Abstract
Oxidative stress occurs in the liver of rats fed alcohol chronically due to ethanol metabolism by CYP2E1, causing liver injury. The proteasome is considered as an antioxidant defense in the cell because of its activity in removing damaged and oxidized proteins, but a growing body of evidence shows that proteasome inhibitor treatment, at a non toxic low dose, provides protection against oxidative stress. In the present study, rats were fed ethanol for 4 weeks and were treated with the proteasome inhibitor PS-341 (Bortezomib, Velcade®). Exposure to proteasome inhibitor elicited the elevation of antioxidative defense by enhancing the levels of mRNA and protein expression transcripts of glutathione reductase (GSR), glutathione synthetase (GSS), glutathione peroxidase 2 (GPX2), and superoxide dismutase 2 (SOD2) in the liver of rats fed ethanol chronically, while ethanol alone did not increase these genes mRNA. Our results also showed that glutamate cysteine ligase catalytic subunit (GCLC), a rate-limiting enzyme in glutathione biosynthesis, was also up regulated in the liver of rats fed ethanol and injected with PS-431. Nrf2 mRNA level was significantly decreased in the liver of ethanol fed rats, as well as in the livers of animal fed ethanol and treated with proteasome inhibitor, indicating that the mechanism by which proteasome inhibitor up regulates the antioxidant response element is not due to regulation of Nrf2. However, ATF4, a major regulator of antioxidant response elements, was significantly up regulated by proteasome inhibitor treatment. The beneficial effects of proteasome inhibitor treatment also reside in the reversibility of the drug because the proteasome activity was significantly increased 72h post treatment. In conclusion, proteasome inhibitor treatment used at a non toxic low dose has potential protective effects against oxidative stress due to chronic ethanol feeding.
Keywords: Ethanol feeding, Oxidative stress, Proteasome inhibitor
Introduction
Marked generation of oxidative stress, associated with ethanol metabolism, is a major cause of liver injury induced by chronic alcohol consumption. CYP2E1 is up regulated to metabolize ethanol, which thus generates a marked increase in lipid peroxidation end-products and reactive oxygen species (ROS) [Bardag-Gorce et al., 2000; Bardag-Gorce et al., 2002; Bardag-Gorce et al., 2006a]. In addition, chronic alcohol feeding has been shown to deplete glutathione (GSH) levels, particularly in the mitochondria, which are usually characterized by high levels of GSH needed to eliminate the ROS generated during activity of the respiratory chain [Fernández-Checa et al., 1997]. Mitochondria cannot synthesize GSH, but imports it from the cytosol using a carrier protein embedded in the membrane surrounding the mitochondria. Alcohol has been reported to interfere with the function of this carrier protein, thereby leading to the depletion of mitochondrial GSH [Fernández-Checa et al., 1997].
The proteasome is considered as an antioxidant defense in the cell because of its activity of removing damaged and oxidized proteins [Friguet et al., 1997; Hill et al., 2008, Marques et al., 2004; Okada et al., 1999]. Although it is now well established that impairment of the ubiquitin proteasome pathway is implicated in the pathogenesis of alcoholic liver disease (ALD), a growing body of evidence shows that proteasome inhibitor treatment provides protection against oxidative stress in the brain [Gilardini et al., 2008; Nencioni et al., 2007; Williams et al., 2006] and in the heart [Lorenz et al., 2009]. Proteasome inhibition treatment is well known because of its efficacy against hyperproliferative diseases, such as cancer. Its inhibition by specific inhibitors induces cell cycle arrest and apoptosis in tumor cells [Hamilton et al. 2005]. Beyond their anti-proliferative and pro-apoptotic properties, proteasome inhibitors have been shown to have potent antioxidation effects.
However, the protective and beneficial effects of a proteasome inhibitor at a non-toxic low dose in the liver are still unknown. Proteasome inhibition is an antioxidative defense, as it leads to enhancement of mRNA transcripts levels of antioxidative enzymes. This is a characteristic that has been discovered in the past few years and merits further investigations. Dreiseitel et al. (2008) have shown that the protective effects of berry fruits reside in the proteasome inhibition by anthocyanins, which are highly concentrated in berry fruits. In the past few years, a growing body of evidence supports the cytoprotective role of the administration of a proteasome inhibitor to block the increased levels of oxidative stress [Du et al., 2009; Hill et al., 2008]. Activation of antioxidative enzymes gene expression has been reported when proteasomes were inhibited with other and even less specific proteasome inhibitors, such as MG132 (lysosome/proteasome inhibitor) [Bieler et al., 2009; Hill et al., 2008; Lee et al., 2004] and lactacystin [Sekhar et al., 2000]. Sekhar et al. showed that the gene GCLC, which encodes the catalytic subunit for γ-glutamylcysteine synthetase, the rate-limiting enzyme for the synthesis of glutathione (GSH), was significantly induced in cells treated with lactacystin [Sekhar et al., 2000].
The induction of GCLC is a consequence of Nrf2 binding to and activation of antioxidative response elements (ARE) [Kobayashi et al., 2006; Li et al., 2009; Satoh et al., 2009; Yang et al., 2005]. Nrf2, a key player in the activation of the antioxidative defense [Cederbaum, 2009; Yang et al., 2005], is a substrate of the proteasome pathway. When the proteasome is inhibited, Nrf2 protein is stabilized and the de novo-synthesized Nrf2 escapes the Kelch-like ECH-associated protein 1 (Keap-1) repression [Satoh et al., 2009], and translocates to the nucleus to regulate the transcriptional activation of a battery of cytoprotective genes [Itoh et al., 2004]. In addition, studies on activating transcription factor 4 (ATF4) null cells revealed that ATF4 regulates a number of genes that are important in preventing from oxidative stress [Harding et al., 2003]. ATF4-null cells also show impaired glutathione biosynthesis [Harding et al., 2003] and proteins involved in the antioxidant response, such as heme oxygenase-1 [Poss and Tonegawa, 1997].
Downregulation of ATF4 using small interfering RNAs (siRNAs), has been shown to confer cell sensitivity to anticancer agents and ATF4-overexpressing cells showed multidrug resistance and marked elevation of intracellular glutathione. Knockdown of ATF4 expression lead to down regulation of glutathione metabolism [Igarashi et al., 2007]. ATF4 is stabilized by proteasome inhibition [Milani et al., 2009; Yang and Karsenty, 2004] and its stabilization significantly contributes to the antioxidative response.
