Summary
Self-perpetuating amyloid-based protein isoforms (prions) transmit neurodegenerative diseases in mammals and phenotypic traits in yeast. Although mechanisms that control species-specificity of prion transmission are poorly understood, studies of closely related orthologs of yeast prion protein Sup35 demonstrate that cross-species prion transmission is modulated by both genetic (specific sequence elements) and epigenetic (prion variants, or “strains”) factors. Depending on the prion variant, the species barrier could be controlled at the level of either heterologous coaggregation or conversion of the aggregate-associated heterologous protein into a prion polymer. Sequence divergence influences cross-species transmission of different prion variants in opposing ways. The ability of a heterologous prion domain to either faithfully reproduce or irreversibly switch the variant-specific prion patterns depends on both sequence divergence and the prion variant. Sequence variations within different modules of prion domains contribute to transmission barriers in different cross-species combinations. Individual amino acid substitutions within short amyloidogenic stretches drastically alter patterns of cross-species prion conversion, implicating these stretches as major determinants of species specificity.
Keywords: amyloid, Saccharomyces bayanus, Saccharomyces cerevisiae, Saccharomyces paradoxus, yeast
Introduction
Transmissible encephalopathies in humans and other mammals (for review, see (Harris & True, 2006; Prusiner, 1998; Weissmann, 2004) and cytoplasmically heritable traits in yeast and other fungi (for review, see Inge-Vechtomov et al., 2007; Wickner et al., 2007a; Wickner et al., 2007b) are controlled by abnormal, self-perpetuating protein isoforms termed prions. Most prions are cross-β polymers (amyloids) thought to propagate by immobilizing normal monomeric protein of the same amino acid (aa) sequence and converting it into a prion form (Lansbury & Caughey, 1995).
The ability of a pre-existing amyloid to convert normal protein into a prion requires a high level of identity between interacting protein sequences. Therefore, transmission of a mammalian prion disease to another mammalian species is usually inefficient due to the so-called species barrier. However, cross-species conversion may overcome the barrier, for example, as in the case of “mad cow” disease (for reviews, see Collinge & Clarke, 2007; Prusiner, 1998). Transmissibility of this disease to humans forced the large-scale extermination of potentially infected cows in Europe in the 1990s and massive beef recalls in the USA in the 2000s. Despite the importance of the species barrier for both practical (prediction of cross-species infectivity) and fundamental (deciphering the molecular basis of amyloid specificity) purposes, its mechanism is still poorly understood. For mammalian prion protein (PrP), a correlation between the results of in vitro or in situ cross-seeding assays and in vivo transmission barriers remains questionable (Chernoff, 2004b; Makarava et al., 2007).
To complicate matters further, a protein of one and the same sequence can form different prion variants or “strains,” distinguishable from each other by both phenotypic characteristics (e. g. incubation periods in mammals, or mitotic stability and level of impairment of the protein function in yeast) and biochemical patterns (e. g. protease digestion profiles, average aggregate size, and proportion of aggregated versus monomeric protein). Furthermore, mammalian prion variants may differ in host specificity (Collinge & Clarke, 2007), and variant-specific patterns are usually maintained during cross-species conversion (Bruce et al., 1994), with some exceptions in cases of prion “adaptation” for more efficient propagation in a new host. This is hypothetically explained by either conformational or kinetic selection (Collinge & Clarke, 2007).
Prion potential of yeast prion proteins is controlled by terminally located and rapidly evolving prion domains (PrDs), that range from 65 to several hundred aa in size and are usually dispensable for the normal cellular function of a respective protein (for review, see (Inge-Vechtomov et al., 2007; Wickner et al., 2007a; Wickner et al., 2007b). Strict prion species barriers were detected between the Saccharomyces cerevisiae prion protein Sup35 and its distantly related orthologs from the yeast Pichia methanolica or Candida albicans, that possessed PrDs with only 30-40% of aa identity to S. cerevisiae (Chernoff et al., 2000; Kushnirov et al., 2000; Santoso et al., 2000). These barriers were controlled by the N-terminal, QN-rich regions and coincided with an inability of the divergent PrDs to coaggregate. Occasional cross-species transmission generated multiple prion variants (Vishveshwara & Liebman, 2009), in contrast to intraspecies transmission that faithfully reproduces patterns of one and the same prion variant. This suggests that the species barrier was crossed due to a non-specific nucleation of the host protein by the aggregated heterologous protein, a phenomenon that is also observed with low frequency for non-homologous prion proteins with similar aa compositions (Derkatch et al., 2001; Osherovich & Weissman, 2001). Relevance of these data to the mammalian species barriers remains unclear due to much lower levels of sequence divergence among mammalian PrPs.
The prion species barrier was also detected for some combinations of more closely related orthologs of the yeast prion protein Ure2, originating from different Saccharomyces species (Baudin-Baillieu et al., 2003; Edskes et al., 2009; Edskes & Wickner, 2002). As in mammals, Ure2 prion variants differed by host specificity; however, variant-specific patterns were faithfully reproduced during cross-species transmission (Edskes et al., 2009).
