage | |||||
| Fragy1¶ | RLG_Athila_GU188675-1 | F. affinis | 2 × LTR, cs | 14 979 | |
| Frigy5 | RLG_Athila_GU188679-1 | F. imperialis | 1 × LTRput, tr_cs | 5442/6644put | |
| Fragy6 | RLG_Athila_GU188680-1 | F. affinis | tr_cs | 3819/4640put | |
| Fragy2 | RLG_Athila_GU188676-1 | F. affinis | 2 × tr_LTR, tr_cs | 9324 | |
| Fragy12 | RLG_Athila_GU188676-2 | F. affinis | tr_cs | 4249 | |
| Ty3/gypsy class: Tat lineage | |||||
| Frigy3¶ | RLG_Tat_GU188678-1 | F. imperialis | 2 × LTR, cs | 11 141 | 81·94 |
| Fragy7 | RLG_Tat_GU188680-1 | F. affinis | tr_cs | 3884/4574put | |
| Frigy8a | RLG_Tat_GU188681-2 | F. imperialis | 2 × LTR, cs | 18 216 | 77·58 |
| Frigy8b | RLG_Tat_GU188681-1 | F. imperialis | 1 × LTR, 2 × tr_cs | 14 260 | |
| Frigy11 | RLG_Tat_GU188682-1 | F. imperialis | tr_cs | 889 | |
| Ty3/gypsy class: chromoviruses | |||||
| Fragy86 | RLG_chromovirus_GU188675-1 | F. affinis | 2 × LTR, cs | 11 054 | 93·87 |
| Fragy87a | RLG_chromovirus_GU188676-1 | F. affinis | 1 × LTR, tr_cs | 5438 | 98·62‡ |
| Fragy87b | RLG_chromovirus_GU188676-2 | F. affinis | 1 × LTR, cs | 7848 | |
| Fragy88a | RLG_chromovirus_GU188677-1 | F. affinis | 1 × LTR, tr_cs | 5706 | 92·54§ |
| Fragy88b | RLG_chromovirus_GU188677-2 | F. affinis | 1 × LTR, cs | 7136 | |
| Fragy88c | RLG_chromovirus_GU188677-3 | F. affinis | 1 × LTR, cs | 7266 | |
| Frigy90 | RLG_chromovirus_GU188679-1 | F. imperialis | 1 × tr_LTR, tr_cs | 5349 | |
| Fragy91 | RLG_chromovirus_GU188680-1 | F. affinis | 2 × LTR, cs | 11 043 | 96·93 |
| Frigy93 | RLG_chromovirus_GU188682-1 | F. imperialis | 2 × LTR, cs | 9099 | |
| Frigy10¶ | RLG_chromovirus_GU188682-1 | F. imperialis | 2 × LTR, cs | 8553 | 88·47 |
| Ty3/gypsy class: not assigned | |||||
| Fragy4¶ | RLG_x_GU188678-1 | F. affinis | 2 × LTR, cs | 8664 | 89·08 |
| Frigy9¶ | RLG_x_GU188681-1 | F. imperialis | 2 × LTR, cs | 8110 | 80·9 |
| Ty1/copia class: Tos 17-like lineage | |||||
| Frico2 | RLG_Tos17_GU188678-1 | F. imperialis | tr_cs | 3335 | |
| Frico2b¶ | RLG_Tos17_GU188678-2 | F. imperialis | 1 × LTRput, tr_cs | 791/4438put | |
| Frico4 | RLG_Tos17_GU188682-1 | F. imperialis | 1 × LTRput, tr_cs | 3184/4659put | |
| Ty1/copia class: Tnt 1-like lineage | |||||
| Fraco3 | RLG_Tnt1_GU188680-1 | F. affinis | tr_cs | 1492/4430put | |
| Ty1/copia class: not assigned | |||||
| Fraco1¶ | RLG_x_GU188677-1 | F. affinis | tr_cs | 628/±3000put | |
We have designated the gypsy elements as Fragy and Frigy, respectively, derived from FRitillaria Affinis/Imperialis GYpsy elements, and Fraco and Frico elements (FRitillaria Affinis/Imperialis COpia elements).
* put, putative; cs, coding sequence; tr_cs, truncated coding sequence.
†Percentage sequence similarity between two LTRs of a single element.
‡Percentage sequence similarity to Fragy87b LTR region.
§Percentage sequence similarity to Fragy88c LTR region.
¶As RT-domain was not identified, the element could not be included in the phylogenetic analysis.