Therefore, proteasome inhibition has potential cytoprotective effects in various pathologies, such as carcinogenesis, chemical toxicity, respiratory distress, and inflammatory diseases. These cytoprotective effects have also already been reported in the brain [Gilardini et al., 2008; Nencioni et al., 2007; Phillips et al., 2000; Williams et al., 2006; Yamamoto et al., 2007].
In the present study, it is postulated that the cytoprotective qualities of a proteasome inhibitor treatment, at a non-toxic dose, may be beneficial for the treatment of hepatocyte dysfunction associated with alcoholic liver disease. This specific proteasome inhibition is different from ethanol-induced dysfunction of the ubiquitin proteasome pathway because chronic ethanol feeding alters the proteasome and its interacting proteins [Bousquet-Dubouch et al., 2009] PS-341 treatment is a reversible inhibition. The role of proteasome inhibitor administration in up regulating the mRNA of antioxidative enzymes and in reducing the oxidative stress generated by ethanol-induced CYP2E1 is the focus of this paper.
Material and Methods
Animals
Male Wistar rats from Harleco (Hollister, CA), weighing between 250–300 g, were used. They were maintained according to the Guidelines of Animal Care, as described by the National Academy of Sciences and published by the Institute of Laboratory Animal Resources Commission on Life Sciences National Research Council.
Ethanol Feeding
Rats were continuously fed intragastrically a liquid diet for 4 weeks, containing ethanol (13 g/kg body weight/day). Controls were pair-fed isocaloric dextrose. Details of the diet and the intragastric model have already been described in Ref. [Bardag-Gorce et al., 2003].
Proteasome Inhibitor Treatment
To investigate the effects of PS-341 in counteracting the effects of ethanol feeding, rats were given proteasome inhibitor PS-341 (LC Laboratories, Woburn, MA), intraperitoneally (IP), at 0.5 mg/kg body weight, one time, and sacrificed 24h later. In the group of rats fed alcohol for one month, the first group of rats were given PS341 at 0.5 mg/kg 24h before sacrifice and the 2nd group of rats fed alcohol were given PS-341 at 0.5 mg/kg once a week, 4 times with ethanol feeding.
Proteasome Chymotrypsin-like Activity Measurement
1 to 5 μg of homogenated by ultra-Turrax total protein from the liver was used. The reaction mixture contained 50 mmol/L Tris–HCl pH 8, 1 mmol/L DTT, and 40 mmol/L Suc-LLVY-AMC substrate for chymotrypsin-like activity. The mixture was incubated for 30 min. at 37°C, and the reaction was then stopped by adding 100 mmol/L monochloroacetate and 30 mmol/L sodium acetate (pH=4.3). Fluorescence was determined by measuring the release of AMC (λ excitation: 370 nm, λ emission: 430 nm), using a Perkin Elmer LS 30 spectrofluorometer.
Microarray Analysis
Fast frozen rat liver tissue was subjected to microarray analysis. Total liver RNA was extracted with UltraspecTM RNA Isolation Systemic (Biotecx Laboratories, Houston, TX), and cleaned with Rneasy columns (Qiagen, Valencia, CA). Five micrograms of total RNA were used for preparing biotin-labeled cRNA. Labeled and fragmented cRNA was subsequently hybridized to Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA). Labeling, hybridization, image scanning, and initial data analysis were performed at the Microarray Core at the Los Angeles Biomedical Research Institute. Sample preparation and loading, hybridization, staining, and microarray data analysis were then performed according to Ref. [Bardag-Gorce et al., 2006b].
Antioxidative Enzymes mRNA Analysis by qRT-PCR
Quantitative RT-PCR was performed to measure the levels of antioxidative enzymes. Total liver RNAs was extracted with Trizol Plus RNA Purification kit (Invitrogen, Carlsbad, CA). Synthesis of cDNAs was performed with 5 μg total RNA, and 50 ng random hexamer primers using SuperScriptIII RNase H-Reverse Transcriptase (Invitrogen). PCR primers were designed using the Primer Express software (Applied Biosystems, Foster City, CA), and have been tested in our lab. The primers for rat GSS, GSR, and GPX2, SOD2 are shown in Table 1. Quantitative PCR was achieved using the SYBR Green JumpStart™ Tag ReadyMix (Sigma, St. Louis, MO) on an ABI PRISM 7700 Sequence Detector System (Applied Biosystems, Foster City, CA). Thermal cycling consists of an initial step at 50°C for 2 min., followed by a denaturation step at 95°C for 10 min., then 40 cycles at 95°C for 15 s and 60°C for 1 min. A single PCR product was confirmed with the heat dissociation protocol at the end of the PCR cycles. Each data point was repeated three times. The target mRNA abundance in each sample was normalized to its 18S level as DCt = Cttarget gene-Ct18S. For each target gene, the highest DCt was assigned as DCtmax.
Table 1. Primers Sequence.
| Glutathione reductase 2 (GSR) | NM_053906 | Forward | ACTGCTCCGCACATCCTGAT |
| Glutathione reductase 2 (GSR) | NM_053906 | Reverse | CCTGGGATCTGGTTCTCATGA |
| Glutathione synthetase (GSS) | NM_012962 | Forward | GGGAATGGAAGCTCCTTTGAG |
| Glutathione synthetase (GSS) | NM_012962 | Reverse | GCTGGCGAGAGGGCAAT |
| Glutathione peroxidase 2 (GPX2) | NM_183403 | Forward | ACCGATCCCAAGCTCATCAT |
| Glutathione peroxidase 2 (GPX2) | NM_183403 | Reverse | TCTCAAAGTTCCAGGACACATCTG |
| Superoxide dismutase 2, mitochondrial (SOD2) | NM_017051 | Forward | CCCTGACCTGCCTTACGACTAT |
| Superoxide dismutase 2, mitochondrial (SOD2) | NM_017051 | Reverse | CTGCATGATCTGCGCGTTA |
| Nrf2 | NM_031789.1 | Forward | GCCAGCTGAACTCCTTAGACTCA |
| Nrf2 | NM_031789.1 | Reverse | GCTCTGCTAGGAAAGCAGAGTAAAATT |
| ATF4 | BC158588.1 | Forward | TGGCCAAGCACTTCAAACC |
| ATF4 | Reverse | ATCCATAGCCAGCCATTCTGA |
Carbonyl Proteins Levels Measurement
Carbonyl protein levels were measured using the Oxyblot kit from Chemicon (Chemicon, Temecula, CA). The carbonyl groups produced by oxidation were detected by derivatization to 2,4-dinotrophenylhydrazone by reacting with 2,4-dinotrophenylhydrasine [Levine et al., 1994]. The dinotrophenyl-derivatized proteins were detected with Western blot using a rabbit antibody.