To study the prion species barrier, we have previously developed a yeast-based experimental model that employs orthologs of the Sup35 prion protein from closely related Saccharomyces species (Chen et al., 2007). The range of divergence among these proteins overlaps the range of divergence among mammalian PrP orthologs. Even though closely related Saccharomyces Sup35 proteins were capable of co-aggregating, transmission of the prion state from one protein to another was impaired, resulting in a species barrier. Our new studies demonstrate that differences between yeast prion variants influence heterologous coaggregation, cross-species transmission, and the ability to faithfully reproduce variant-specific patterns via a heterologous protein. Our data also identify potential aa stretches within the Sup35 PrD that are involved in the control of species specificity.
Results
Saccharomyces model for cross-species prion conversion
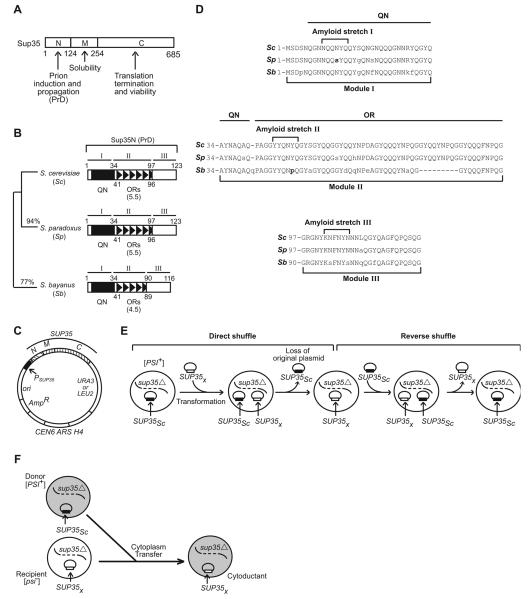
The Saccharomyces cerevisiae Sup35 protein can be divided into three major domains as follows (Fig. 1A): 1) a N-proximal prion-forming domain (Sup35N), or PrD; 2) a middle domain (Sup35M) promoting protein solubility; and 3) a C-proximal release factor domain (Sup35C) essential for translational termination and cell viability (for review, see Chernoff, 2004a; Chernoff, 2004c). This PrD can be further subdivided into three regions (for review, see Chernoff, 2004a): 1) a QN-rich region (QN), located before aa position 40; 2) a region of 5.5 imperfect oligopeptide repeats (ORs) with the consensus sequence PQGGYQQYN (positions 41-96); 3) region 97-123, that lacks any obvious sequence pattern. PrDs of S. paradoxus and S. bayanus, two species with completely sequenced genomes that are closely related to S. cerevisiae (Cliften et al., 2003; Kellis et al., 2003), show respectively 94% and 77% aa identity to S. cerevisiae (Fig. 1B; for sequence alignment, see Fig. 1D) and maintain the same structural organization, except that one OR unit is missing in S. bayanus (Chen et al., 2007; Cliften et al., 2003; Jensen et al., 2001; Kellis et al., 2003). We previously confirmed the existence of a species barrier in the transmission of a prion form of Sup35 (termed [PSI+]) from S. cerevisiae to S. paradoxus or S. bayanus, and implicated the PrD as the major determinant of the barrier (Chen et al., 2007). Here, we employ the same experimental strategy to further decipher the role of prion variants and various PrD modules in cross-species prion transmission. Low copy plasmids were constructed as described previously (Chen et al., 2007), as described below in Experimental procedures, and in Supplement (Fig. S7, and Tables SI and SII). They contained either complete SUP35 genes of various origins (S. cerevisiae, S. paradoxus or S. bayanus), or artificially assembled constructs with PrDs of various species or chimeric origins (see below), fused to the S. cerevisiae SUP35MC region. All constructs were expressed from the endogenous S. cerevisiae SUP35 promoter (Fig. 1C). Experiments were performed in a S. cerevisiae strain lacking chromosomal SUP35 and maintained alive by SUP35 on a plasmid. The various SUP35 constructs were introduced and exchanged by transformation and plasmid shuffle (Fig. 1E). This approach was in some cases supplemented by cytoduction, or cytoplasmic transfer to the S. cerevisiae strain with heterologous or chimeric Sup35 proteins (Fig. 1F). Presence of [PSI+] was monitored by partial loss of the Sup35 translation termination function as a result of prion formation, which leads to translational readthrough (suppression) of the reporter UGA allele ade1-14. This can be detected by growth on –Ade medium and white or pink (as opposed to red) color on the complete YPD medium (Chernoff et al., 2002). Prion protein can also be detected by its ability to form aggregates composed of SDS-resistant polymers (Chernoff et al., 2002; Kushnirov et al., 2006).
Figure 1. Experimental system for studying cross-species prion conversion.