Ty1/copia retrotransposons included only three sequences possessing RT-coding domains. Phylogenetic analyses of respective protein sequences showed that they belonged to two distinct lineages (Supplementary Data Fig. S2, available online). Frico2 and Frico4 shared significant similarity and appeared phylogenetically close to the rice element Tos17. Fraco3 shared homology with a lineage containing the previously described element Tnt1 from tobacco (Grandbastien et al., 1989) and Angela from Triticum monococcum (Wicker et al., 2001).
Identification of a satellite repeat constituting heterochromatic bands of F. affinis chromosomes
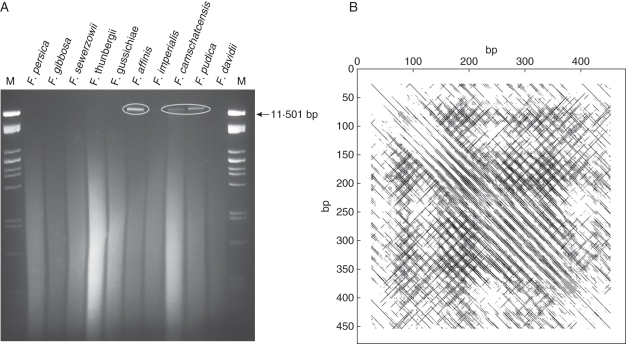
Although the New World species differ from the Eurasian ones by containing a higher proportion of heterochromatin (La Cour, 1951; present study – see Discussion), an analysis of the highly repeated genomic elements in the F. affinis fosmid clones failed to identify any repetitive elements associated with heterochromatic regions. A search for satellite repeats, which typically constitute heterochromatic chromosomal bands, was therefore undertaken by restriction digestion of genomic DNA from ten Fritillaria species selected from both phylogeographical clades (Fig. 4A). None of the 21 restriction endonucleases tested generated a ladder-like pattern of genomic DNA fragments indicative of the presence of highly amplified tandem repeats (data not shown). However, a high-molecular-weight ‘relic’ band was observed in F. affinis, F. camschatcensis and F. pudica after digestion with several restriction enzymes (Fig. 4A). These enzymes mostly recognize 4-bp restriction sites, indicating that the relic band comprises low-complexity sequences. The band was isolated from a gel of MboI-digested F. affinis DNA and amplified by a modified DOP-PCR to facilitate its cloning by increasing the amount of isolated DNA and proportion of shorter fragments (see Methods). Following cloning and sequencing of the amplified DNA, one clone (c1063) was identified as a major component of the relic band based on its hybridization to the genomic DNA digests and the structure of its sequence. The clone possessed a relatively irregular structure of tandemly arranged palindromic motifs which were extremely AT-rich (87 % A + T), explaining the lack of recognition sites for most restriction enzymes (Fig. 4B). To exclude the possibility that this sequence structure is an artefact of the PCR-based isolation procedure, we constructed and screened a short-insert F. affinis library with c1063 to isolate corresponding genomic sequences. Sequencing of four positive clones confirmed the high similarity of c1063 with genomic sequences of this repeat (Supplementary Data Fig. S3), which was named FriSAT1. The irregular structure of FriSAT1 was also confirmed by Southern hybridization experiments using genomic DNA of F. affinis digested with various restriction endonucleases, including a partial digestion with DraI and MseI, which have AT-rich target sequences. Hybridization of the c1063 probe to these DNA samples failed to produce ladder-like signals; instead, a smear with only a few stronger bands was visible (data not shown).
Fig. 4.
Identification and sequence characterization of the FriSAT1 satellite repeat in species of subgenus Liliorhiza. (A) MboI digestion of genomic DNA of various Fritillaria species (M = marker). ‘Relic’ band corresponding to the FriSAT1 repeat encircled. (B) A dot-plot sequence analysis of the DOP-PCR-amplified c1063 clone against itself.
Estimating the copy numbers of repeat sequences in different Fritillaria species
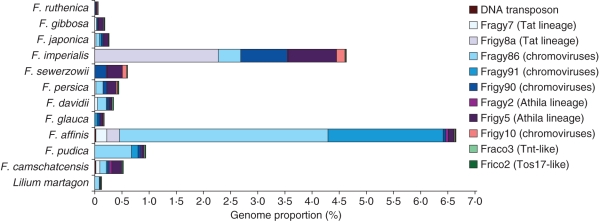
Representatives of all identified dispersed repeats were selected for dot-blot quantification in a range of Fritillaria species (Fig. 5). The most abundant dispersed repeats identified in F. affinis were the Ty3/gypsy elements from the chromovirus lineage – Fragy86 (3·85 % of the genome) and Fragy91 (2·1 %; Supplementary Data Table S1A, available online). In other Liliorhiza species, Fragy86 was also the predominant chromovirus-like retrotranspon in F. pudica (0·7 %) but comprised less than 0·1 % of the genomes of the other Liliorhiza species analysed. The other Ty/gypsy lineages (i.e. Athila and Tat) comprised 0·1 % or less of the F. pudica genome, which was similar to the frequency of all analysed Ty3/gypsy elements in the genomes of F. glauca and F. camschatcensis.
