Abstract
Background
Genetic testing for long QT syndrome (LQTS) has diagnostic, prognostic, and therapeutic implications. Hundreds of causative mutations in 12 known LQTS-susceptibility genes have been identified. Genetic testing that includes the 3 most commonly mutated genes is available clinically. Distinguishing pathogenic mutations from innocuous rare variants is critical to the interpretation of test results. We sought to quantify the value of mutation type and gene/protein region in determining the probability of pathogenicity for mutations.
Methods and Results
Type, frequency, and location of mutations across KCNQ1 (LQT1), KCNH2 (LQT2) and SCN5A (LQT3) were compared between 388 unrelated “definite” (clinical diagnostic score > 4 and/or QTc > 480 ms) cases of LQTS and over 1300 healthy controls for each gene. From these data, estimated predictive values (EPV, meaning the percent of mutations found in definite cases that would be LQTS-causing) were determined according to mutation type and location. Mutations were 10× more common in cases than controls (0.58/case vs 0.06/control). Missense mutations were the most common, accounting for 78%, 67%, and 89% of mutations in KCNQ1, KCNH2, and SCN5A in cases and >95% in controls. Non-missense mutations have an EPV >99% regardless of location. In contrast, location appears to be critical for characterizing missense mutations. Relative frequency of missense mutations between cases and controls ranged from ~1:1 in the SCN5A interdomain linker (IDL) to infinity in KCNH2’s pore (P), transmembrane (TM), and linker (L). These correspond to EPVs ranging from 0% in the IDL of SCN5A to 100% in the TM/L/P regions of KCNH2. EPV is also high in KCNQ1’s L, P, TM, and C-terminus and the TM/L of SCN5A.
Conclusions
Distinguishing pathogenic mutations from rare variants is of critical importance in the interpretation of genetic testing in LQTS. Mutation type, mutation location, and ethnic specific background rates are critical factors in predicting pathogenicity of novel mutations. Novel mutations in low-EPV regions, such as the IDL of SCN5A, should be viewed as variants of uncertain significance (VUS) and prompt further investigation to clarify the likelihood of disease causation. However, mutations in regions such as the TM, L, and P of KCNQ1 and KCNH2 may be defined confidently as high probability LQTS-causing mutations. These findings will have implications for other genetic disorders involving mutational analysis.
Keywords: Genetics, Long-QT Syndrome, Ion Channels
INTRODUCTION
The modern era of genetic testing has afforded clinicians the opportunity to test for many genetically transmitted diseases. As genetic tests make their transition from “investigational” tests to bona fide, clinically indicated and reimbursed tests, it is likely that increasing numbers of patients will undergo genetic analysis, either for specific diseases or eventually of their entire genome, potentially revealing numerous rare mutations of unknown clinical significance, otherwise known as “variants of uncertain significance” (VUS). Currently, nearly 1000 genetic tests (www.genetests.org) are available clinically and are offered through more than 600 diagnostic laboratories worldwide.[1]
Interpreting the significance of a genetic test result in the context of insufficient clinical evidence for a particular disease phenotype (or pre-test probability of disease) is challenging for medical geneticists, disease subspecialists, and primary physicians.[2] A recent editorial by Hunter et al. detailed some of the challenges with genetic testing and the associated diagnostic implementation.[3] For sudden death predisposing diseases like congenital long QT syndrome (LQTS), in which highly effective medical-, surgical-, and device-related therapies are available, proper interpretation of the genetic test is critical considering the diagnostic, prognostic, and therapeutic implications of a positive LQTS test.[4]
Since the sentinel discovery of the first LQTS-susceptibility locus on chromosome 11 in 1991[5] and the discovery of mutations in key cardiac potassium and sodium channels in 1995[6,7], LQTS has been understood as a cardiac channelopathy. To date, hundreds of LQTS-causing mutations in at least 12 LQTS-susceptibility genes have been described, and a litany of genotype-phenotype studies have revealed relatively gene-specific electrocardiographic patterns, arrhythmogenic triggers, risk for sudden death, and responses to pharmacotherapy.[8–14] Recognizing the essential role of LQTS genetic testing in the evaluation and management of patients, pseudo-clinical testing was conducted in a handful of research laboratories from 1995 – 2004. In 2004, LQTS genetic testing for 5 LQTS-susceptibility genes, including the three major genes [KCNQ1 (LQT1), KCNH2 (LQT2), and SCN5A (LQT3)], became a commercially available genetic test in North America.
Mutations in the 3 major LQTS-susceptibility genes account for approximately 70–75% of congenital LQTS cases while the 9 other minor genes contribute approximately an additional 5%.[15] The remaining 20–25% of LQTS continues to be genetically elusive. Genetic variants identified through genetic testing may represent known polymorphisms; known disease-causing mutations; radical mutations, including splice, in-frame, nonsense and frame-shift mutations; silent variants; or rare, non-synonymous single nucleotide polymorphisms (nsSNPs)/missense mutations that have not been identified or characterized previously.
In addition to the published and web-available compendia of putative LQTS-causing mutations, two compendia of rare potassium channel and sodium channel variants discovered among over 800 ostensibly healthy volunteers have been published.[16–17] These control data established not only the ethnicity-dependent frequencies for common polymorphisms but also revealed that approximately 5% of otherwise healthy individuals (3–4% of Caucasians and 6–8% of African Americans) may have what could be interpreted as a “positive” genetic test.[16–17] In other words, nearly 1 in 20 controls hosts a rare (<0.5% allelic frequency, typically unique) genetic variant that alters the amino acid sequence of one of the channel proteins. Considering that LQTS has an estimated incidence of 1 in 2500 persons, the vast majority of these rare variants found in controls must be innocuous, having no clinical relevance and little or no functional difference from wild-type.
This so-called “background genetic noise”, now quantified for the LQTS genes, is almost certain to be present for virtually every disease-susceptibility gene and serves as a powerful reminder that genetic tests, like most diagnostic tests, are probabilistic rather than binary. Distinguishing pathogenic mutations from otherwise “just rare, just there” mutations is of paramount importance. Heterologous expression studies of every ion channel variant to assess in vitro whether the mutation confers a perturbed molecular/cellular phenotype is prohibitively time consuming, sometimes not translatable from heterologous system to human host, and thus not suitable for regulated, clinical testing. To date, < 20% of all published LQTS-associated mutations have been characterized functionally. New variants are being discovered at a far greater rate than characterization experiments can be performed currently.
Therefore, in an effort to enhance the diagnostic interpretability of the next novel genetic variant, we conducted a large multi-center case-control study examining the properties of mutations derived from high probability clinical cases compared with those similarly rare variants derived from ostensibly healthy volunteers. Here, we have expanded on previous observations to quantify the signal-to-noise ratio in distinguishing a pathogenic mutation from an innocuous one based on mutation type and location.
METHODS
Case-Control Study Design
From 1997 to 2007, over 1300 patients were referred to either Mayo Clinic’s Windland Smith Rice Sudden Death Genomics Laboratory in Rochester, Minnesota, USA, the Cardiogenetics Clinic at the Academic Medical Center in Amsterdam, The Netherlands, or PGxHealth in New Haven, CT for comprehensive LQTS genetic testing following written, informed consent.