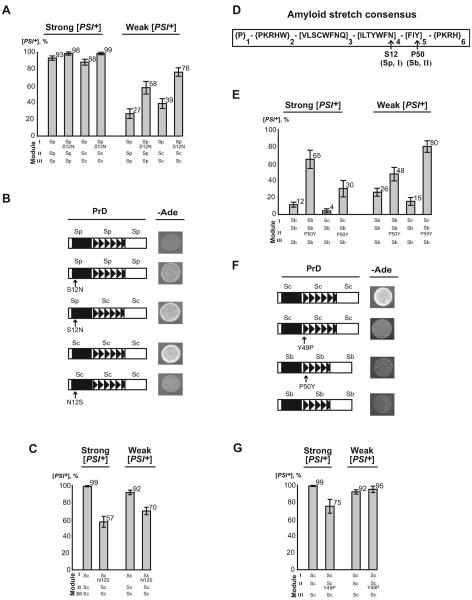
(A ) Structural and functional organization of the S. cerevisiae Sup35 protein. N, M and C refer to Sup35N, Sup35M and Sup35C domains, respectively. Numbers correspond to amino acid (aa) positions. (B ) Comparison of the Sup35N regions (prion domains) from different Saccharomyces species. I, II and III refer to modules of the Sup35N region that were used in constructing chimeric PrDs (see below). QN and ORs refer to the QN-rich stretch and oligopeptide repeats, respectively. Sc – Saccharomyces cerevisiae, Sp - S. paradoxus, Sb - S. bayanus. Aa identities (in %) to S. cerevisiae Sup35N are shown. (C ) Prototype yeast plasmid used. PSUP35, CEN6, ARS H4 and ori refer to the endogenous S. cerevisiae SUP35 promoter, centromere, autonomously replicating sequence, and bacterial origin of replication, respectively; N, M and C – domains of SUP35, that could be of various origins; URA3 and LEU2 – yeast selectable markers; AmpR – bacterial selectable marker (ampicillin resistance). For the construction strategy and plasmid list, see Supplement Fig. S7 and Table SI, respectively. (D ) Alignment of PrD sequences of S. cerevisiae (Sc), S. paradoxus (Sp) and S. bayanus (Sb). Numbers refer to aa positions. Boundaries of the modules I, II and III (see Fig. 1B and below, Fig. 5A) and locations of amyloid stretches (see below, Fig. 6D) are indicated. The aa residues that are changed in S. paradoxus and S. bayanus compared to S. cerevisiae are shown in lowercase. The aa residues that were altered in further experiments are shown in bold. (E) Scheme of direct and reverse plasmid shuffle. SUP35X refers to SUP35 genes of various origins, or chimeric constructs with PrDs of various origins or sequences. See comments in the text. (F) Cytoduction scheme. Designations are as above. Shaded area denotes cytoplasm that is transferred from donor to recipient. See more detailed description in Experimental procedures.
Sup35 protein of one and the same aa sequence can produce various prion “strains” or “variants” (Derkatch et al., 1996). While our previous work (Chen et al., 2007) employed a strong S. cerevisiae [PSI+] variant that grows well on –Ade medium, is light-pink on YPD, is 100% stable in mitotic divisions and contains almost all Sup35 protein in the aggregated state, our current study is also extended to the isogenic weak [PSI+] variant that grows slower on –Ade medium, is dark-pink on YPD, exhibits detectable prion loss in mitotic divisions and contains some soluble Sup35 protein, in addition to aggregates.
Prion variants influence polymerization of heterologous Sup35 proteins
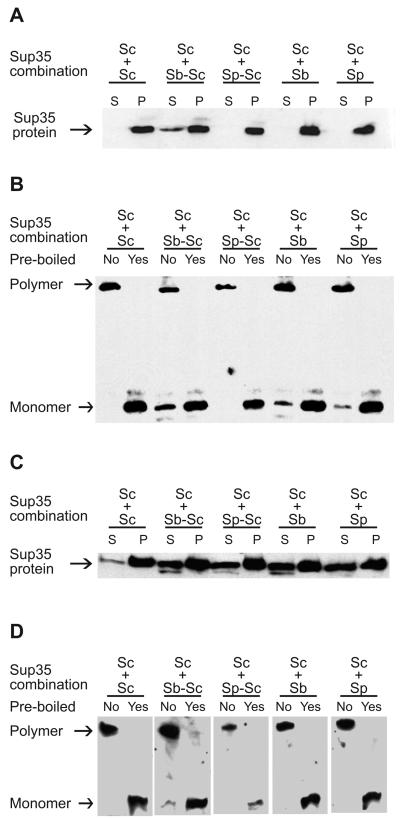
It has been shown previously (Chen et al., 2007) that the strong S. cerevisiae [PSI+] strain expressing both endogenous Sup35 and either S. paradoxus or S. bayanus Sup35 contains essentially all detectable Sup35 protein (that is, including a heterologous protein) in the aggregated state. Although a more detailed analysis (to be reported elsewhere) indicates that distribution of aggregates by sizes somewhat depends on the growth phase of the culture, we have confirmed that practically all Sup35-reactive material is precipitated at 39,000 g from exponential cultures producing either S. cerevisiae Sup35 alone, or S. cerevisiae Sup35 in combination with either S. paradoxus or S. bayanus Sup35 (Fig. 2A). Our new data also show that all Sup35 protein is precipitated in these conditions from the strong S. cerevisiae [PSI+] strain coexpressing the chimeric protein, with PrD of S. paradoxus and most of the Sup35 protein is precipitated from the strong S. cerevisiae [PSI+] strain coexpressing the chimeric protein with PrD of S. bayanus (Fig. 2A). (In each chimeric construct, heterologous PrD was fused to the Sup35MC region of S. cerevisiae.) To determine if co-aggregated proteins are co-polymerized into SDS-resistant prion polymers, we analyzed Sup35 aggregates by using the “boiled gel” approach (Kushnirov et al., 2006; see below, Material and methods), that is based on the inability of polymerized protein to enter the polyacrylamide gel without boiling. After interruption of electrophoresis and boiling of the gel, polymers are denatured and can enter the gel if electrophoresis is continued. Aggregates isolated from the strong [PSI+] strain bearing only endogenous S. cerevisiae Sup35 protein were composed entirely of SDS-resistant polymers. However, a fraction of the non-polymerized Sup35 protein was observed in the presence of S. paradoxus Sup35, S. bayanus Sup35, or chimeric Sup35 protein with S. bayanus PrD (Fig. 2B). As the S. bayanus Sup35 protein is shorter than S. cerevisiae Sup35 due to deletions in both PrD (Fig. 1B and D) and Sup35M (not shown), we have rerun the respective sample on a gel with a lower concentration of polyacrylamide and confirmed that the non-polymerized band has a lower molecular weight expected for the S. bayanus Sup35 protein (Supplement Fig. S8). This indicates that at least a portion of the aggregate-associated heterologous protein is not converted into polymers. Notably, a non-polymerized fraction was not detected for the chimeric protein with S. paradoxus PrD (Fig. 2B).