Fig. 5.
Proportion of dispersed repetitive elements estimated using quantitative dot-blot hybridization in 11 Fritillaria species and in Lilium martagon.
In F. imperialis, Frigy8a (Tat lineage) was identified as the most abundant gypsy element (2·3 %). In the remaining Eurasian species, Ty3/gypsy retrotransposons from the Athila lineage were identified as the most abundant repetitive elements, although in all cases they represented less than 1 % of the genomes analysed (Fig. 5 and Supplementary Data Table S1A).
From the four Ty1/copia-type retrotransposons identified (Supplementary Data Fig. S1, Table 2), elements Frico2 and Fraco3 were selected for quantification. Both repeats represented less than 0·1 % of Fritillaria genomes (max. 1200 copies per Cx in F. ruthenica, F. sewerzowii, F. affinis and F. camschatcensis). In total, we quantified approx. 6·65 and 4·65 % of the F. affinis and F. imperialis genome, respectively (Fig. 5, Supplementary Data Table S1A), with only 2 % of these repetitive sequences found to be shared by F. affinis and F. imperialis.
To verify the dot-blot copy number estimates, we hybridized the same set of probes to fosmid genomic libraries of F. affinis and F. imperialis (Supplementary Data Table S1B). Eighty per cent of the copy number estimates were within a two- to three-fold range of the dot-blot estimations. For 64 % of the analysed dispersed repeats, the copy number estimates were lower than those obtained by the dot-blot analysis. Largely deviating (four- to 13-fold difference to dot-blot results) estimates were found for Frigy90, Fraco3, Frico2 and the DNA transposon. These differences may be explained by the low abundance of these elements in the respective genomes (<2000 copies per Cx) and therefore in corresponding fosmid libraries. The abundance of the identified DNA transposon was estimated using both methods as less than 1 % per Cx in all analysed species (Supplementary Data Table S1A, B).
Copy numbers of the FriSAT1 satellite repeat reflected the differential hybridization of genomic DNA from F. affinis and F. imperialis to analysed species (Fig. 1, Supplementary Data Table S1A). FriSAT1 was the most abundant repeat component identified in F. glauca (6 %), F. affinis (26 %) and F. falcata (36 %). However, the repeat comprised only 0·64 and 1 % of the F. camschatcensis and F. pudica genome, respectively. In all other species not belonging to subgenus Liliorhiza, the proportion of FriSAT1 was below 0·05 % (Supplementary Data Table S1A).
Chromosomal localization of Fritillaria repetitive elements
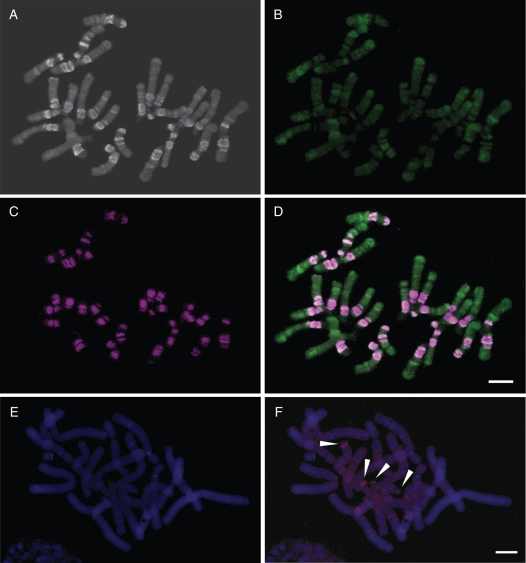
FISH of the FriSAT1 satellite repeat to chromosomes of species belonging to subgenus Liliorhiza showed its exclusive co-localization, with distinct heterochromatic bands visible as intensely DAPI-stained chromosomal regions (Figs 6 and 7A, D). The number of FriSAT1 hybridization signals differed between species and generally reflected the varying proportions of the repeat estimated from dot-blot hybridizations (Supplementary Data Table S1A). Whereas we observed large blocks of the repeat in F. affinis (approx. 26 % of the genome made up by FriSAT1) co-localized with most (but not all) heterochromatic bands (Figs 6A and 7D), in F. pudica (approx. 1 % of FriSAT1) the hybridization signals were far more restricted (Fig. 6B). In F. camschatcensis (approx. 0·6 % of FriSAT1), despite a large number of DAPI-stained heterochromatic bands, the signals were restricted to just a few bands (Fig. 6C).