In an effort to polarize the cohorts (cases versus controls) as cleanly as possible, only “clinically definite” unrelated index cases were examined, amounting to 388 of the total referral population, which increases the a priori or pre-test probabilities that a case-derived mutation is indeed pathogenic. For the purpose of this case-control mutation analysis, “clinically definite” cases were defined as those with a clinical diagnostic score (“Schwartz score”) > 4 or a corrected QT interval (QTc) greater than 480 ms. These criteria have been shown to correlate with a high clinical probability for LQTS.[18] All cases were unrelated, and demographics including ethnicity, age, and sex were recorded at the time of sequencing.
The spectrum and prevalence of case mutations as well as mutation type and mutation location were compared with the characteristics of the genetic variants found among controls. The controls consisted of 744 unrelated seemingly healthy volunteers analyzed previously for K channel variants[16] and the 829 healthy volunteers analyzed previously for Na channel variants[17] along with over 500 additional non-LQTS volunteers sequenced for this study by PGxHealth (Table 1). DNA samples were obtained from the Human Genetic Cell Repository sponsored by the National Institute of General Medicine Sciences and the Coriell Institute for Medical Research (Camden, NJ) [16–17] as well as anonymized blood donors. An electrocardiogram evidencing a normal QT interval was not a pre-requisite for subjects comprising this control cohort. Table 1 summarizes the ethnic distribution of all cases and controls.
Table 1.
Ethnic Distribution of Cases and Controls
| Ethnicity | Cases | Controls | ||
|---|---|---|---|---|
| KCNQ1 | KCNH2 | SCN5A | ||
| African American | 7 | 372 | 372 | 368 |
| Caucasian | 326 | 559 | 559 | 647 |
| Asian | 6 | 172 | 172 | 131 |
| Hispanic | 13 | 155 | 180 | 148 |
| Unknown/other | 36 | 86 | 86 | 86 |
| Total | 388 | 1344 | 1369 | 1380 |
LQTS Genetic Testing
The genomic DNA of all cases and controls was analyzed for mutations in the 60 translated exons and the splice site regions of the three major LQTS-susceptibility genes: KCNQ1, KCNH2, and SCN5A, responsible for LQT1, LQT2, and LQT3 respectively. Mutation analyses were performed using polymerase chain reaction and either denaturing high-performance liquid chromatography followed by automated DNA sequencing, or direct high throughput DNA sequencing.[19]
Mutation sequence analysis
Only genetic variants predicted to alter the open reading frame thereby affecting the primary amino acid composition of the cardiac channel were included. To be considered a possible pathogenic mutation in this study, the particular genetic variant found in a case must not have been observed in our control population. Variants seen in controls more than once were considered polymorphisms for purposes of this study and were excluded from analysis. Accordingly, common amino acid substitutions, a.k.a. “common polymorphisms,” such as KCNH2-K897T and SCN5A-H558R did not influence the analyses of mutation type and topological locations in this study. The term “mutation” in this paper is not meant to imply pathogenicity or even functional abnormality, merely rarity and predicted protein alteration as defined above. Mutations were then classified using standard nomenclature.[20] Splice site, nonsense, frame-shift, and in-frame insertion/deletions were grouped together as “radical” mutations.
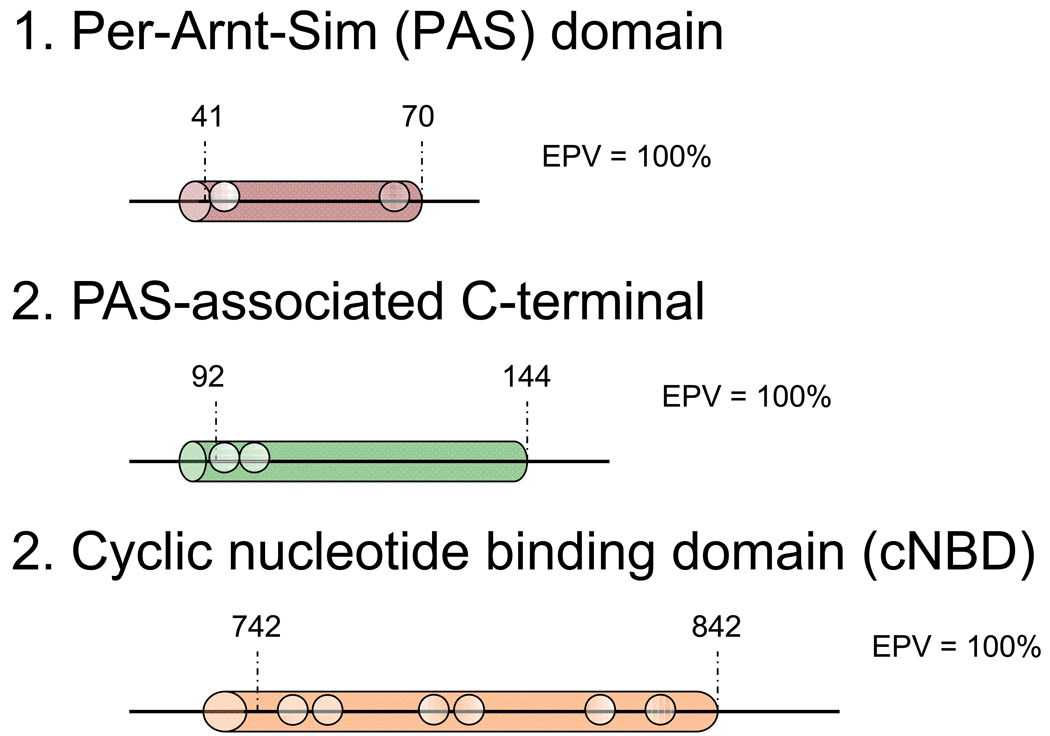
Mutations were localized to exon and specific gene-encoded protein regions according to a combination of Swissprot (http://ca.expasy.org/uniprot/) and recent studies of genomic and protein organization for the 3 genes.[21–23] The Swissprot database offers generally accepted residue ranges corresponding with each region and specialized sub-regions of the ion channel, used here with slight modifications. For KCNQ1 and KCNH2, mutations were characterized as being located in the N-terminus (KCNQ1, amino acids (aa) 1–121; KCNH2, aa 1–403), the region between the N- and C- termini comprising the transmembrane (TM), linker (L), and Pore (P) (KCNQ1, aa 122–348; KCNH2 aa 404–659; or the C-terminus (KCNQ1, aa 349–676; KCNH2, aa 660–1159).[14] For SCN5A, mutations were characterized as being located in the N- terminus (aa 1–126), inter-domain linker (IDL I-II, aa 416–711, IDL II–III, aa 940–1200, and IDL III–IV, aa 1471–1523), transmembrane/linker (Domain I, aa 127–415, Domain II, aa 712–939, Domain III, aa 1201–1470, and Domain IV, aa1524–1772), or C-terminus (aa 1773–2016). Further sub-categorizations were used for four specialized sub-regions: subunits assembly domain (SAD, aa 589–620) of the C-terminus of KCNQ1, the Per-Arnt-Sim (PAS, aa 41–70) and PAS-Associated C-terminal (PAC, aa 92–144) sub-domains of the N-terminus of KCNH2, and the cyclic nucleotide binding domain (cNBD, aa 742–842) of KCNH2.
Statistical Analysis
Variables were analyzed using Fisher’s exact test. Frequencies of each type of mutation were computed for all subjects as well as a Caucasian-only subset analysis. Further analyses were performed within particular channel regions. Each mutation was counted every time it was seen, because the statistics concern frequencies of people carrying particular classes of mutation.