Figure 2. Aggregation and polymerization of heterologous and chimeric Sup35 proteins in the S. cerevisiae [PSI+] strains.
Species designations are as on Fig. 1. Sp-Sc and Sb-Sc refer to the chimeric proteins bearing PrDs of S. paradoxus and S. bayanus, respectively, in conjunction with the MC region of S. cerevisiae. (A) and (C) Sup35 aggregation in the strong and weak [PSI+] strain, respectively. Protein extracts were fractionated by centrifugation at 39,000 g. Pellets (P) and supernatants (S) were boiled in 2% SDS, run on SDS-PAGE and analyzed by Western blotting with the Sup35C antibody. (B) and (D) Inclusion of aggregated Sup35 into SDS-insoluble polymers in the strong and weak [PSI+] strains, respectively. Pellets obtained by centrifugation as shown on panels (A) and (C) were solubilized in 2% SDS, either unboiled or pre-boiled as indicated, and run on SDS-PAGE, so that only non-polymerized protein enters the gel. After 1 hr, electrophoresis was interrupted and polymers absorbed in the wells were denatured by boiling the gel as described in Experimental procedures. Upon continuation of electrophoresis, denatured polymers entered the gel and are seen as the upper band. For identification of the heterologous protein in a monomeric fraction in case of S. bayanus Sup35, see Supplement Fig. S8.
In contrast to the strong prion strain, a weak S. cerevisiae [PSI+] strain, containing the S. paradoxus protein, S. bayanus protein, or chimeric protein with either S. paradoxus or S. bayanus PrD, exhibited a significant increase in the supernatant Sup35 fraction, in comparison to the same strain bearing only the S. cerevisiae protein (Fig. 2C). This indicates that either coaggregation of a heterologous protein with the weak prion is impaired, or the size of these co-aggregates is smaller, and at least some of them are not precipitated in the same conditions as in case of the strong [PSI+] strain. The precipitated fraction of the weak prion strain usually did not contain non-polymerized Sup35 protein, except for trace amounts observed in the case of a protein with S. bayanus PrD (Fig. 2D).
Prion variants influence cross-species conversion
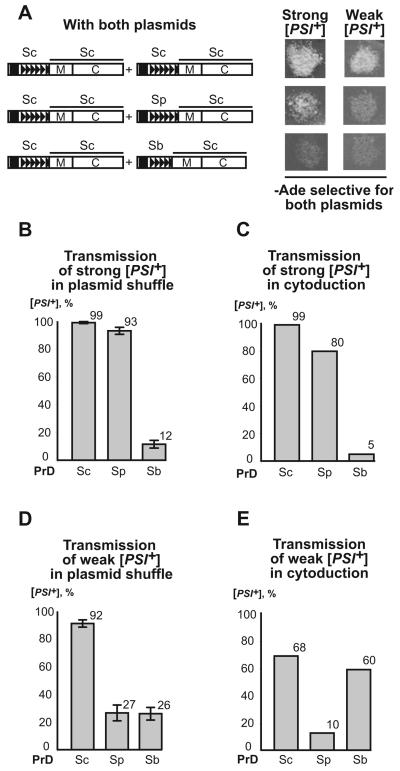
Next, we compared transmission of the strong and weak S. cerevisiae prion variants from the S. cerevisiae Sup35 protein to the chimeric proteins, bearing the PrDs of S. paradoxus or S. bayanus, at the phenotypic level by using plasmid shuffle (see above, Fig. 1E) and cytoduction (Fig. 1F). The strong S. cerevisiae [PSI+] variant showed only a slight decrease in transmission to S. paradoxus PrD but exhibited a clear transmission barrier with S. bayanus PrD (Fig. 3A, B, C and Supplement Tables SIII and SIV). In contrast, the weak S. cerevisiae [PSI+] variant exhibited a transmission barrier with S. paradoxus PrD in both variants of the experiment, but showed a clear barrier with S. bayanus PrD only in plasmid shuffle (Fig. 3A, D, E and Supplement Tables SV and SVI). Even in this case, the barrier was not as severe as for strong [PSI+]
Figure 3. Transmission of different S. cerevisiae [PSI+] variants to the Sup35 proteins with different PrDs.
Designations are as on Fig. 1. (A) Growth of the transformants containing both the original and newly introduced plasmids on –Ade medium selective for both plasmids. (B) and (D) Results of direct plasmid shuffle performed as shown on Fig. 1E. Here and further, standardized errors are indicated. For exact numbers, see Supplement Tables SIII and SV respectively. (C) and (E) Results of cytoduction experiments performed as shown on Fig. 1F. For exact numbers, see Supplement Tables SIV and SVI, respectively. Errors were not calculated for cytoduction experiments, as our procedure does not guarantee that all cytoductants, obtained for a given construct, were independent of each other.