Fig. 6.
FISH localization of the FriSAT1 satellite repeat in three species from subgenus Liliorhiza: (A) F. affinis, (B) F. pudica and (C) F. camschatcensis. On DAPI-stained chromosomes (left), heterochromatin domains and bands are visible as intensely stained, dark regions; hybridization sites of the FriSAT1 repeat are shown as violet signals (right). Arrowheads indicate heterochromatin bands (A) and whole chromosomes without the FriSAT1 repeat (B). Scale bar = 10 µm.
Fig. 7.
FISH localization of repetitive DNAs in Fritillaria affinis (A–D) and F. imperialis (E, F). Chromosomes of F. affinis stained with DAPI (A), probed with Ty3/gypsy repeat Fragy86 (green hybridization signal; B) and reprobed with FriSAT1 satellite repeat (purple hybridization signal; C). Images of (B) and (C) are overlaid in (D). DAPI-stained chromosomes of F. imperialis (E) probed with the Ty1/copia repeat Frico2 LTR retrotransposons (F; red hybridization signal). Arrows indicate strong bands of hybridization on two acrocentric chromosomes. Scale bars = 10 µm.
The chromosomal organization of Ty3/gypsy and Ty1/copia elements was also analysed by FISH. Fragy86 (a Ty3/gypsy element isolated from F. affinis) showed a generally homogeneous dispersed distribution pattern along the chromosomes although there was stronger hybridization in some regions (Fig. 7B, D). Nevertheless, these did not coincide with the heterochromatic bands visible with DAPI (Fig. 7A) where hybridization was reduced. Indeed, rehybridization of the same slide using FriSAT1 as a probe (Fig. 7C) confirmed that regions depleted of Fragy86 corresponded to FriSAT1 arrays residing at the heterochromatic bands (Fig. 7D). Frico2, a copia retrotransposon isolated from F. imperialis, generally showed a dispersed but homogeneous distribution pattern along the chromosomes of F. imperialis, although a stronger band of hybridization was observed on the short arm of two acrocentric chromosome pairs (Fig. 7F).
DISCUSSION
Genome size variation
Genome sizes were determined for all but one species (F. davidii). The largest genome was diploid F. japonica (subgenus Japonica) with a monoploid genome size of 1Cx = 87·3 pg. This is similar to the estimate for F. koidzumiana (1Cx = 77·4 pg) belonging to the same subgenus (Leitch et al., 2007). Both these large diploid genomes stand out in contrast to the monoploid genome sizes reported in other subgenera, not only in the present work (Fig. 1) but also in Leitch et al. (2007). Such very large genomes may well be characteristic of subgenus Japonica, as these two species together with F. amabilis have similar sized karyotypes (Noda, 1964, 1968). Indeed, these are the largest known diploid genomes in the plant kingdom, as the genome size estimate for the diploid F. davisii (1C = 1Cx = 89·48 pg) listed in the Plant DNA C-values database (Bennett and Leitch, 2005) and estimated by McLeish and La Cour (listed in Bennett and Smith, 1976 as a pers. comm. from 1971) has been suggested to be an overestimate (see Leitch et al., 2007; Zonneveld, 2010). This is further supported by a more recent genome size estimate and chromosome count for this species of 1Cx = 62·42 pg and 2n = 2x = 24 at the Royal Botanic Gardens, Kew (L. Hanson, pers. comm.).
At the other end of the scale, the smallest genomes in the current study were found in three species belonging to subgenus Liliorhiza, i.e. F. maximowiczii (1Cx = 35·6 pg), F. pudica (1Cx = 38·3 pg) and F. camschatcensis (1Cx = 38·4 pg), values which were similar to those reported in Leitch et al. (2007). Phylogenetically, these species occupy the early diverging branches of subgenus Liliorhiza (Rønsted et al., 2005) with the more derived species being characterized by larger genomes (up to 1Cx = 53·6 pg in F. glauca). Thus, viewed within a phylogenetic context the direction of genome size evolution within this subgenus has been one of increase rather than decrease.