In order to estimate the likelihood of disease causation, a modification of the positive predictive value was used. We made the logical assumption that the incidence of benign, background mutations is the same for the case and control populations. We further made the simplifying assumption that all mutations found in controls are benign, background mutations, a reasonable assumption for the purpose of these analyses given the low prevalence of LQTS. On this basis, the case frequency of mutations in excess of the control frequency should represent the frequency of disease-causing mutations in cases. Employing these principles along with the genetic testing results of our study populations, we then calculated estimated predictive values (EPVs), the probability of pathogenicity of a mutation found in a case, where EPV = (case frequency − control frequency)/case frequency. The 95% confidence intervals (CI) were calculated for all EPVs and are included in brackets after the EPV. Upper and lower bounds of the 95% confidence intervals were calculated using the formula CI = 1−1/{e^[(ln (RR) ± z*(SE(log RR))]}, where RR = relative ratio (mutation frequency in cases/mutation frequency in controls); z = 1.959964 for 1-alpha = 95%; and SE(log(RR)) is the standard error around the log of RR. The EPVs calculated in this paper are specific to clinically definite cases as defined above.
RESULTS
Frequency of mutations in cases and controls
In total, 248 mutations (180 distinct: 129 missense mutations, 51 radical) were found among 224 of the 388 LQTS cases (58%). In contrast, only 79 unique mutations (77 missense) were found across the three genes among the > 1300 controls, (Figure 1 and Supplemental Table). While each control had no more than one mutation, 22 cases had two mutations across the three LQTS-causing genes, and one had three. Nine cases had two mutations in KCNQ1, two had two mutations in KCNH2, and one had two mutations in SCN5A, whereas the remaining multiples included more than one gene. In addition, 31 single amino acid substitutions, 2 inframe deletions and 1 inframe duplication were seen more than once in the controls (Figure 1 and Supplemental Table).
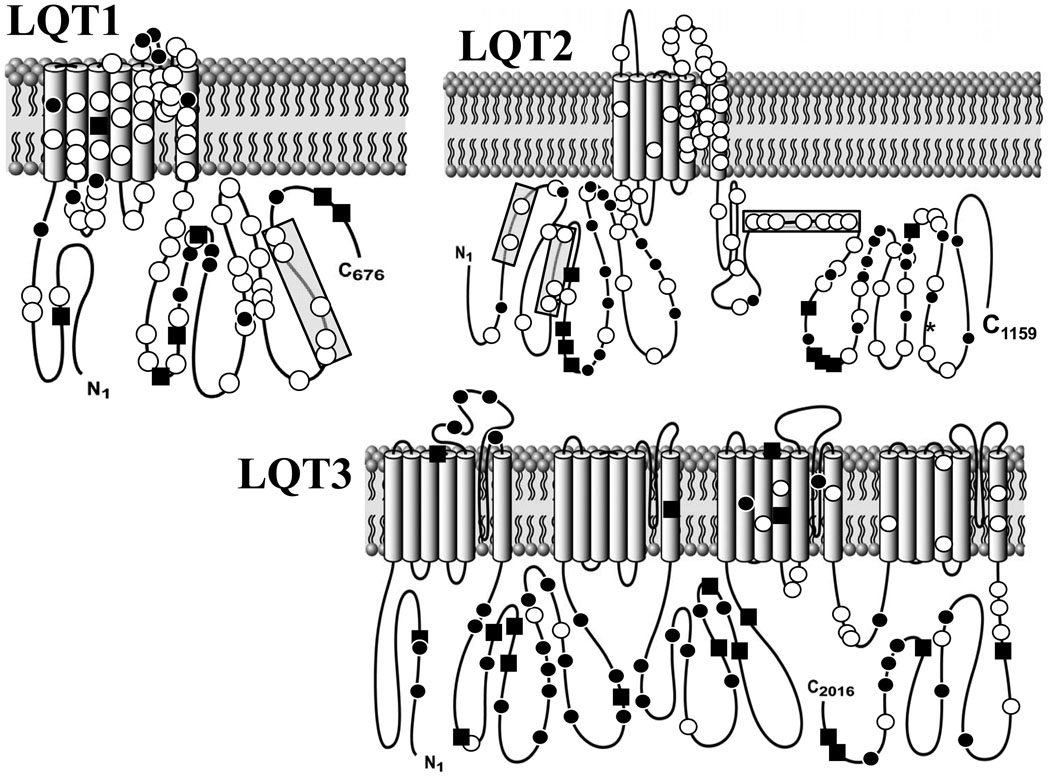
Figure 1. Topological Depiction of All Protein-Altering Mutations.
White circles represent case mutations. Black circles depict rare (each observed only once) genetic variants observed among the > 1300 healthy volunteers while black squares depict the genetic variants/polymorphisms observed more than once among the controls. All 155 distinct mutations, both radical and missense types, are included. Shaded regions: (1) SAD (subunits assembly domain); (2) PAS; (3) PAC; (4) cNBD (cyclic nucleotide binding domain).
Mutations were significantly more common in cases than controls in each of the three genes, but to different degrees (KCNQ1: 112/388 cases (0.29/subject) vs 14/1344 controls (0.01/subject), p = 1.24 × 10−62; KCNH2: 108/388 cases (0.28/subject) vs 28/1369 controls (0.02/subject), p = 1.41 × 10−50; SCN5A: 28/388 cases (0.07/subject) vs 37/1380 controls (0.03/subject), p = 9.71 × 10−5). Table 2 shows how these mutation rates break down by ethnicity, with typically lower mutation rates among Caucasian cases and controls than non-Caucasian cases and controls. The biggest ethnic difference was the greater mutation rate in non-Caucasian controls (0.08/subject) than Caucasian controls (0.04/subject, p = 0.0025).
Table 2.
Ethnic Specific Differences in Mutation Rates for Cases and Controls.
| Gene | Caucasians (%) | Non-Caucasians (%) | ||
|---|---|---|---|---|
| Case | Control | Case | Control | |
| KCNQ1 | 95/326 (0.29) | 4/559 (0.007) | 9/26 (0.35) | 10/699 (0.015) |
| KCNH2 | 89/326 (0.27) | 11/559 (0.02) | 9/26 (0.35) | 16/724 (0.022) |
| SCN5A | 25/326 (0.077) | 8/647 (0.012) | 2/26 (0.077) | 29/647 (0.045) |
Data are listed as the number of mutations seen/the number of subjects tested, with rate (mutations/subject) in parentheses. Those of race defined as “unknown” or “other” (N=36, cases; N=86, controls) were not included in either group.
While over 25% of all mutations in cases were considered radical mutations, such mutations were almost never observed amongst controls. In fact, the presence of a radical mutation (splice site, nonsense, frame-shift, or in-frame insertion/deletions in KCNQ1 and KCNH2; in-frame insertions/deletions in SCN5A) confers an EPV >99% regardless of gene or gene region. The presence of a radical mutation in KCNQ1 confers an EPV of 100% (CI 91–100), in KCNH2 an EPV of 99% (CI 94–100), and in SCN5A an EPV of 91% (CI 10–99).