Western Blots
Proteins from the cytosolic fraction were separated by SDS-PAGE electrophoresis using 12% polyacrylamide gels. Proteins were transferred to a PVDF membrane (Bio-Rad, Hercules, CA) for 1 Hr. in 25 mM Tris–HCl (pH 8.3), glycine 192 mM and 20% methanol. Antibodies against GPX2 (R&DSystems, Inc. Minneapolis, MN) and GCLC (Aviva system Biology, San Diego, CA) were used. Goat anti-mouse and goat anti rabbit antibody (Bio-Rad, Hercules, CA) were used as the second antibody. Immunodetection was done using an alkaline phosphatase kit from Bio-Rad or ECL plus (Amersham Bioscience Corp., Piscataway, NJ). Densitometric measurements of the bands were done using the GS-700 imaging densitometer (Bio-Rad, Hercules, CA).
TUNEL Staining
To quantify the effects of PS-341 and ethanol feeding on the hepatocellular apoptosis, the terminal deoxynucleotidyl transferasemediated deoxyuridine triphosphate nick-end labeling (TUNEL) assay was performed on liver slices. DNA fragmentation detection was the indicator for apoptosis.
Statistical Analysis
Data were obtained from at least three separate experiments. Bars represent mean values ± SEM. P values are determined by one-way ANOVA and Student-Newman Keuls for multiple group comparisons (Sigma-Stat software, San Francisco, CA). Statistical significance is set at p= < 0.5.
Results
Chronic ethanol feeding causes a significant increase of oxidative stress in rats liver cells. This is caused by the induction of CYP2E1 activity that metabolizes ethanol [Bardag-Gorce et al., 2006a]. It is our hypothesis that proteasome inhibitor treatment is responsible for the up regulation of antioxidative stress response elements (ARE), which, in turn, is expected to counteract the ethanol-induced oxidative stress
Microarray analysis of liver of rats fed ethanol chronically did not show an increase of ARE mRNA levels, even if the levels of ROS were significantly increased by ethanol (data not shown). However, data mining of a microarray analysis performed on the liver of rats given the proteasome inhibitor PS-341 only, one time, IP, at 0.5 mg/kg body weight (bw), and sacrificed 24h later, showed a significant increase in the mRNA transcripts levels of several important antioxidative enzymes (Table 2), which play an important role in recycling glutathione (GSH) and in reducing the level of ROS (Figure 1A). This dosing protocol also caused a significant inhibition of proteasome activity (Figure 1B).
Table 2.
Data Mining of Microarray Analysis Comparing Antioxidative Enzymes mRNA Levels in the Livers of Rat Given PS-341 and its Control.
| Gene Title | Gene Symbol | Fold Change Ratio PS-341/control | Gene Ontology |
|---|---|---|---|
| Glutathione peroxidase 2 | GPX2 | +18.4 | Responsible for glutathione-dependent hydrogen peroxide-reducing activity in the epithelium of the gastrointestinal tract |
| Superoxide dismutase 2, mitochondrial | SOD2 | +8 | Responsible for ROS and superoxide removal |
| Glutathione reductase | GSR | +4 | Maintains high levels of reduced glutathione in the cytosol |
| Glutathione synthetase | GSS | +2 | Activates glutathione biosynthesis |
Figure 1.
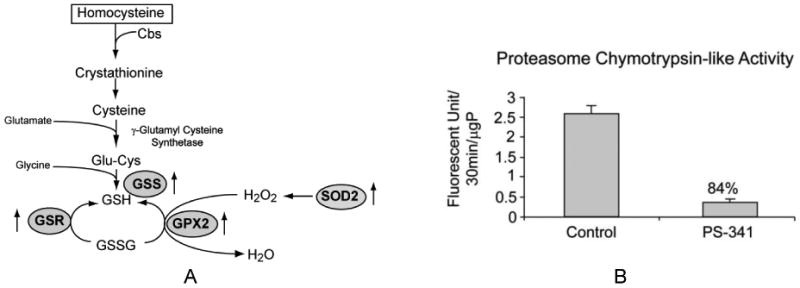
A. Transsulfuration pathway and the role of antioxidative enzymes in recycling glutathione (GSH). Note that GSR, GSS, GPX2, and SOD2 enzymes were up regulated by the proteasome inhibitor (↑ arrows). B. Proteasome chymotrypsin-like activity was measured in the liver of rats given PS-341 one time and sacrificed 24 hr. later.
qRT-PCR experiments were performed and results confirmed the data obtained in the microarray analysis: the gene expression of these antioxidative stress enzymes was significantly induced when proteasome was inhibited by PS-341 (Figure 2).
Figure 2.