Overall, combining the data presented here with those reported by Leitch et al. (2007) has extended the range of genome sizes encountered in Fritillaria from 2·5- to 2·8-fold. Generally, we found good agreement between the C-values acquired by flow cytometry in the present study and those obtained using Feulgen densitometry (Leitch et al., 2007), although flow cytometric values tended to be slightly higher (Fig. 1, Table 1). We assume this difference is caused in part by the differential effects of chromatin condensation on the staining kinetics of Feulgen and propidium iodide (for flow cytometry), something which is likely to be particularly acute for species with such large genomes and lots of heterochromatin. This is currently being analysed in greater detail and will be reported in a subsequent paper (P. Bureš et al., in preparation).
There was no clear relationship between the amount of heterochromatin visible as DAPI- or C-bands and genome size in either subgenus Liliorhiza (compare C-values in Fig. 1 and DAPI-bands on chromosomes in Fig. 6) or in various species in subgenera Petilium, Theresia and Rhinopetalum based on the percentage of heterochromatin reported by Bakhshi Khaniki (2004). For example, F. persica (1Cx = 42·0 pg) and F. gibbosa (1Cx = 42·4 pg) have similar genome sizes and yet the percentage of heterochromatin in the karyotype of F. gibbosa (8·1 %) was estimated to be nearly twice that of F. persica (4·5 %) (Bakhshi Khaniki, 2004). In addition, F. sewerzowii has no C-bands at all (La Cour, 1978) yet its genome is also similar in size (1Cx = 42·3 pg). These results suggest that genome size variation in the North American clade, and probably in other subgenera as well, is not only caused by variable amounts of heterochromatin and tandem repeats (e.g. FriSAT1) but also by other classes of repetitive DNA.
Global repeat similarity
Parallel to the construction of fosmid libraries in F. affinis and F. imperialis, we analysed global repeat similarities between these species and 12 others using dot-blot hybridization (Fig. 1). Using F. affinis genomic DNA as a probe clearly differentiated between all but one of the Liliorhiza species [hybridization intensity was 40–100 % except for F. maximowiczii (<10 %)] and the remaining subgenera where hybridization intensity ranged from 5 to 20 %. Interestingly, F. maximowiczii is the only Liliorhiza species analysed which occurs solely in Eurasia (China, south-east Russia). It has been suggested that subgenus Liliorhiza may have originated in Asia followed by a later dispersal to North America (Rønsted et al., 2005). If so, this could explain the difference in hybridization strength observed when genomic DNA from F. affinis was used as a probe, as this species is phylogenetically one of the more derived species of subgenus Liliorhiza. Alternatively, the placement of F. maximowiczii may need to be revised in light of further phylogenetic studies. Indeed, it perhaps shares closer genomic similarity with F. davidii, which also occurs solely in China, than with the North American Liliorhiza species (called the core Liliorhiza species thereafter).
Subsequent quantification of the FriSAT1 satellite repeat (Supplementary Data Table S1A; see below) indicated that this tandem repeat is exclusively present in the Liliorhiza species analysed (no data for F. maximowiczii), albeit in varying amounts. We expect that the strong genomic cross-hybridization found between F. affinis and F. falcata (Fig. 1) is predominantly caused by the FriSAT1 satellite repeat comprising approx. 26 and 36 % of their genomes, respectively (see Supplementary Data Table S1A). Nevertheless, the weaker hybridization of the F. affinis genomic probe to the remaining Liliorhiza species analysed (Fig. 1) indicates that their repetitive DNA profiles have diverged from those of F. affinis and F. falcata. This is supported by the fact that the FriSAT1 tandem repeat also comprises a significantly smaller proportion of their genomes (i.e. F. glauca, 6 %; F. pudica, 1·1 %; F. camschatcensis, 0·6 %), although the amounts of heterochromatin in these species are comparable.
Hybridization using genomic DNA of F. imperialis showed a quite different picture (Fig. 1). In contrast to F. affinis, DNA of F. imperialis hybridized to at least 30 % of the genomic sequences in all species analysed, including Liliorhiza species and Lilium martagon. As observed in the F. affinis data, lower hybridization intensity was observed in F. ruthenica (subgenus Fritillaria) and F. davidii (subgenus Davidii) compared with the other species, suggesting that there has been considerable repetitive sequence divergence between the two subgenera and subgenus Petilium (F. imperialis). However, our data are scarce and limited only to a single species of the species-rich subgenus Fritillaria (approx. 50 species), and thus extrapolating these results across the subgenus is not possible at present. Hybridization intensities close to 70 % were observed in F. camschatcensis and F. maximowiczii, while significantly lower intensities were seen in the core Liliorhiza species (35–45 %). This corroborates the closer phylogenetic and geographical relationship of the two species with the Eurasian subgenera than with the core Liliorhiza species (Rønsted et al., 2005).