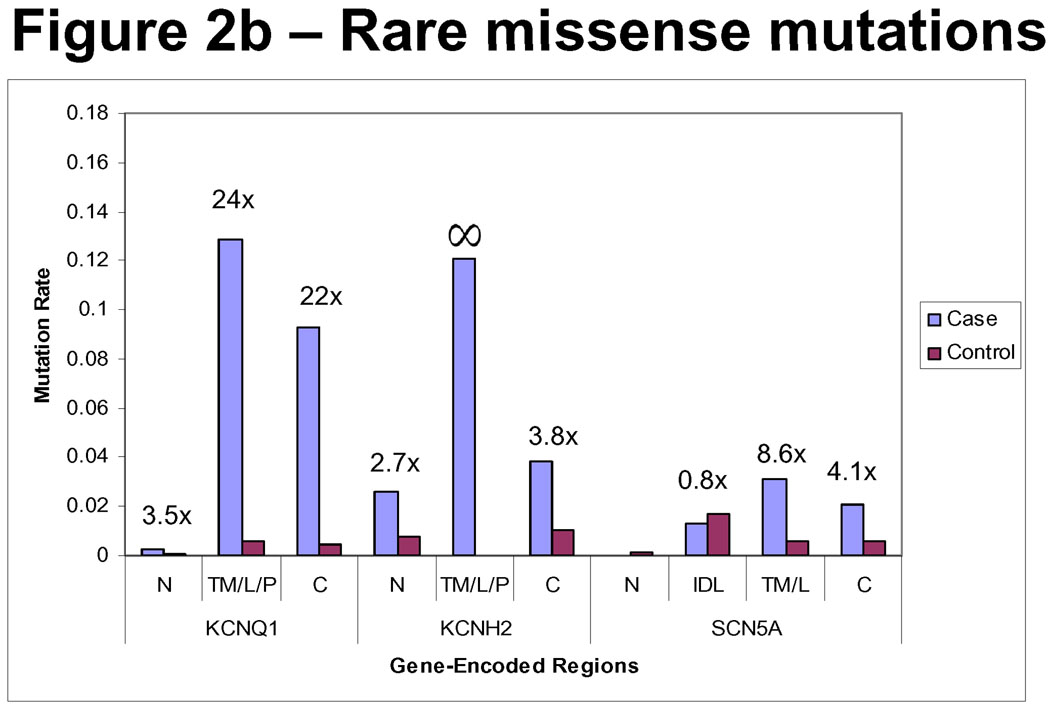
Missense mutations were the most common type of mutation, representing 74% (184/248) of all case mutations, including 78% (87/112) of KCNQ1, 67% (72/108) of KCNH2, 89% (25/28) of SCN5A mutations, and > 95% (77/79) of the control mutations. In contrast to radical mutations, the probability of pathogenicity, as captured in EPVs, for missense mutations is highly dependent on gene and protein region in which the mutation is discovered, as detailed in the next section.
Gene-specific estimated predictive value (EPV) analysis for missense mutations
KCNQ1 (LQT1-susceptibility gene)
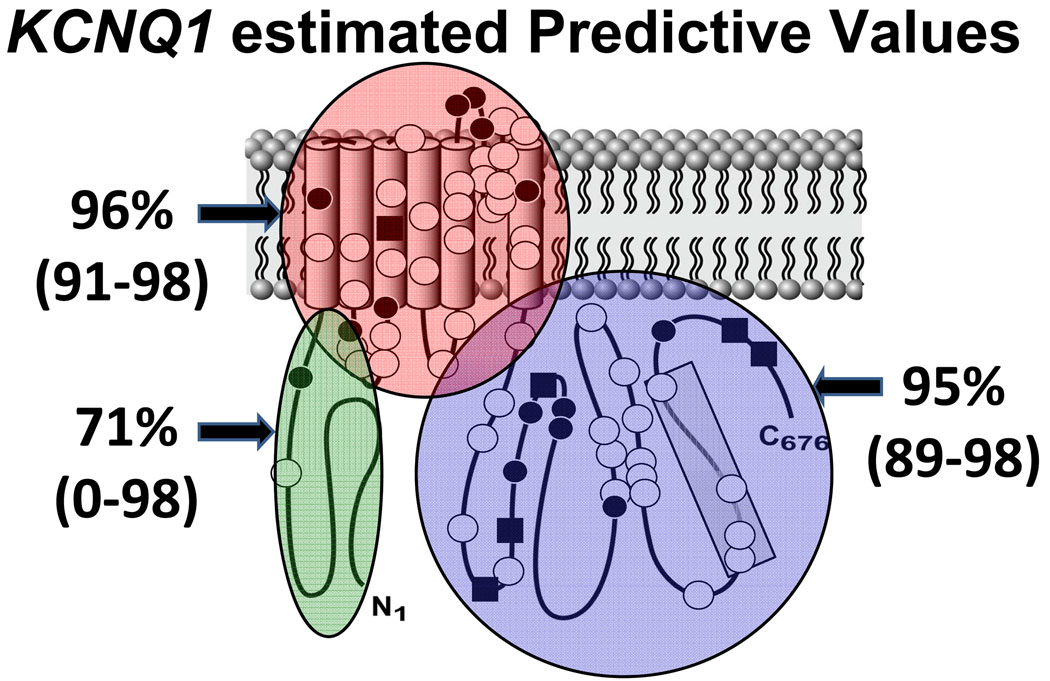
Figure 2 depicts the relative incidence of missense mutations in cases versus controls, divided on the basis of location. For all regions within the KCNQ1-encoded Kv7.1 potassium channel, there is a higher relative frequency of missense mutations in cases than in controls for all ethnicities (3.5 times (×) higher in cases than controls in the N-terminus, 24× in the transmembrane/linker/pore spanning region, and 21× in the C-terminus). The EPV for a mutation localizing anywhere in KCNQ1 was 96% (CI 94–98) and for a missense mutation was 95% (CI 92–97).
Figure 2. Mutation Rates Across Gene-Encoded Regions.
Depicted are the relative frequencies of mutations of all types (2a) and missense only (2b) in cases compared with controls. Note the relatively higher rate of mutations of all types across most regions in cases than controls. Furthermore, in most regions, there is a relatively higher frequency of missense mutations in cases than controls. The exceptions are the N-terminus and inter-domain linker (IDL) regions of SCN5A. N = N-terminus. T/L/P = Transmembrane/Linker/Pore. C = C-terminus. IDL = Inter-domain linker. T/L = Transmembrane/Linker.
While the EPV, or probability for pathogenicity, for a missense mutation localizing to the N-terminus of KCNQ1 was only 71% (CI 0–98), an EPV > 90% was suggested for missense mutations localizing to the transmembrane/linker/pore and C-terminus domains of KCNQ1 (Table 3). There were no control mutations localizing to the specialized subunits assembly domain (SAD) of the KCNQ1 C-terminus, suggesting a high probability of pathogenicity of mutations in this region (nominal EPV = 100%). The EPVs of missense mutations localizing to each region in KCNQ1 are shown in Figure 3.
Table 3.