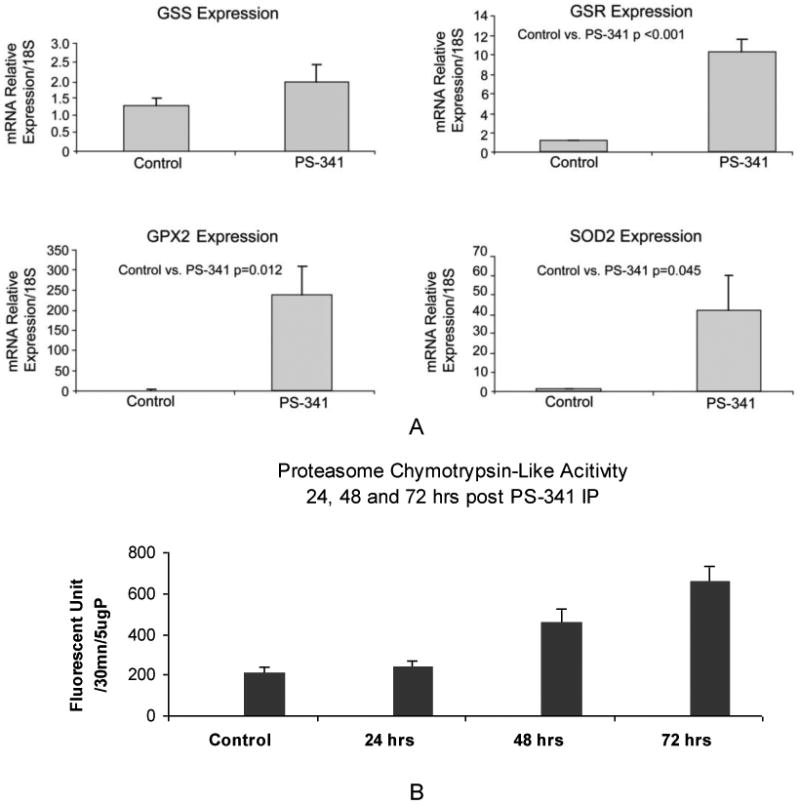
A. qRT-PCR analysis of GSS, GSR, GPX2, and SOD2 enzymes gene expression showed a significant increase of these antioxidative enzymes. Mean ± SEM, n=3. B. Proteasome chymotrypsin-like activity. rats were treated with PS-341 at a lower dose (0.25mg/kg) and sacrificed 24h, 48h and 72h post IP injection.
To evaluate the cytoprotective effects of proteasome inhibitor treatment, we hypothesized that the administration of proteasome inhibitor to rats fed ethanol may enhance the antioxidant response and play a protective role against ethanol. Therefore, rats were fed ethanol for 1 month and injected with proteasome inhibitor PS-341 IP, once a week, 4 times, at 0.5 mg/kg body weight (bw). Proteasome activity was then measured and a significant inhibition of proteasome activity was found (Figure 3A). This marked inhibition was similar to the inhibition observed with PS-341 alone (Figure 1B), indicating that proteasome inhibition by PS-341 is different from proteasome dysfunction caused by ethanol feeding, which suggests that there is no accumulative inhibitory effects of PS-341 administration and ethanol feeding. In our previous study [Bousquet-Dubouch et al., 2009], we demonstrated that chronic ethanol feeding causes a dysfunction of the ubiquitin proteasome pathway altering, not only the binding of the 20S proteasome to its regulatory complexes, but also the proteasome interacting proteins, which is totally different from the drug-induced proteasome inhibition. PS-341 is a reversible proteasome inhibitor and there is a stimulation of the proteasome activity 48h post injection (Figure 2B). These results indicate that proteasome inhibitor beneficial effects resides in this reversibility.
Figure 3.
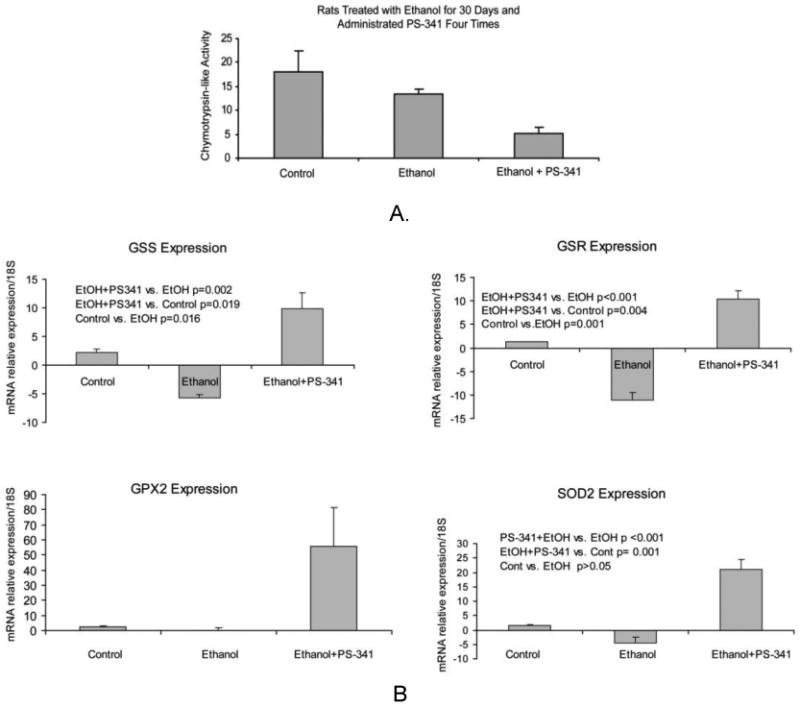
A. Proteasome chymotrypsin-like activity in the liver of rats fed ethanol and given PS-341. B. qRT-PCR analysis of GSS, GSR, GPX2, and SOD2 enzymes gene expression in the liver of rats fed ethanol and rats fed ethanol and treated with PS-341. Proteasome inhibitor treatment significantly increased their gene expression. Mean ±SE, n=3.
As expected, we found a significantly up regulation of the antioxidative enzymes mRNA in the liver of rats fed ethanol and treated with PS-341. On the other hand, ethanol feeding alone did not increase the gene expression of these enzymes (Figure 3B). These results thus confirm that ethanol-induced proteasome inhibition is different from that of PS-341 since ethanol alone has no effects in up regulating the antioxidative response. The mechanisms are yet to be determined.
Rats fed ethanol tolerated well the once a week PS-341 0.5 mg/kg treatment. ER stress proteins were analyzed to examine the potential side effects of this 4 times/PS-341 administration and sacrifice 4h post injection. We did not find any changes in ER stress markers Grp78 and PDI protein levels, indicating that this proteasome inhibitor treatment did not cause ER stress (data not shown). However, there was an increase in the polyubiquitinated proteins levels because of the proteasome activity inhibition (Figure 4A). The effect of proteasome inhibitor treatment in increasing apoptosis was evaluated using the TUNEL method. The results showed an increase in DNA fragmentation, indicating that the administration of proteasome inhibitor at 0.5 mg/kg caused an increase in apoptosis (Figure 4B). However, since PS-341 is a reversible proteasome inhibitor, and proteasome activity rebounds higher than the control 48h and 72h post injection (Fig2B), it is expected the polyubiquitinated protein to be cleared up by the stimulated proteasome activity. We believe that this reversibility helped the rats fed ethanol for 4 weeks to tolerate the 0.5 mg/kg, once a week, 4 times dosing. PS-341 increased apoptosis was not greatly different from the ethanol increased apoptosis, which, we believe, indicates that a lower dose of PS-341 is certainly more beneficial.