Heterochromatin and FriSAT1 satellite repeat
The dot-blot hybridization data and FISH localization showed that the FriSAT1 repeat is restricted only to Liliorhiza species. The interspecific variation in the abundance of FriSAT1 (0·6–35·8 %) was related to the variable amount of heterochromatin across the subgenus Liliorhiza (La Cour, 1951). In two (F. pluriflora and F. purdyi) of seven North American species no heterochromatin was observed, whereas approx. 50 % of heterochromatin was reported for F. falcata (La Cour, 1951). High heterochromatin amount in F. falcata was corroborated by the highest copy number of FriSAT1 (35·8 %) among all species analysed. Interestingly, despite numerous heterochromatin bands visible in F. camschatcensis only a few harbour the FriSAT1 satellite (Fig. 6C). This suggests that heterochromatin in this species is composed of other tandem repeats. As F. camschatcensis is sister to the core Liliorhiza species (Rønsted et al., 2005) we assume that its heterochromatin is composed of an ancestral form(s) of the FriSAT1 satellite, not detectable at the level of the stringency used in the current work. Our data on the distribution of the FriSAT1 repeat corroborate conclusions drawn by Rønsted et al. (2005), namely that Fritillaria and Lilium species originated in Eurasia and only later migrated over the Aleutian Islands to North America. A negligible amount of the FriSAT1 repeat in F. camschatcensis apparently supports its position as an ancestral link between the Eurasian subgenera and Liliorhiza species (Rønsted et al., 2005). Our data do not support the conclusion of Vinnersten and Bremer (2001) that Fritillaria originated in North America and later expanded to Eurasia. The origin of FriSAT1 associated with the radiation of Liliorhiza species in North America, after their isolation from the Eurasian ancestor(s), is a more plausible scenario.
Are the genome processes generating obese genomes in fritillarias different from those operating in species with smaller genomes?
Sequence analysis of fosmid clones selected for their repetitive nature showed they were predominantly composed of LTR retrotransposons, with the gypsy-like elements far more abundant than copia-like retrotransposons (Fig. 2). A similar prevalence of gypsy elements has also been reported in many other plant genomes (Kentner et al., 2003; Zuccolo et al., 2007; Sun et al., 2008), although there appears to be no correlation between the dominant repeat type and genome size across plant species (Hawkins et al., 2006, 2008; Vitte and Bennetzen, 2006; Moisy et al., 2008). Nevertheless, it is likely that as the proportion of repetitive DNA grows with increasing genome size, the abundance of individual repeat families increases as well. There are several examples showing that increases in genome size have arisen through the amplification of one or a few specific repeats. In maize nearly 25 % of the genome consists of just five major classes of LTR retrotransposons (SanMiguel et al., 1996). In five Louisiana iris species the IRRE retroelement accounts for 6–10 % of the genome (Kentner et al., 2003) and in Vicia pannonica a single family of gypsy elements is responsible for expanding the genome by 50 % (Neumann et al., 2006). Given these observations, it is therefore perhaps surprising that the most abundant dispersed repeats isolated from the fosmid clones make up such a small proportion of the Fritillaria genomes and that all the repeats identified in the fosmid clones together represent only 4·7 and 6·7 % of the F. imperialis and F. affinis genomes, respectively (Fig. 5 and Supplementary DataTable S1A). Nevertheless, it should be noted that in terms of genomic copy numbers, the isolated elements rank among the most amplified families described in plants, reaching 4·4 × 104(Frigy5 in F. imperialis) and 1·5 × 105 (Fragy86 in F. affinis) copies per 1C (Supplementary Data Table S1A).