Relatively efficient transmission of strong [PSI+] variant to the chimeric construct with S. paradoxus PrD contrasted with the previously detected barrier in the transmission of this prion variant to intact S. paradoxus Sup35 (Chen et al., 2007). This agrees with our observation that the strong S. cerevisiae [PSI+] variant efficiently converts all of the chimeric construct but not all of the intact S. paradoxus protein into the SDS-insoluble polymers (see above, Fig. 2B).
Asymmetry and infidelity of cross-species prion conversion
Even when the parental S. cerevisiae [PSI+] variant was strong, prion isolates resulting from cross-species transmission to proteins with S. paradoxus or S. bayanus PrDs were phenotypically weak (Fig. 4A). This was similar to both our previous observations for complete S. paradoxus and S. bayanus proteins (Chen et al., 2007) and some previous reports on cross-species transmission of mammalian prions (Collinge & Clarke, 2007). To determine whether such an alteration of the variant-specific patterns is reversible, we performed a “reverse shuffle” (see Fig. 1E), thus transmitting the prion state back to the S. cerevisiae Sup35 protein. In line with our previous observations for intact proteins (Chen et al., 2007), the prion state was efficiently transmitted from protein with S. paradoxus PrD to the S. cerevisiae protein, confirming asymmetry of cross-species prion transmission in this combination (Fig. 4B and Supplement Tables SVII and SVIII). In the case of the S. bayanus PrD, asymmetry of cross-species prion transmission was also observed, but it was less pronounced for strong S. cerevisiae prion variant.
Figure 4. Reproduction and switch of prion variants during cross-species transmission.
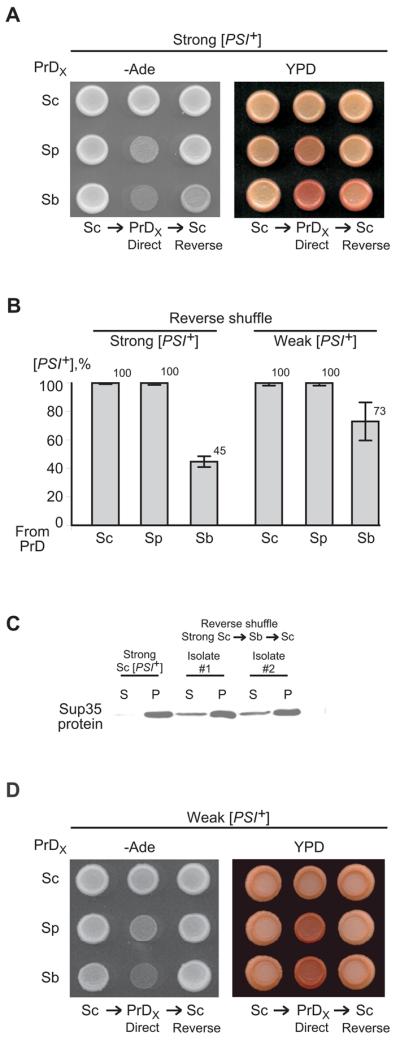
(A) and (D) Patterns of [PSI+] isolates obtained from strong and weak S. cerevisiae prion variant, respectively, via direct shuffle to the control S. cerevisiae Sup35 protein and chimeric proteins with either S. paradoxus or S. bayanus PrDs, followed by reverse shuffle back to S. cerevisiae Sup35 (as shown on Fig. 1E). PrDX refers to PrDs of various origins as indicated. –Ade plates are photographed after 6 ds (A) and 7 ds (D); YPD plates are photographed after 3 ds of incubation followed by 4 ds in the refrigerator (A and D). (B) Frequencies of [PSI+] transmission in reverse shuffle. Species designations are as on Fig. 1. For exact numbers, see Supplement Tables SVII and SVIII. (C) Centrifugation analysis of Sup35 aggregation in two different isolates obtained via shuffle from strong S. cerevisiae [PSI+] strain to the chimeric protein with S. bayanus PrD, followed by a reverse shuffle to the S. cerevisiae protein as shown on Fig. 4A. Extract of the strong S. cerevisiae strain that has not been propagated through a heterologous protein (“Strong Sc”) is shown as a control. Protein extracts were fractionated by centrifugation at 39,000 g. Supernatants (S) and pellet (P) fractions were boiled in 2% SDS, run on SDS-PAGE and analyzed by Western blotting with the Sup35C antibody. The prion isolates obtained from reverse shuffle contain more Sup35 protein in supernatant, compared to the control strong prion strain. This confirms irreversible change in the prion variant patterns during propagation through a heterologous stage,
Alteration of phenotypic patterns of the strong prion variant propagated via S. paradoxus PrD was reversible, as strong [PSI+], phenotypically indistinguishable from the parental S. cerevisiae variant, was recovered after the reverse shuffle to S. cerevisiae Sup35 protein (Fig. 4A). In contrast, [PSI+] isolates produced by the reverse shuffle from the protein with S. bayanus PrD to the S. cerevisiae protein were weaker (Fig. 4A) and produced more protein in the soluble state (Fig. 4C), compared to the strong S. cerevisiae [PSI+] variant that has not been propagated through the heterologous PrD. These isolates were confirmed by plasmid isolations and subsequent DNA analysis to contain the unaltered S. cerevisiae SUP35 gene (data not shown), thus excluding the possibility that they might originate from any recombination events during the period of coexistence of the S. cerevisiae and S. bayanus genes within the same cell. Therefore, our data demonstrate that variant-specific prion patterns could be altered irreversibly during cross-species transmission involving S. bayanus PrD, so that the resulting prion may keep a “memory” of being transiently propagated via a heterologous PrD. Moreover, various [PSI+] isolates obtained from reverse shuffle exhibited different levels of suppression, even though none of them could match the original strong S. cerevisiae [PSI+] variant in suppression efficiency (Supplement Fig. S9). This indicates that heterologous conversion could be imprecise and generate multiple variants of a prion. Such an infidelity in prion transmission was not detected with the weak S. cerevisiae [PSI+] variant, which produced even weaker prion isolates while propagated via a heterologous protein but was restored after the reverse shuffle (Fig. 4D). Therefore, S. bayanus PrD can faithfully propagate weak S. cerevisiae prion despite a temporary change in its phenotypic expression, but irreversibly alters patterns of the strong S. cerevisiae prion.