Figure 4.
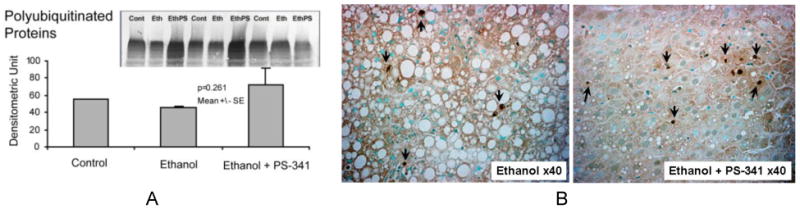
A. Polyubiquitinated levels analysis in the liver homogenates of rats fed ethanol for 4 weeks and given proteasome inhibitor 4 times once a week. Note that polyubiquitinated proteins significantly accumulated with proteasome inhibitor treatment. B. Morphology analysis of the effects of proteasome inhibitor treatment of rats fed ethanol chronically for 4 weeks. TUNEL staining was performed on liver sections. Note that proteasome inhibitor treatment induced a significant apoptosis (arrows) and significantly reduced the macrovesicular fat formation compared to the ethanol alone
To verify that proteasome inhibitor treatment was efficient in reducing the ethanol-induced oxidative stress at this dosing, we analyzed the protein levels of the antioxidative enzymes GPX2 and GCLC. Figures 5A and 5B show that a significant increase of the levels of these proteins occurred.
Figure 5.
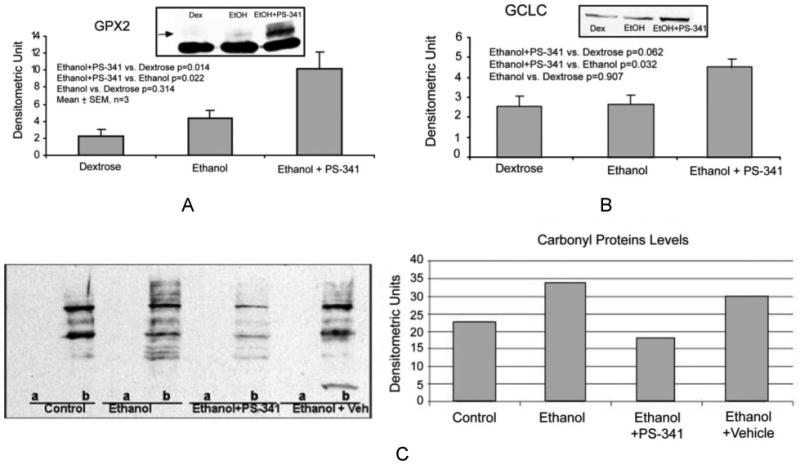
Western blot analysis of GPX2 (A) and GCLC (B) levels in the liver of rats fed ethanol and rats fed ethanol+PS-341. Proteasome inhibitor treatment increased the protein levels of these antioxidative enzymes. (C) Left: Immunodetection of oxidized protein levels was performed on control derivatized samples (A), negative control) and on DNP-derivatized sample (B). Right: Densitometric Unit measurement of the carbonyl proteins smear. Ethanol feeding increased the amount of oxidized proteins (smear), and proteasome inhibitor PS-341 treatment prevented this increase.
Carbonyl proteins levels were also measured using Western blot analysis. The results show that proteasome inhibitor administration to rats fed ethanol prevented the protein carbonyl formation (Figure 5C).
Nrf2 stabilization in the cytoplasm was assessed by immunohistochemistry analysis to investigate the mechanism of proteasome inhibitor treatment in up regulating the antioxidative enzymes gene expression. Nrf2 is the transcription factor that plays a key role in the activation of cellular responses to oxidative stress [Kobayashi et al., 2002, 2006]. Figure 6A shows that the gene expression of Nrf2 was surprisingly decreased by the combination of ethanol feeding and proteasome inhibitor treatment. The protein levels and the nuclear extract were also found down regulated (data not shown). It has been documented that when Nrf2 levels are high, the de novo synthesized Nrf2 translocates to the nucleus, and not only activates the ARE gene expression, but also autoregulates its own gene expression [Kwak et al., 2002].
Figure 6.
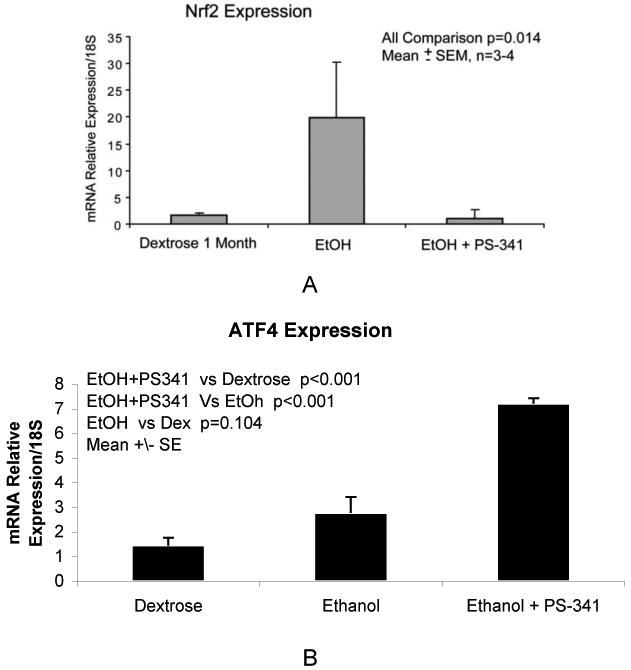
A: (A) qRT-PCR analysis of Nrf2 gene expression, in the livers of rats fed ethanol and given PS-341. The combination of ethanol feeding and proteasome inhibitor treatment decreased the gene expression of Nrf2. Mean ±SE, n=3. B : qRT-PCR analysis of ATF4 gene expression in the liver of rats fed ethanol and rats fed ethanol and rats fed ethanol and treated with PS-341. Mean ±SE, n=3.