Estimated Predictive Values for Missense Mutations in Clinically Definite LQTS Cases
| Gene | Location | Mutation Positive | Estimated Predictive Value (95% CI) | |
|---|---|---|---|---|
| Case | Control | |||
| KCNQ1 | N-terminus | 1/388 | 1/1344 | 71 (0–98) |
| Transmembrane/Linker/Pore | 50/388 | 7/1344 | 96 (91–98) | |
C-terminus
|
36/388
|
6/1344
|
95 (89–98)
|
|
| KCNH2 | N-terminus
|
10/388
|
13/1369
|
63 (17–84)
|
| Transmembrane/Linker/Pore | 47/388 | 0/1369 | 100 (95–100) | |
C-terminus
|
15/388
|
14/1369
|
74 (46–87)
|
|
| SCN5A | N-terminus | 0/388 | 2/1380 | 0 (0–95) |
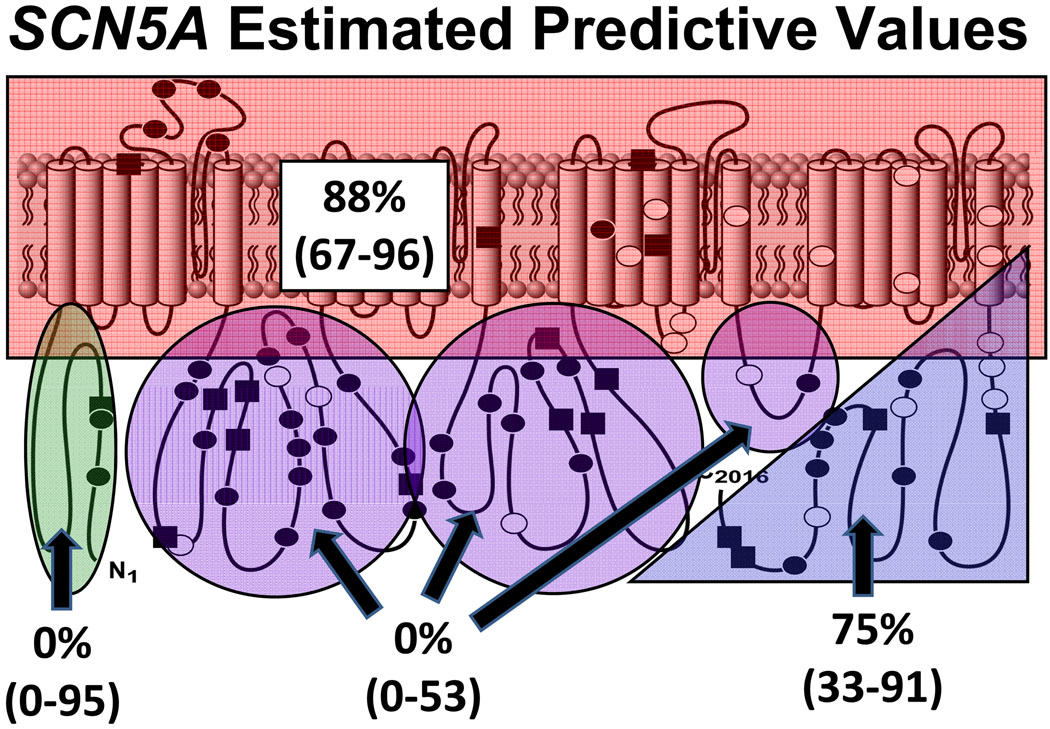
| Transmembrane/Linker | 12/388 | 5/1380 | 88 (67–96) | |
| Inter-domain linker (IDL) | 5/388 | 22/1380 | 0 (0–53) | |
| C-terminus | 8/388 | 7/1380 | 75 (33–91) | |
EPV = (Case rate − Control rate)/Case rate
Figure 3. Estimated Predictive Values for KCNQ1.
Estimated predictive values in clinically definite cases for a missense mutation occurring in each major structure-function domain in the KCNQ1-encoded Kv7.1 potassium channel. The 95% confidence intervals are provided in parentheses. White circles represent case mutations. Black circles depict rare genetic variants observed only once among healthy volunteers while black squares depict the genetic variants/polymorphisms observed more than once among the controls. Shaded region (1) represents the SAD (subunits assembly domain).
KCNH2 (LQT2-susceptibility gene)
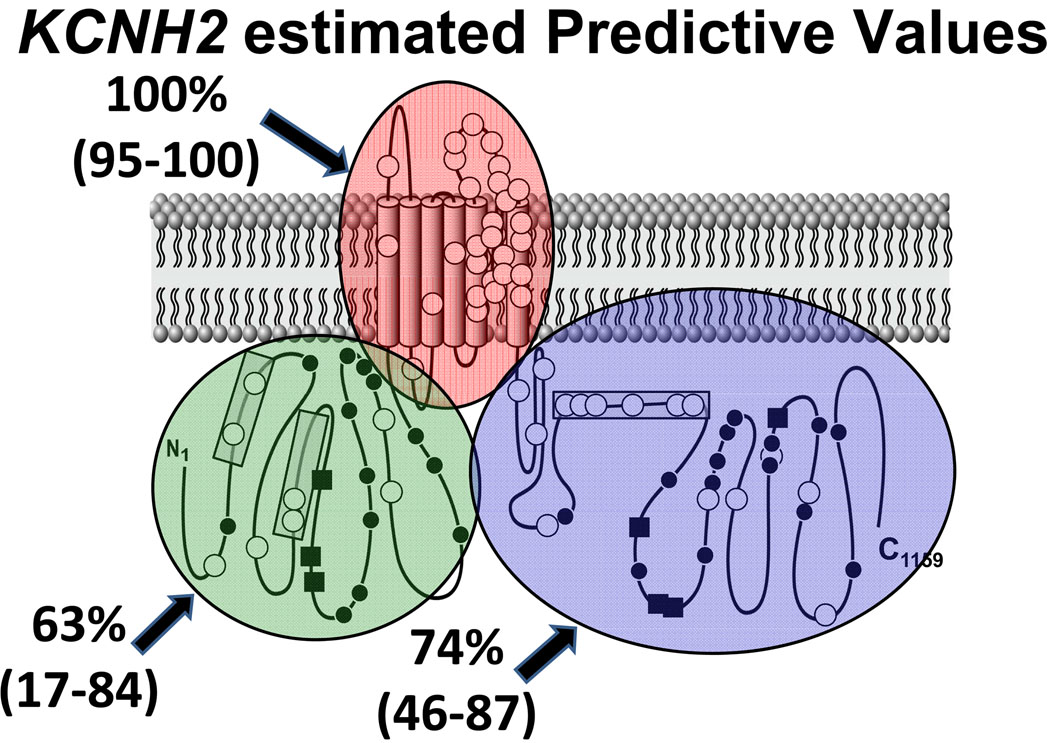
Mutations were discovered in cases throughout all regions of the KCNH2-encoded Kv11.1 potassium channel. In contrast, missense mutations discovered in controls localized only to Kv11.1’s N- and C-termini. The relative frequency of mutations discovered in the N-terminus and C-terminus in cases versus controls was 2.7× and 3.8× respectively. The EPV for any type of mutation localizing anywhere in KCNH2 was 93% (CI 89–95) and for a missense mutation was 89% (CI 84–93). There was an extremely high probability (EPV = 100% [CI 95–100]) of pathogenicity for missense mutations localizing to the linker, transmembrane, and pore regions of KCNH2, but lower EPVs for missense mutations in the N-terminus (63% [CI 17–84]) or C-terminus of KCNH2 (74% [CI 46–87]). There were no control mutations localizing to the specialized PAS/PAC regions of the N-terminus or the cyclic nucleotide binding domain (cNBD) region of the C-terminus, suggesting a very high EPV (100% [CI 78–100]) for those sub-regions, but reduced EPV for the remainder of the N-terminal (26% [CI 0–74]) and C-terminal (56% [CI 0–81]) regions. These EPVs are summarized in Table 3 and Figure 4.
Figure 4. Estimated Predictive Values for KCNH2.