Construction of Sup35 proteins with the chimeric prion domains
To determine which specific region of Sup35N is responsible for the species barrier, we constructed a set of chimeric SUP35 genes as described in Supplement (Fig. S7). The convenient location of conserved restriction sites enabled us to divide the Sup35N-coding region of the SUP35 gene into 3 exchangeable modules, designated as modules I, II and III (Fig. 1BD). Module I includes most of the QN region up to (and including) position 33, encompassing the whole fragment 8-27 with the maximal percentage of QN residues, which is primarily responsible for the species barrier in the Saccharomyces-Candida combination (Santoso et al., 2000). Module II includes the very end of the QN region and the whole ORs region, while module III includes the remaining portion of Sup35N. There is no difference in aa sequence between S. cerevisiae and S. bayanus within the “tail” of the QN region that falls into module II (positions 34 to 40), and there is only one aa substitution within this region in S. paradoxus (Fig. 1D).
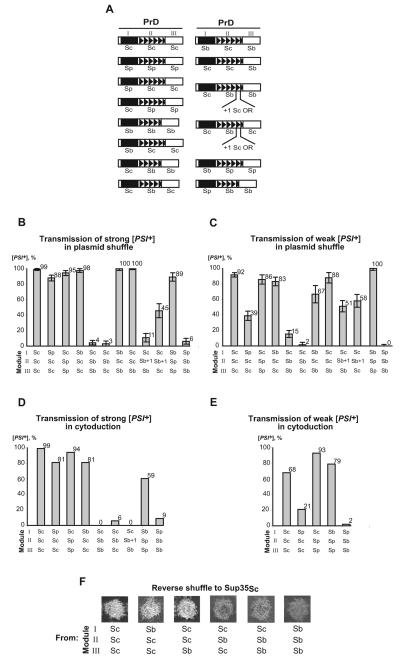
We generated a set of SUP35 genes with chimeric SUP35N domains, combining modules I, II and III of S. cerevisiae, S. paradoxus or S. bayanus in various combinations (Fig. 5A). Chimeric PrDs were fused in frame to the SUP35MC region from S. cerevisiae and placed under control of the endogenous S. cerevisiae SUP35 promoter (PSUP35), located on a low-copy (centromeric, or CEN) plasmid with the URA3 selectable marker. All constructs were proven to maintain viability of S. cerevisiae in the absence of endogenous Sup35, and remained completely functional in translation termination, as confirmed by inability of the [psi−] sup35Δ strain, bearing each of these constructs, to grow on –Ade medium, that is, to read through the ade1-14 reporter. Each chimeric construct tested was expressed at the same level as S. cerevisiae Sup35 when placed on a plasmid of the same structure (Supplement Fig. S10). For most chimeric proteins, we also show that the protein can be induced into a prion ([PSI+]) state by transient overproduction of the same chimeric construct and/or at least one of the parental SUP35 genes (Supplemental Materials and methods). All the constructs could generate both strong and weak prion strains, with the exception of constructs containing module II of S. bayanus, that produced preferentially weak variants (data not shown).
Figure 5. Identification of PrD modules responsible for the species barrier.
(A) Chimeric PrDs constructed in this study. Exchangeable modules are designated by Roman numerals. For other designations, see Fig. 1. (B) and (C) Frequency of transmission of the strong and weak [PSI+], respectively, to the Sup35 proteins with chimeric PrDs by plasmid shuffle. Direct plasmid shuffle was performed as shown on Fig. 1E. Data for the control S. cerevisiae construct reproduce those shown on Fig. 3B and 3D. For exact numbers, see Supplement Tables SIII and SV. (D) and (E) Frequency of transmission of the strong and weak [PSI+], respectively, to the Sup35 proteins with chimeric PrDs by cytoduction. Cytoduction was performed as shown on Fig. 1F. Data for the control S. cerevisiae construct reproduce those shown on Fig. 3C and 3E. (F) Phenotypic patterns of strong prion variant are switched in the chimeric constructs bearing module II of S. bayanus, as detected after reverse shuffle to the S. cerevisiae Sup35 protein, performed as shown on Fig. 1E. –Ade plates were photographed after 6 ds.