Analyses of repeat diversity in species with truly obese genome sizes comparable with those of Fritillaria species are scarce. In the genome of Lilium henryi (approx. 32 000 Mb, Bennett and Smith, 1976), a Ty3/gypsy retrotransposon del was estimated to be present in over 13 000 copies per genome (Sentry and Smyth, 1989) comprising approximately just 0·4 % of the genome, while and a non-LTR retrotransposon del2 was shown to occupy 4 % of the L. speciosum genome (Leeton and Smyth, 1993). Given the low percentage of the genome represented by the LTR retrotransposons found in the present study it seems likely that they represent just a small fraction of the LTR families present in the Fritillaria genomes. This is supported by the observed dramatic increase in hybridization signals in quantitative dot-blot hybidizations when relaxed hybridization and washing stringency was used (data not shown). Thus, although the stringent hybridization conditions used for repeat quantification presented in the Results provided strictly family-specific estimates, there is a large number of related but partially diverged families that make up most of the genome. If so, then the evolution of truly obese genomes may be largely determined by the failure of mechanisms responsible for repeat elimination which operate effectively in species with smaller genomes to counteract genome expansion.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Fig. S1: Detailed annotation of the fosmid clones (GenBank accession numbers GU188675 to GU188682). Fig. S2: A phylogenetic tree of RT-domains isolated from Ty1/copia-type retrotransposons. Fig. S3: Multiple sequence alignment of the FriSAT1 satellite repeat (GenBank accession numbers GU182252 to GU182256). Table S1: (A) Results of dot-blot quantification of selected Ty3/gypsy and Ty1/copia class LTR retrotransposons, and a DNA transposon performed under 85 % stringency in 11 Fritillaria species and Lilium martagon. (B) Quantification of selected repeats estimated using fosmid libraries hybridization performed under 85 % stringency in two model Fritillaria species.
ACKNOWLEDGEMENTS
We thank K. Strange (RBG, Kew) and Laurence Hill for helpful discussions and providing plant material, and J. Latalova and H. Stepancikova for technical assistance. K.A. was supported by a SYNTHESYS grant. This study was supported by research grants from the Czech Science Foundation (no. 521/07/0284), the Czech Ministry of Education (nos. MSM0021622415, MSM0021622416, LC06073 and LC06004), Academy of Sciences of the Czech Republic (no. AVOZ50510513) and the Natural Environment Research Council (NERC) UK, (NE/G020256/1).
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshi Khaniki G. Giemsa C-banding and karyological studies in species of Rhinopetalum (Liliaceae) National Academy of Science Letters. 2004;27:399–411. [Google Scholar]
- Bennett MD, Leitch IJ. Plant DNA C-values database. 2005 doi: 10.1007/978-1-0716-3389-2_9. (release 4·0, October 2005). http://data.kew.org/cvalues/ [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London. B, Biological Sciences. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Darlington CD. The internal mechanics of the chromosomes I.-The nuclear cycle in Fritillaria. Proceedings of the Royal Society of London-Series B. 1935;118:33–59. [Google Scholar]
- Darlington CD. Recent Advances in Cytology. London: Churchill; 1937. [Google Scholar]
- Darlington CD, La Cour LF. The detection of inert genes. Journal of Heredity. 1941;32:115–121. [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113–113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel OH. The causal sequence of meiosis. I. Chiasma formation and the order of pairing in Fritillaria. Journal of Genetics. 1940;41:9–34. [Google Scholar]
- Grandbastien MA, Spielmann A, Caboche M. Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature. 1989;337:376–380. doi: 10.1038/337376a0. [DOI] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Research. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Hu G, Rapp RA, Grafenberg JL, Wendel JF. Phylogenetic determination of the pace of transposable element proliferation in plants: copia and LINE-like elements in Gossypium. Genome. 2008;51:11–18. doi: 10.1139/g07-099. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and Genome Research. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kentner EK, Arnold ML, Wessler SR. Characterization of high-copy-number retrotransposons from the large genomes of the Louisiana iris species and their use as molecular markers. Genetics. 2003;164:685–697. doi: 10.1093/genetics/164.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. doi:10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cour LF. Heterochromatin and the organization of nucleoli in plants. Heredity. 1951;5:37–50. doi: 10.1038/hdy.1951.2. [DOI] [PubMed] [Google Scholar]
- La Cour LF. The constitutive heterochromatin in chromosomes of Fritillaria sp., as revealed by Giemsa Banding. Philosophical Transactions of the Royal Society of London Series B. 1978;285:61–71. doi: 10.1098/rstb.1978.0094. [DOI] [PubMed] [Google Scholar]