Estimated predictive values in clinically definite cases for a missense mutation occurring in each major region in the KCNH2-encoded Kv11.1 potassium channel. The 95% confidence intervals are provided in parentheses. White circles represent case mutations. Black circles depict rare genetic variants observed only once among healthy volunteers while black squares depict the genetic variants/polymorphisms observed more than once among the controls. Shaded regions (2) and (3) are the PAS and PAC specialized regions, respectively, and shaded region (4) represents the specialized cNBD (cyclic nucleotide binding domain).
SCN5A (LQT3-susceptibility gene)
The greatest prevalence of missense mutations observed in the control population occurred in SCN5A, for which 3% of controls were “positive” for a mutation. The EPV for any type of mutation localizing anywhere in the SCN5A-encoded NaV1.5 sodium channel was 63% (CI 40–77) and for a missense mutation was 60% (CI 33–75). Although clustering in the three interdomain linkers (IDL), control mutations were scattered throughout all gene regions, highlighting a significantly greater degree of genetic background “noise” in SCN5A than in either KCNQ1 or KCNH2. Moreover, given that the case frequency of missense mutations is lowest in SCN5A, the relative frequency of missense mutations found in cases compared to controls was quite low (Figure 2B). In fact, for both the N-terminus and the IDLs, the control mutation rates exceeded those of the case mutation rates, making the calculated point estimates of EPV for these regions less than the logical limit of 0%, presumably due to sampling error and perhaps the greater non-Caucasian population among the controls compared to the cases. Given that the third IDL region (DIII–DIV) may have different biophysical properties making mutations there more likely to be pathogenic, we re-calculated the EPV specific to the DIII–DIV region and the remainder of the interdomain linkers separately. This did not significantly change the EPV conferred by mutations localizing to IDL1 (DI–DII) or IDL2 (DII–DIII), which was still below the logical limit of 0% (CI, 0–49). Similarly, the EPV of a mutation localizing to IDL3 (DIII–DIV), while 72%, had a large CI with a lower bounds below 0% and an upper bounds of 98%, likely secondary to small sample size. EPV was moderate for the C-terminus region (75% [CI 32–91]) (Figure 5). The region in SCN5A with the greatest association between mutation discovery and disease causation was the transmembrane/linker regions, in which the EPV was 88% (CI 67–96).
Figure 5. Estimated Predictive Values for SCN5A.
Estimated predictive values in clinically definite cases for a missense mutation occurring in each major region in the SCN5A-encoded Nav1.5 sodium channel. The 95% confidence intervals are provided in parentheses. White circles represent case mutations. Black circles depict rare genetic variants observed only once among healthy volunteers while black squares depict the genetic variants/polymorphisms observed more than once among the controls.
DISCUSSION
Part of the clinical utility of a diagnostic test is the ability to recognize whether or not results of a particular clinical test fit within a “normal range” defined by population studies of presumably non-diseased patients. However, further clinical studies may redefine the normal range when taking into account other measures of clinical likelihood. One hallmark example of this is the cholesterol panel, the acceptable results of which have come to be dependent on the clinical likelihood of myocardial disease.[24] Thus, the test has limited value as a stand alone or screening measure of risk but is invaluable in the management of at-risk patients. It is becoming increasingly clear that these concepts extend to genetic testing as well, since the presence of a mutation alone needs to be considered in clinical and epidemiologic contexts. Although once hoped to be “binary” in interpretation and still often incorrectly viewed as such, genetic tests must be viewed fundamentally as probabilistic tests.
Indeed, genetic screening raises several new questions regarding patient management because genetic testing is meant variably to verify clinical suspicion or to assess susceptibility to disease. Certain mutations, in particular radical mutations, may suggest a high likelihood of disease, regardless of other clinical evidence. However, some uncharacterized missense mutations may be difficult to interpret. Recent efforts in various diseases have concentrated on methods for determining the probability of mutation pathogenicity.[2, 6] Very few studies address the interpretability of novel, rare genetic variants in the clinical expression of genetically associated diseases.
One of the largest population studies of this, done on the BRCA1 and BRCA2 genes for breast and ovarian cancer, suggested that disease causation may be difficult to assess and depends, in part, on the availability of a large control population to assess the relative frequency of novel missense mutations.[25] However, for diseases such as LQTS in which disease incidence is smaller and phenotypic expression can be surreptitious, outcomes of over-interpretation may lead to unnecessary invasive therapies and under-interpretation may, though rarely, lead to death. Therefore, the ability to accurately distinguish pathogenic mutations from innocuous mutations is vital. One of the main limitations in achieving this level of differentiation for individual mutations is the relative paucity of independent validations of pathogenicity with heterologous expression studies. In fact, less than 20% of all published “LQTS-causative” mutations have been studied functionally in this manner by all of the channel function research laboratories throughout the world combined to date. Furthermore, these studies may be hard to control for due to different cellular expression systems and the many other variables that may affect interpretation of findings. Thus, by design, all such functional testing was explicitly ignored in our study to focus instead on developing evaluative criteria for probability of pathogenicity based solely on mutation type and location.
Many mutations encountered during ongoing LQTS genetic testing continue to represent novel missense mutations (data not shown). As we have shown, the occurrence of this type of mutation does not necessarily imply disease causation given the “background noise rate” of 4% among Caucasians and up to 8% among non-Caucasians gleaned here from an examination of the three main genes in over 1300 controls. Thus, even when the pre-test probability of LQTS is high, other factors still need to be taken into account when interpreting the genetic test and counseling the patient.
The results of our study raise several key issues regarding genetic testing in general and LQTS genetic testing in particular. First, there exists “background genetic noise” in the general population and the largest component of this noise consists of very rare missense mutations. The only way to estimate the amount of “noise” is through complete testing of large numbers of ostensibly healthy controls. To our knowledge, the only other disease-susceptibility gene that has undergone this level of scrutiny among ostensibly healthy subjects is the BRCA1 breast cancer-susceptibility gene. It would be expected that the same frequency of “noise” in the general population should also exist in patients with disease, and thus, the presence of a mutation alone, despite a high index of clinical suspicion, may not be sufficient to establish pathogenicity.
In the case of LQTS, the established yield of genetic testing among clinically irrefutable cases of LQTS is approximately 70 – 75%.[26] This implies that even among absolute cases, there is an estimated 6% (4%/70%) chance among Caucasian subjects that a “positive” genetic test is a “false positive”. In addition, this background noise rate is ethnicity dependent and there is a greater chance (up to 10%) for “false positives” to occur among African Americans receiving the same LQTS genetic test. As the a priori or pre-test clinical probability for a diagnosis of LQTS decreases, the probability that the identified mutation is simply “just there, just rare” rather than being an LQTS-causative mutation increases. In other words, estimated predicted values (EPVs or probability of pathogenicity) for mutations found in lower-probability patients are correspondingly lower, to the extreme of effectively 0%, except perhaps for the most critical protein regions, if testing were done as part of a universal screening program for this 1:2500 disease. However, the higher the certainty of disease, the more likely it is that a mutation is causative and the EPVs provide an upper bounds on those probabilities.
In calculating the estimated predictive value for mutations occurring in each gene region, we sought to provide a means of estimating whether a variant discovered in a proband may be causative, though this does not equate to “diagnosing” the proband. Ideally, strong linkage analysis and good functional studies would be available for every mutation, but this is usually not possible and certainly not at the moment of first discovery. Thus, the EPV is helpful in guiding initial decision making in the absence of more conclusive data on each particular mutation.