Identification of PrD modules responsible for the species barrier
In order to determine which module of Sup35N controls the species specificity of prion conversion from S. cerevisiae Sup35 to the other S. sensu stricto Sup35 proteins, we performed the plasmid shuffle experiments (Fig. 1E) with each of the chimeric SUP35 constructs. Our results unambiguously demonstrated that module I of S. paradoxus is responsible for the transient decrease in [PSI+] phenotypic stringency (data not shown) and for the species barrier in prion transmission (Fig. 5B, C, and Supplement Tables SIII and SV), while the region encompassing modules II and III of S. paradoxus exhibits little or no effect. In contrast, module II of S. bayanus was responsible for the species barrier, while modules I and III of S. bayanus exhibited little or no effect (Fig. 5B, C and Supplement Tables SIII and SV). Even in this case, barrier was not as severe as for strong [PSI+]. These results were generally confirmed by cytoduction experiments (Fig. 5D, E, and Supplement Tables SIV and SVI).
Notably, insertion of an additional OR unit of S. cerevisiae origin into the S. bayanus module II somewhat increased cross-species prion conversion but did not completely eliminate the barrier in plasmid shuffle (Fig. 5B, C), and did not show any effect on the barrier in cytoduction (Fig. 5D). This indicates that while the size of module II plays a certain role in cross-species prion transmission, its specific sequence features are also important.
Importantly, module II of S. bayanus was both required and sufficient for switching the strong [PSI+] variant to the weaker variant as detected in reverse shuffle (Fig. 5F). This demonstrates that in addition to controlling the frequency of cross-species prion conversion in the S. cerevisiae / S. bayanus combination, module II also controls fidelity of reproduction of the variant-specific prion patterns via a heterologous stage.
Based on the observed differential effects of PrD modules, we predicted that artificial PrD composed of module I of S. bayanus and modules II and III of S. paradoxus would show “promiscuous” behavior, while the reciprocal artificial PrD, composed of S. paradoxus module I and S. bayanus modules II and III would exhibit a very stringent species barrier in all combinations. Indeed, our data confirmed this prediction (Fig. 5B, C, D, and E).
Role of amyloid stretches in cross-species prion conversion
Within module I, more aa substitutions are found between S. cerevisiae and S. bayanus (5 out of 33 positions) than between S. cerevisiae and S. paradoxus (only 3, see Fig. 1D). This seems to disagree with our observation that module I of S. paradoxus is sufficient for the species barrier while module I of S. bayanus is not (see above, Fig. 5). However, all three aa substitutions in the S. paradoxus sequence are located between positions 12 and 20, while S. bayanus has only 2 substitutions within this fragment (Fig. 1D). It therefore appears that identity of the fragment 12-20 rather than that of the whole module I is crucial for prion transmission and stringency. Moreover, 2 out of 3 variable positions within this region are changed in both S. paradoxus and S. bayanus, so that only asparagine (N) to serine (S) substitution at position 12 is specific to S. paradoxus. We changed the codon for S12 into a codon for N in the S. paradoxus sequence, and found that such a substitution significantly increased both cross species transmission of weak S. cerevisiae [PSI+] from either complete S. paradoxus PrD or chimeric PrD containing S. paradoxus module I (Fig. 6A), and phenotypic stringency of the strong prion variant in a heterologous protein background (Fig. 6B). Next, we mutated the codon for N12 (S. cerevisiae version) into a codon for S (S. paradoxus version) in the otherwise intact S. cerevisiae SUP35 gene, and demonstrated that transmission of weak S. cerevisiae prion to the mutant protein is decreased, even though not to such an extent as in the case of the substitution of the whole module I by its S. paradoxus counterpart (Fig. 6C). In the case of the strong prion variant, N12S substitution impaired prion transmission more severely than did the whole module I of S. paradoxus (Fig. 6A) and decreased the phenotypic stringency of the prion maintained by a heterologous protein (Fig. 6B). This was not due to inability of the mutant protein to maintain a phenotypically strong prion variant in principle, as it could be induced into a strong prion variant de novo by overproduction of the S. cerevisiae Sup35N fragment (data not shown). Taken together, our results show that a single aa substitution at position 12 of the Sup35 protein plays an important role in both specificity of prion transmission and stringency of the prion isolates obtained from cross-species conversion, even though it is not solely responsible for the specificity.
Figure 6. Role of amyloid stretches and individual amino acid substitutions in cross-species prion conversion.
Construction of the mutant SUP35 derivatives is described in Supplement (Fig. S11 and Fig. S12). (A) Frequencies of the S. cerevisiae prion transmission in direct shuffle to the derivatives containing S. paradoxus module I with mutated residue 12. (B) Residue 12 controls phenotypic expression of the strong S. cerevisiae prion in the S. cerevisiae / S. paradoxus combination, as seen on –Ade plates photographed after 6 ds. (C) Frequencies of the S. cerevisiae prion transmission in direct shuffle to the S. cerevisiae protein with mutated residue 12. (D) Amyloid stretch hexapeptide consensus (Pastor et al., 2007). Numbers correspond to aa positions within the stretch. Residues within { } are forbidden at a given position, while residues within [ ] are the only ones allowed at a given position. Alterations of the amyloid stretch consensus by substitutions at position 12 of S. paradoxus and position 50 of S. bayanus are shown. (E) and (G) Frequencies of the S. cerevisiae prion transmission in direct shuffle to the S. bayanus PrD derivatives with altered position 50 and to the S. cerevisiae protein with altered position 49, respectively. Data for the control Sp-Sp-Sp (A), Sc-Sc-Sc (C and G), and Sb-Sb-Sb (E) constructs reproduce those shown on Fig. 3B and D. Data for the Sp-Sc-Sc (A) and Sc-Sb-Sb (E) constructs reproduce those shown on Fig. 5B and 5C. Species designations are as on Fig. 1. Shuffle was performed as shown on Fig. 1E. Exact numbers are shown in Tables SIII and SV. Species designations are as on Fig. 1. Shuffle was performed as shown on Fig. 1E. Exact numbers are shown in Supplement Tables SIII and SV. (F) Effect of the Y49P and P50Y substitutions on the phenotypic expression of strong [PSI+] in direct shuffle.