Therefore, it was clear that different mechanism was involved in the beneficial effects of proteasome inhibitor treatment against the oxidative stress. ATF4 is a transcriptional factor with a well documented role in up regulating the antioxidative stress response. Figure 6B shows that ATF4 gene expression was significantly increased when the rats were fed ethanol and treated with PS-341. However, ethanol feeding alone did not significantly increase ATF4 gene expression, indicating that proteasome inhibitor treatment stabilized ATF4, which would stimulate the antioxidative stress response.
Discussion
This study shows that exposure to proteasome inhibitor PS-341 (Bortezomib, Velcade®) produces significant increase in gene expression of antioxidative enzymes. We hypothesized that the elevation of these antioxidative enzymes levels by proteasome inhibitor treatment would be beneficial against the oxidative stress observed in the liver of rats fed ethanol chronically. Therefore, we examined the effects of proteasome inhibitor PS-341 treatment in the liver of rats fed ethanol for one month. The 0.5 mg/kg bw dose was chosen as it is the maximum dose that rats could tolerate. Higher doses were lethal. The results showed an up regulation in the mRNA levels of antioxidative enzymes GPX2, GSS, GSR, when rats were exposed to the proteasome inhibitor, while ethanol feeding for 1 month alone did not up regulate the levels of mRNA transcripts of these antioxidative enzymes. For the first time, the present study documents the potential beneficial effects of a proteasome inhibitor treatment against oxidative stress increased in experimental alcoholic liver disease.
We demonstrate here that proteasome inhibition has different actions than does ethanol. The ethanol-induced proteasome dysfunction is different from the PS-341-induced proteasome inhibition, because it did not up regulate the antioxidative enzymes gene expression, and because it mainly alters the interaction of the proteasome with its associating proteins that help regulating the specificity of the proteasome system [Bardag-Gorce, 2010; Bousquet-Dubouch et al., 2009]. In addition, PS-341 is a reversible inhibitor. 48h to 72h post treatment, the proteasome activity recovered and our results showed that it was enhanced, which strongly supports the cytoprotective effects of proteasome inhibitor treatment. It is not an added burden of proteasome inhibition along with the ethanol-mediated decrease of proteasome function, because rats, fed ethanol chronically did, indeed tolerate very well the proteasome inhibitor treatment at 0.5 mg/kg, once a week, or even at lower dose administered more than once a week (data not shown).
In an in vitro study [Bardag-Gorce et al., 2006a], we showed that ethanol treatment causes a decrease in proteasome chymotrypsin-like activity, which lead to an accumulation of oxidized and altered proteins in their way to be degraded by the proteasome. Our results showed no polyubiquitinated proteins accumulation in the liver of rat fed ethanol chronically and showed an increase in polyubiquitinated proteins levels when proteasome activity was inhibited by PS-341. However, 48h to 72h post injection PS-341 was no longer inhibiting the proteasomes. And our results showed a significant increase of proteasome activity, which would digest the accumulated polyubiquitinated proteins.
The present study brings additional evidence that proteasome inhibition, obtained with the specific proteasome inhibitor PS-341, has different mechanisms of action than that of proteasome inhibition obtained with chronic ethanol feeding. Our present results show that proteasome inhibition, induced by chronic ethanol feeding, does not up regulate the mRNA levels of antioxidative enzymes. It is well known that proteasome inhibition obtained by PS-341 treatment for a long period of time and at high doses, causes cell death. This is the concept of proteasome inhibition used in the cancer research field. Therefore, no cytoprotective effects are obtained, but rather cytotoxic effects to suppress the tumor cells. The dosing protocol is the key to successful cytoprotective effects of proteasome inhibitor treatment in alcoholic liver disease. In addition, we believe that the specificity of the proteasome inhibitor is also an important factor in obtaining this cytoprotection. The literature reports in in vitro studies that treatment with lactacystin or MG132 proteasome inhibitors produced the same beneficial effects, but at a specific dosing protocol [Bradford et al., 2008; Sekhar et al., 2000; Yamamoto et al., 2007]. PS-341 is currently used in humans as an antitumor drug [Gilardini et al., 2008], and represents a potential drug treatment in the alcoholic liver disease as well, since chronic ethanol exposure is reported to decrease the level of GSH [Das et al., 2007; Zeng et al., 2008]. Proteasome inhibitor is thus a promising treatment to reduce ROS production due to ethanol metabolism by up regulating the antioxidative enzymes, thus protecting against ethanol-induced oxidative stress that causes liver injuries.
Glutathione peroxidase 2 (GPX2), which is responsible for glutathione-dependent hydrogen peroxide-reducing activity in the epithelium of the gastrointestinal tract, was significantly induced in the liver of rats treated with proteasome inhibitor treatment. Glutathione synthetase (GSS), which activates glutathione biosynthesis, glutathione reductase, which maintains high levels of reduced glutathione in the cytosol, and superoxide dismutase 2, a mitochondrial that is responsible for ROS and superoxide removal, were all up regulated by proteasome inhibitor treatment in the liver of rats fed ethanol. Glutamate cysteine ligase catalytic subunit (GCLC), which is the rate limiting enzyme in the synthesis of glutathione, showed significantly increased levels in the liver of rats fed ethanol and given the proteasome inhibitor PS-341. These up regulations were observed in two different dosing protocols (one and four administrations of proteasome inhibitor), which indicates the strong effects of proteasome inhibitor treatment in inducing the antioxidative enzymes.