Second, the nature of the detected mutation matters greatly in predicting the probability of pathogenicity. In particular, “radical” mutations, specifically nonsense, frameshift, and splice site mutations, have a near 100% estimated predictive value for disease pathogenicity involving the two major potassium channels implicated in LQTS. In this study, one fourth of the case mutations were of this variety, enabling high-confidence positive test interpretations for this subset. For LQTS genetic testing, the challenge lies in the discernment of rare, single amino acid substitutions (missense mutations), which comprise 75% of the “positive” test results.
Third, the probability of pathogenicity for a particular missense mutation may be impacted profoundly by its location in the protein.[27] In the case of LQTS genetic testing, a mutation’s specific location in the complex structure of the voltage-gated ion channel confers markedly different probabilities of pathogenicity. Thus, knowledge about the specific gene-encoded regions (i.e. structure-function domains) is a critical determinant underlying the probability of pathogenicity. In this study, missense mutations represent high probability disease mutations when localized to a channel’s pore, transmembrane-spanning domains and other critical sub-domains, while extraordinary caution is necessary when interpreting the potential significance of similarly rare single amino acid substitutions in other locations, such as the IDL I–II of the SCN5A-encoded NaV1.5 sodium channel. Thus far, secondary analyses involving amino acid conservation, inspection of physico-chemical properties and in silico predictions of impact on secondary structure have not helped to further refine these estimates of pathogenicity in low EPV domains such as Kv11.1’s C-terminus or NaV1.5’s DI–II and DII–III linkers (data not shown).
Limitations
The cases in this study represented a referral population not necessarily evaluated clinically at either LQTS specialty center. Previous studies have demonstrated a 70–75% yield from LQT1-3 genetic testing among single center-derived, definite cases.[26] The case yield in this study was 58%, indicating that some of the cases may not have been of the highest a priori clinical probability. It can also be assumed that not all mutations found in our cases are pathogenic. In fact, this is a central premise of and motivation for our study. Specifically, the overall yield of 6% among the entire control cohort implies that approximately 10% (6%/58%) of the case mutations elucidated in this study may be “false positives”. This estimate may be high for two reasons. First, our case population has a much higher representation of Caucasians, in whom the background rates are lower but overall case mutation yield is similar. This same skew in ancestry also suggests that the EPVs in this paper are relatively underestimated for Caucasians and overestimated in non-Caucasians. To estimate these ethnic-specific EPVs would require significantly larger sample sizes than are currently available. Second, the 10% “false positive” estimate above may be overestimated, and our EPVs underestimated, because this paper is based on an historical control population. The next 1300 controls would presumably have considerably fewer novel variants, as many of the rare variants would have been observed before in controls, or even in cases.
Second, the 79 rare mutations found in the control cohort are not guaranteed to all be 100% benign. Because a normal 12-lead electrocardiogram with a normal QTc was not a prerequisite for the ostensibly healthy volunteers who comprised the control cohort, it is possible that some of the controls may have LQTS and therefore, their particular rare variant could be falsely considered part of the “background genetic noise”. For example, we recently found in an expanded cohort that the background rate may be less than 3% among Caucasians (data not shown). While acknowledging that some mutations amongst “controls” may represent incompletely penetrant mutations that may be pathogenic in another patient, the average penetrance of these variants must be extremely low. Otherwise, the prevalence of LQTS must be far greater than believed, or is plausible. Based on an incidence of 1 in 2500 of LQTS among the general population, we can be 98% confident that no more than two mutations listed in the control compendia are strongly pathogenic mutations rather than background noise. Among the 79 mutations found in the 1300 controls, one that is extremely suspicious for possible “pathogenic” status is the KCNQ1-A300T missense mutation that localizes to the pore-forming region. In addition, the radical 4299 + 2 T>A splice mutation in SCN5A might lead to loss of channel function, which might be predicted to confer a Brugada syndrome phenotype.
Despite representing arguably the second largest and most expansive examination of “background genetic noise” among any of the clinically available genetic tests, perhaps the biggest limitation of this study is still the relatively small sample size (> 2500 reference alleles) of the controls. Given the overall low control mutation level, upon which genetic testing generally draws its strength, the EPVs of some regions may be strongly influenced by small numbers, as is reflected in the sometimes broad confidence intervals. Given that a timely functional assay does not exist to assess whether a particular VUS can be upgraded or downgraded based upon its degree of channel perturbation, further comparative genomic investigations of cases and controls, among both Caucasian and non-Caucasian populations, are needed to improve further the diagnostic accuracy of the LQTS genetic test.
Despite these limitations, this study demonstrates clearly that ethnicity, mutation type, and mutation location, in particular channel structure-function domains are critical determinants of a particular mutation’s probability of pathogenicity in the three major LQTS-causing genes.
Conclusions
When evaluating the results of any genetic test for any disease, factors including location, the possible role of ethnicity, and the possible existence of mutations in other, untested genes should be weighed in the balance. Genetic testing has the potential to offer valuable clinical information that can facilitate early clinical intervention. In the case of LQTS, the diagnostic, prognostic, and therapeutic implications of genetic testing are now viewed as fundamental to the clinical evaluation.[28–29] However, the presence of a genetic mutation alone cannot supplant clinical evidence and, even in robust cases of LQTS, a positive genetic test result must be carefully reviewed. Indeed, genetic tests, in most cases, must be viewed by clinicians as probabilistic tests, not binary (yes/no, positive/negative) tests, which places genetic testing in good company with virtually every diagnostic test that we order.
Supplementary Material
ACKNOWLEDGMENTS
FUNDING SOURCES
The comparative genomic analyses were performed in Dr. Ackerman’s research program with support from the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program. Dr. Wilde’s research program is supported by Interuniversity Cardiology Institute the Netherlands (ICIN) project 27 and a Leducq programm grant “Alliance against sudden cardiac death.”
Footnotes
Journal Subject Codes: [109] Clinical Genetics, [89] Genetics of Cardiovascular Diseases
CONFLICT OF INTEREST DISCLOSURES
SK: None. DJT: None. BAS and CHK are employees of PGxHealth, which offers the FAMILION® LQTS Test, and stockholders of the parent company, Clinical Data.
MSP: None. MA: None. AAMW: None. MJA is a consultant for PGxHealth and chairs their FAMILION Medical/Scientific Advisory Board (approved by Mayo Clinic’s Medical-Industry Relations Office and Conflict of Interests Review Board). In addition, “cardiac channel gene screen” and “know-how relating to long QT genetic testing” license agreements, resulting in consideration and royalty payments, were established between Genaissance Pharmaceuticals (now PGxHealth) and Mayo Medical Ventures (now Mayo Clinic Health Solutions) in 2004.