- Leeton PR, Smyth DR. An abundant LINE-like element amplified in the genome of Lilium speciosum. Molecular and General Genetics. 1993;237:97–104. doi: 10.1007/BF00282789. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Beaulieu JM, Cheung K, Hanson L, Lysak MA, Fay MF. Punctuated genome size evolution in Liliaceae. Journal of Evolutionary Biology. 2007;20:2296–2308. doi: 10.1111/j.1420-9101.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- Li R, Shang Z. The chromosome observation on five species of rare plants of China. Journal of Wuhan Botanical Research. 1989;7:217–220. [Google Scholar]
- Llorens C, Fares MA, Moya A. Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evolutionary Biology. 2008;8:276. doi: 10.1186/1471-2148-8-276. doi:10.1186/1471-2148-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navrátilova A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. doi:10.1186/1471-2164-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae) Plant Cell. 2008;20:2559–2570. doi: 10.1105/tpc.108.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant CJ, Macfarlane RM. Chromosome polymorphism in triploid populations of Fritillaria lanceolata Pursh (Liliaceae) in California. Botanical Journal of the Linnean Society. 1980;81:135–154. [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Research. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisy C, Garrison KE, Meredith CP, Pelsy F. Characterization of ten novel Ty1/copia-like retrotransposon families of the grapevine genome. BMC Genomics. 2008;9:469. doi: 10.1186/1471-2164-9-469. doi:10.1186/1471-2164-9-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Koblizkova A, Navratilova A, Macas J. Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics. 2006;173:1047–1056. doi: 10.1534/genetics.106.056259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda S. Cytology in the genus Fritillaria. I. Variations in karyotypes and B-chromosomes in F. amabilis. Bulletin of the Osaka Gakuin University. 1964;2:125–132. [Google Scholar]
- Noda S. Cytology in the genus Fritillaria. II. Karyotypes and B-chromosomes in F. japonica var. japonica and var. koidzumiana. Bulletin of the Osaka Gakuin University. 1968;10:127–141. [Google Scholar]
- Patterson TB, Givnish TJ. Phylogeny, concerted convergence, and phylogenetic niche conservatism in the core Liliales: insights from rbcL and ndhF sequence data. Evolution. 2002;56:233–252. doi: 10.1111/j.0014-3820.2002.tb01334.x. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society. 2010;164:10–15. [Google Scholar]
- Peruzzi L, Leitch IJ, Caparelli KF. Chromosome diversity and evolution in Liliaceae. Annals of Botany. 2009;103:459–475. doi: 10.1093/aob/mcn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfosser M, Amon A, Lelley T, Heberle-Bors E. Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat-rye addition lines. Cytometry. 1995;21:387–393. doi: 10.1002/cyto.990210412. [DOI] [PubMed] [Google Scholar]
- Rix EM. Fritillaria. A revised classification. The Fritillaria Group of the Alpine Garden Society, UK. 2001 [Google Scholar]
- Rønsted N, Law S, Thornton H, Fay MF, Chase MW. Molecular phylogenetic evidence for the monophyly of Fritillaria and Lilium (Liliaceae; Liliales) and the infrageneric classification of Fritillaria. Molecular Phylogenetics and Evolution. 2005;35:509–527. doi: 10.1016/j.ympev.2004.12.023. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Sentry JW, Smyth DR. An element with long terminal repeats and its variant arrangements in the genome of Lilium henryi. Molecular and General Genetics. 1989;215:349–354. doi: 10.1007/BF00339741. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Kalitsis P, Joseph JL, Sentry JW. Plant retrotransposon from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. The Proceedings of the National Academy of Sciences USA. 1989;86:5015–5019. doi: 10.1073/pnas.86.13.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Staden R. The Staden sequence analysis package. Molecular Biotechnology. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Sun HY, Dai HY, Zhao GL, et al. Genome-wide characterization of long terminal repeat-retrotransposons in apple reveals the differences in heterogeneity and copy number between Ty1-copia and Ty3-gypsy retrotransposons. Journal of Integrative Plant Biology. 2008;50:1130–1139. doi: 10.1111/j.1744-7909.2008.00717.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Hof J. Relationships between mitotic cycle duration, S period duration and the average rate of DNA synthesis in the root meristem cells of several plants. Experimental Cell Research. 1965;39:48–58. doi: 10.1016/0014-4827(65)90006-6. [DOI] [PubMed] [Google Scholar]
- Vinnersten A, Bremer K. Age and biogeography of major clades in Liliales. American Journal of Botany. 2001;88:1695–1703. [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. The Proceedings of the National Academy of Sciences USA. 2006;103:17638–17643. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. The Plant Journal. 2001;26:307–316. doi: 10.1046/j.1365-313x.2001.01028.x. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Zonneveld BJM. New record holders for maximum genome size in eudicots and monocots. Journal of Botany. 2010;2010:527357. doi:10.1155/2010/527357. [Google Scholar]
- Zonneveld BJM, Leitch IJ, Bennett MD. First nuclear DNA amounts in more than 300 angiosperms. Annals of Botany. 2005;96:229–244. doi: 10.1093/aob/mci170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo A, Sebastian A, Talag J, et al. Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evolutionary Biology. 2007;7:152. doi: 10.1186/1471-2148-7-152. doi:10.1186/1471-2148-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.