These phase II detoxification enzymes genes are targeted by Nrf2 activation [Gong et al., 2006; de Vries et al., 2008]. Nrf2 is known to be induced in the liver of rodents fed ethanol chronically. Fed ethanol knockout rodent for Nrf2 showed dramatic mortality [Lamle et al., 2008], which was also found in our model of alcoholic liver disease. Nrf2 turnover is controlled by the ubiquitin proteasome pathway [Kobayashi et al., 2002; Stewart et al., 2003], thus proteasome inhibition would stabilizes Nrf2 levels. The results showed that proteasome inhibition by PS-341 did not induce Nrf2 gene expression, in contrast it was significantly decreased. These results indicate that other mechanisms of antioxidative response elements (ARE) activation are involved in the stimulatory effects of proteasome inhibitor treatment, and that Nrf2 may not be the sole regulator of ARE activation. ATF4 is a major transcription factor involved in the ER unfolded protein response and integrated stress response and is up regulated by PS-341 [Milani et al., 2009]. ATF4 is a key player in regulating the activation of glutathione synthesis because knockdown of ATF4 impairs the expression of genes involved in amino acid import, glutathione biosynthesis, and resistance to oxidative stress [Harding et al., 2003]. Our results confirm what has been reported in the literature and indicated that ATF4 was stabilized by proteasome inhibitor treatment, thus showing that ATF4 is the mechanism by which proteasome inhibitor treatment confers protective effects against the oxidative stress and glutathione depletion caused by chronic ethanol feeding. As proteasome inhibition contributes to ARE activation by a limited degradation of a transcriptional factors recruited for the target gene expression, and because proteasome inhibitor PS-341 is a reproducible and potent stimulatory agent of the antioxidant response elements, we believe it is necessary to further investigate the dosing protocol in order to address safety aspects of the treatment for a perspective translational/clinical research.
Acknowledgments
The authors would like to thank Emmanuel Gorce for drawing illustrations and word processing and editing this manuscript.
Financial Support: NIH/NIAAA Grant 8116 and the USC Research Center for Alcoholic liver and Pancreatic Disease (ALPD) and cirrhosis Pilot Project funding, and Morphologic Core.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bardag-Gorce F, Yuan QX, Li J, French BA, Fang C, Ingelman-Sundberg M, French SW. The effect of ethanol-induced cytochrome p4502E1 on the inhibition of proteasome activity by alcohol. Biochem Biophys Res Commun. 2000;279(1):23–29. doi: 10.1006/bbrc.2000.3889. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, Li J, French BA, French SW. Ethanol withdrawal induced CYP2E1 degradation in vivo, blocked by proteasomal inhibitor PS-341. Free Radic Biol Med. 2002;32(1):17–21. doi: 10.1016/s0891-5849(01)00768-7. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, French BA, Nan L, Song H, Nguyen SK, Yong H, Dedes J, French SW. CYP2E1 induced by ethanol causes oxidative stress, proteasome inhibition and cytokeratin aggresome (Mallory body-like) formation. Exp Mol Pathol. 2006a;3:191–201. doi: 10.1016/j.yexmp.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, French BA, Dedes J, Li J, French SW. Gene expression patterns of the liver in response to alcohol: in vivo and in vitro models compared. Exp Mol Pathol. 2006b;80:241–251. doi: 10.1016/j.yexmp.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F. Effects of ethanol on the proteasome interacting proteins. World J Gastroenterol. 2010;16(11):1349–1357. doi: 10.3748/wjg.v16.i11.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler S, Meiners S, Stangl V, Pohl T, Stangl K. Comprehensive proteomic and transcriptomic analysis reveals early induction of a protective anti-oxidative stress response by low-dose proteasome inhibition. Proteomics. 2009;9(12):3257–3267. doi: 10.1002/pmic.200800927. [DOI] [PubMed] [Google Scholar]
- Bousquet-Dubouch MP, Nguen S, Bouyssié D, Burlet-Schiltz O, French SW, Monsarrat B, Bardag-Gorce F. Chronic ethanol feeding affects proteasome-interacting proteins. Proteomics. 2009;9:3609–3622. doi: 10.1002/pmic.200800959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederbaum A. Nrf2 and antioxidant defense against CYP2E1 toxicity. Expert Opin Drug Metab Toxicol. 2009;5(10):1223–1244. doi: 10.1517/17425250903143769. [DOI] [PubMed] [Google Scholar]
- Das SK, Vasudevan DM. Alcohol-induced oxidative stress. Life Sci. 2007;81(3):177–187. doi: 10.1016/j.lfs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- de Vries HE, Witte M, Hondius D, Rozemuller AJ, Drukarch B, Hoozemans J, van Horssen J. Nrf2-induced antioxidant protection: a promising target to counteract ROS-mediated damage in neurodegenerative disease? Free Radic Biol Med. 2008;45(10):1375–1383. doi: 10.1016/j.freeradbiomed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Dreiseitel A, Schreier P, Oehme A, Locher S, Rogler G, Piberger H, Hajak G, Sand PG. Inhibition of proteasome activity by anthocyanins and anthocyanidins. Biochem Biophys Res Commun. 2008;372(1):57–61. doi: 10.1016/j.bbrc.2008.04.140. [DOI] [PubMed] [Google Scholar]
- Du ZX, Zhang HY, Meng X, Guan Y, Wang HQ. Role of oxidative stress and intracellular glutathione in the sensitivity to apoptosis induced by proteasome inhibitor in thyroid cancer cells. BMC Cancer. 2009:56. doi: 10.1186/1471-2407-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Checa JC, Kaplowitz N, García-Ruiz C, Colell A, Miranda M, Marí M, Ardite E, Morales A. GSH transport in mitochondria: defense against TNF-induced oxidative stress and alcohol-induced defect. Am J Physiol. 1997;273(1):G7–G17. doi: 10.1152/ajpgi.1997.273.1.G7. [DOI] [PubMed] [Google Scholar]
- Friguet B, Szweda LI. Inhibition of the multicatalytic proteinase (proteasome) by 4-hydroxy-2-nonenal cross-linked protein. FEBS Lett. 1997;405:21–25. doi: 10.1016/s0014-5793(97)00148-8. [DOI] [PubMed] [Google Scholar]
- Gilardini A, Marmiroli P, Cavaletti G. Proteasome inhibition: a promising strategy for treating cancer, but what about neurotoxicity? Curr Med Chem. 2008;15(29):3025–3035. doi: 10.2174/092986708786848622. [DOI] [PubMed] [Google Scholar]
- Gong P, Cederbaum AI. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology. 2006;43:144–153. doi: 10.1002/hep.21004. [DOI] [PubMed] [Google Scholar]