REFERENCES
- 1.GeneTests: Medical Genetics Informational Resource (database online) Seattle: University of Washington; 2007. pp. 1995–2007. Copyright. [Google Scholar]
- 2.Evans JP, Skrzynia C, Burke W. The complexities of predictive genetic testing. BMJ. 2001;322:1052–1056. doi: 10.1136/bmj.322.7293.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter DJ, Hunter DJ, Drazen JM. Letting the genome out of the bottle--will we get our wish? N Engl J Med. 2008;358:105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu W. The long QT syndrome: therapeutic implications of a genetic diagnosis. Cardiovasc Res. 2005;67:347–356. doi: 10.1016/j.cardiores.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Keating M, Atkinson D, Dunn C, Timothy K, Vincent GM, Leppert M. Linkage of a cardiac arrhythmia, the long QT syndrome, and the Harvey ras-1 gene. Science. 1991;252:704–706. doi: 10.1126/science.1673802. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 7.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmias: HERG mutations cause long QT syndrome. Cell. 1995;80:795–804. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 8.Zareba W, Moss AJ, Schwartz PJ, Vincent GM, Robinson JL, Prior SG, Benhorin J, Locati EH, Towbin JA, Keating MT, Lehmann MH, Hall WJ, Andrews ML, Napolitano C, Timothy K, Zhang L, Medina A, MacCluer JW. Influence of the genotype on the clinical course of the Long QT Syndrome. N Engl J Med. 1998;339:960–965. doi: 10.1056/NEJM199810013391404. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz PJ, Prior SG, Spazzolini C, Moss AJ, Vincent GM, Napolitano C, Denjoy I, Guicheney P, Breithardt G, Keating MT, Towbin JA, Beggs AH, Brink P, Wilde AAM, Toivonen L, Zareba W, Robinson JL, Timothy KW, Corfield V, Wattanasirichaigoon D, Corbett C, Haverkamp W, Schulze-Bahr E, Lehmann MH, Schwartz K, Coumel P, Bloise R. Genotype-phenotype correlation in the long QT syndrome: Gene specific triggers for life threatening arrhythmias. Circulation. 2001;103:89–95. doi: 10.1161/01.cir.103.1.89. [DOI] [PubMed] [Google Scholar]
- 10.Priori SG, Schwartz PJ, Napolitano C, Bloise R, Ronchetti E, Grillo M, Vincentini A, Spazzolini C, Nastoli J, Bottelli G, Folli R, Cappelletti D. Risk stratification in the long- QT syndrome. N Engl J Med. 2003;348:1866–1874. doi: 10.1056/NEJMoa022147. [DOI] [PubMed] [Google Scholar]
- 11.Zareba W. Genotype-specific ECG patterns in long QT syndrome. J Electrocardiology. 2006;39:S101–S106. doi: 10.1016/j.jelectrocard.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Napolitano C, Bloise R, Priori SG. Gene-specific therapy for inherited arrhythmogenic disorders. Pharmacology & Therapeutics. 2006;110:1–13. doi: 10.1016/j.pharmthera.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Tan HL, Bardai A, Shimizu W, Moss AJ, Schulze-Bahr E, Noda T, Wilde AAM. Genotype-specific onset of arrhythmias in congenital Long QT syndrome: possible therapy implications. Circulation. 2006;114:2096–2103. doi: 10.1161/CIRCULATIONAHA.106.642694. [DOI] [PubMed] [Google Scholar]
- 14.Moss AJ, Shimizu W, Wilde AAM, Towbin JA, Zareba W, Robinson JL, Qi M, Vincent M, Ackerman MJ, Kaufman ES, Hofman N, Seth R, Kamakura S, Miyamoto Y, Goldenberg I, Andrews ML, McNitt S. Clinical aspects of type-1 long QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115:2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tester DJ, Ackerman MJ. Novel gene and mutation discovery in congenital Long QT syndrome: Let's keep looking where the street lamp standeth. Heart Rhythm. 2008;5:1282–1284. doi: 10.1016/j.hrthm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackerman MJ, Tester DJ, Jones GS, Will ML, Burrow CR, Curran ME. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clin Proc. 2003;78:1479–1487. doi: 10.4065/78.12.1479. [DOI] [PubMed] [Google Scholar]
- 17.Ackerman MJ, Splawski I, Makielski J, Tester D, Will M, Timothy K, Keating M, Jones G, Chadha M, Burrow C. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: Implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1:600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome. Circulation. 1993;88:782–784. doi: 10.1161/01.cir.88.2.782. An update. [DOI] [PubMed] [Google Scholar]
- 19.Tester DJ, Will ML, Ackerman MJ. Mutation detection in congenital long QT syndrome: Cardiac channel gene screen using PCR, dHPLC, and direct DNA sequencing. Methods in Molecular Medicine. 2007;128:181–207. doi: 10.1385/1-59745-159-2:181. [DOI] [PubMed] [Google Scholar]
- 20.Nomenclature Working Group. Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 21.Splawski I, Shen J, Timothy KW, Vincent M, Lehmann MH, Keating MT. Genomic structure of three long QT syndrome genes: KVLQT1, HERG, and KCNE1. Genomics. 1998;51:86–97. doi: 10.1006/geno.1998.5361. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Li Z, Shen J, Keating MT. Genomic organization of the human SCN5A gene encoding the cardiac sodium channel. Genomics. 1996;34:9–16. doi: 10.1006/geno.1996.0236. [DOI] [PubMed] [Google Scholar]
- 23.Neyroud N, Richard P, Vignier N, Donger C, Denjoy I, Demay L, Shkolnikova M, Pesce R, Chevalier P, Hanique B, Coumel P, Schwartz K, Guicheney P. Genomic organization of the KCNQ1 K+ channel gene and identification of C-terminal mutations in the long- QT syndrome. Circulation Research. 1999;84:290–297. doi: 10.1161/01.res.84.3.290. [DOI] [PubMed] [Google Scholar]
- 24.Grundy S, Cleeman J, Bairey Merz C, Brewer H, Jr, Clark L, Hunninghake D, Pasternak R, Smith S, Jr, Stone N. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. J Am Coll Cardiol. 2004;44:720–732. doi: 10.1016/j.jacc.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, Goldgar DE. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tester DJ, Will ML, Haglund C, Ackerman MJ. Effect of clinical phenotype on yield of long QT-syndrome genetic testing. J Am Coll Cardiol. 2006;47:764–768. doi: 10.1016/j.jacc.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 27.Worth CL, Bickerton GR, Schreyer A, Forman JR, Cheng TM, Lee S, Gong S, Burke DF, Blundell TL. A structural bioinformatics approach to the analysis of nonsynonymous single nucleotide polymorphisms (nsSNPs) and their relation to disease. J Bioinform Comput Biol. 2007;5:1297–1318. doi: 10.1142/s0219720007003120. [DOI] [PubMed] [Google Scholar]
- 28.Lehnart SE, Ackerman MJ, Benson DW, Jr, Brugada R, Clancy CE, Donahue JK, George AL, Jr, Grant AO, Groft SC, January CT, Lathrop DA, Lederer WJ, Makielski JC, Mohler PJ, Moss A, Nerbonne JM, Olson TM, Przywara DA, Towbin JA, Wang LH, Marks AR. Inherited arrhythmias: a National Heart, Lung, and Blood Institute and Office of Rare Diseases workshop consensus report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation. 2007;116:2325–2345. doi: 10.1161/CIRCULATIONAHA.107.711689. [DOI] [PubMed] [Google Scholar]
- 29.Zipes DP, Camm AJ, Borggrefe M, Buxton AE, Chaitman B, Fromer M, Gregoratos G, Klein G, Moss AJ, Myerburg RJ, Priori SG, Quinones MA, Roden DM, Silka MJ, Tracy C. ACC/AHA/ESC 2006 guidelines for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: A report of the American College of Cardiology/American Heart Association task force and the European Society of Cardiology committee for practice guidelines (Writing Committee to Develop Guidelines for Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death): Developed in collaboration with the European Heart Rhythm Association and the Heart Rhythm Society. 2006;114:e385–e484. doi: 10.1161/CIRCULATIONAHA.106.178233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.