- Hamilton AL, Eder JP, Pavlick AC, Clark JW, Liebes L, Garcia-Carbonero R, Chachoua A, Ryan DP, Soma V, Farrell K, Kinchla N, Boyden J, Yee H, Zeleniuch-Jacquotte A, Wright J, Elliott P, Adams J, Muggia FM. Proteasome inhibition with bortezomib (PS-341): a phase I study with pharmacodynamic end points using a day 1 and day 4 schedule in a 14-day cycle. J Clin Oncol. 2005;23(25):6107–6116. doi: 10.1200/JCO.2005.01.136. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko C, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hill B, Haberzettl P, Hamed Y, Srivastava S, Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem J. 2008;410:525–534. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- Igarashi T, Izumi H, Uchiumi T, Nishio K, Arao T, Tanabe M, Uramoto H, Sugio K, Yasumoto K, Sasaguri Y, Wang KY, Otsuji Y, Kohon K. Clock and ATF4 transcription system regulates drug resistance in human cancer cell lines. Oncogene. 2007;26:4749–4760. doi: 10.1038/sj.onc.1210289. [DOI] [PubMed] [Google Scholar]
- Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 2004;36:1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cel Biol. 2006;1:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: Role of antioxidant response element-like sequences in the Nrf2 promoter. Mol Cel Biol. 2002;22(9):2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamle J, Marhenke S, Borlak J, von Wasielewski S, Eriksson CJ, Geffers R, Manns MP, Yamamoto M, Vogel A. Nuclear factor-eythroid 2–related factor 2 prevents alcohol-induced fulminant liver injury induce in ALD. Gastroenterol. 2008;134:1159–1168. doi: 10.1053/j.gastro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Lee CS, Tee LY, Warmke T, Vinjamoori A, Cai A, Fagan AM, Snider BJ. A proteasomal stress response: pre-treatment with proteasome inhibitors increases proteasome activity and reduces neuronal vulnerability to oxidative injury. J Neurochem. 2004;91(4):996–1006. doi: 10.1111/j.1471-4159.2004.02813.x. [DOI] [PubMed] [Google Scholar]
- Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- Li J, French BA, Fu P, Bardag-Gorce F, French SW. Mechanism of the alcohol cyclic pattern: role of catecholamines. Am J Physiol Gastrointest Liver Physiol. 2003;285(2):G442–G448. doi: 10.1152/ajpgi.00093.2003. [DOI] [PubMed] [Google Scholar]
- Li L, Chiu JF, Kelsen A, Lu SC, Fukagawa NK. Identification and characterization of an Nrf2-mediated ARE upstream of the rat glutamate cysteine ligase catalytic subunit gene (GCLC) J Cel Biochem. 2009;107:944–954. doi: 10.1002/jcb.22197. [DOI] [PubMed] [Google Scholar]
- Lorenz M, Wilck N, Meiners S, Ludwig A, Baumann G, Stangl K, Stangl V. Proteasome inhibition prevents experimentally-induced endothelial dysfunction. Life Sci. 2009;84(25-26):929–934. doi: 10.1016/j.lfs.2009.04.016. [DOI] [PubMed] [Google Scholar]
- Marques C, Pereira P, Taylor A, Liang JN, Reddy VN, Szweda L, Shang F. Ubiquitin-dependent lysosomal degradation of the HNE-modified proteins in lens epithelial cells. FASEB J. 2004;18:1424–1426. doi: 10.1096/fj.04-1743fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, Harris AL. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69(10) doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- Nencioni A, Grünebach F, Patrone F, Ballestrero A, Brossart P. Proteasome inhibitors: antitumor effects and beyond. Leukemia. 2007;21:30–36. doi: 10.1038/sj.leu.2404444. [DOI] [PubMed] [Google Scholar]
- Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, Yamamoto M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci USA. 2010;107(7):2842–2847. doi: 10.1073/pnas.0914036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K. 4-Hydroxy-2-nonenal-mediated impairment of intracellular proteolysis during oxidative stress. Identification of proteasomes as target molecules. J Biol Chem. 1999;274:23787–23793. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- Phillips JB, Williams AJ, Adams J, Elliott PJ, Tortella FC. Proteasome inhibitor PS519 reduces infarction and attenuates leukocyte infiltration in a rat model of focal cerebral ischemia. Stroke. 2000;31:1686–1693. doi: 10.1161/01.str.31.7.1686. [DOI] [PubMed] [Google Scholar]
- Poss KD, Tonegawa S. Reduced stress defense in heme oxygenase 1-deficient cells. PNAS. 1997;94(20):10925–10930. doi: 10.1073/pnas.94.20.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Harada N, Hosoya T, Tohyama K, Yamamoto M, Itoh K. Keap1/Nrf2 system regulates neuronal survival as revealed through study of keap1 gene-knockout mice. Biochem Biophys Res Commun. 2009;380(2):298–302. doi: 10.1016/j.bbrc.2009.01.063. [DOI] [PubMed] [Google Scholar]
- Sekhar KR, Soltaninassab SR, Borrelli MJ, Xu ZQ, Meredith MJ, Domann FE, Freeman ML. Inhibition of the 26S proteasome induces expression of GLCLC, the catalytic subunit for g-glutamylcysteine synthetase. Biochem Biophys Res Comm. 2000;270:311–317. doi: 10.1006/bbrc.2000.2419. [DOI] [PubMed] [Google Scholar]
- Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J Biol Chem. 2003;278(4):2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Dave JR, Tortella FC. Neuroprotection with the proteasome inhibitor MLN519 in focal ischemic brain injury: relation to nuclear factor kappaB (NF-kappaB), inflammatory gene expression, and leukocyte infiltration. Neurochem Int. 2006;49:106–112. doi: 10.1016/j.neuint.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Sawada H, Izumi Y, Kume T, Katsuki T, Shimohama S, Akaike A. Proteasome inhibition induces glutathione synthesis and protects cells from oxidative stress. J Biol chem. 2007;282(7):4364–4372. doi: 10.1074/jbc.M603712200. [DOI] [PubMed] [Google Scholar]
- Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem. 2004;279(45):47109–47114. doi: 10.1074/jbc.M410010200. [DOI] [PubMed] [Google Scholar]
- Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, Lu SC. Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kB and AP-1. Mol Cel Biol. 2005;14:5933–5946. doi: 10.1128/MCB.25.14.5933-5946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T, Zhang CL, Zhu ZP, Yu LH, Zhao XL, Xie KQ. Diallyl trisulfide (DATS) effectively attenuated oxidative stress-mediated liver injury and hepatic mitochondrial dysfunction in acute ethanol-exposed mice. Toxicology. 2008;252(1-3):86–91. doi: 10.1016/j.tox.2008.07.062. [DOI] [PubMed] [Google Scholar]