Location of residue 12 is quite remarkable, as it falls within the only sequence found in module I (see above, Fig. 1D) that satisfies requirements for the “amyloid stretch” (Fig. 6D), a consensus hexapeptide detected in most proteins efficiently forming amyloids in vitro (Lopez de la Paz & Serrano, 2004; Pastor et al., 2007). Moreover, the N12S substitution breaks this consensus (Fig. 6D). Despite a relatively high level of flexibility allowed at some positions of the amyloid stretch (Fig. 6D), the S. cerevisiae Sup35N region contains only two more hexapeptides satisfying the consensus requirements, at positions 45-50 within module II, and 102-107 within module III (see above, Fig. 1D). Both stretches are conserved in S. paradoxus; however, they contain respectively one and two aa substitutions in S. bayanus. Substitutions within module III do not break the amyloid stretch consensus, but substitution of tyrosine (Y) to proline (P) at position 49 (S. bayanus position 50) within module II does (Fig. 6D). We mutated P50 into Y in the S. bayanus sequence and found that this substitution significantly increased cross-species transmission of both strong and weak S. cerevisiae prions to the mutated protein (Fig. 6E). In the case of weak [PSI+], the species barrier was essentially eliminated when mutated module II of S. bayanus was combined with module I of S. cerevisiae origin. Although P50Y substitution did not restore the phenotypic stringency of a heterologous prion (Fig. 6F), and did not completely restore the fidelity of reproduction of the variant-specific prion patterns during reverse shuffle in case of strong [PSI+] (Supplement Fig. S9), it altered the spectrum of the prion variants obtained after reverse transmission to the S. cerevisiae Sup35 protein, so that at least some isolates now matched the original strong S. cerevisiae prion in suppression efficiency.
Reciprocal substitution Y49P within the S. cerevisiae Sup35N domain moderately decreased transmission of the strong S. cerevisiae prion but had no detectable effect on transmission of the weak S. cerevisiae prion to a mutated protein, indicating that disruption of the amyloid stretch II consensus by itself is not sufficient for the species barrier (Fig. 6G) Notably, this mutation decreased phenotypic stringency of the strong prion (Fig. 6F), although this was not sufficient for the irreversible switch of a prion variant, as stringency was restored after the reverse shuffle to the non-mutant S. cerevisiae protein (data not shown). Taken together, our data point to the important even though not exclusive role of amyloid stretches in control of the species specificity and fidelity of cross species prion transmission.
Discussion
Relationship between coaggregation, polymerization and prion transmission
We previously demonstrated that even when a heterologous Sup35 protein can coaggregate with the endogenous S. cerevisiae Sup35 prion, this coaggregation does not necessarily lead to efficient conversion of a heterologous protein into a prion (Chen et al., 2007). Indeed, our new data confirm that some of the Sup35 protein associated with aggregates in a heterologous combination might not undergo conversion into the SDS-resistant polymers capable of propagating prion properties (Fig. 2B). Interestingly, efficiency of coaggregation, copolymerization and prion conversion of the heterologous Sup35 protein does not appear to be entirely determined by its PrD. Transmission of the strong S. cerevisiae prion variant to the intact S. paradoxus Sup35 protein is inefficient at both phenotypic level (Chen et al., 2007) and level of SDS-resistant polymers (this study, Fig. 2B), while chimeric protein bearing only PrD of S. paradoxus is susceptible to polymerization (Fig. 2B) and shows only a weak barrier in prion transmission (Fig. 3A, B, and C). The latter result somewhat contradicts our previous observation of a strict species barrier in transmission of strong S. cerevisiae [PSI+] to the chimeric protein with S. paradoxus PrD made for a small sample of colonies (Chen et al., 2007). It is possible that we have either dealt with a statistical fluctuation previously, or more likely, overlooked the [PSI+] colonies appearing after heterologous transmission, as Sup35 PrD from S. paradoxus significantly decreases suppression efficiency of [PSI+] generated by transmission from S. cerevisiae protein, thus requiring longer time for detection of [PSI+] by suppression (for example, see Fig. 6B). In any case, our new data unequivocally confirm that while PrD of S. paradoxus is sufficient to generate a strong transmission barrier for the weak [PSI+], it causes only a slight decrease in transmission of strong [PSI+]. However, it should be stressed that PrD of Sup35 remains the major region responsible for the species barrier between S. cerevisiae and S. bayanus, and at least in case of weak [PSI+], between S. cerevisiae and S. paradoxus.
As Sup35C domains of S. paradoxus and S. cerevisiae are 100% identical to each other, the differences in behavior of complete S. paradoxus protein and chimeric protein with S. paradoxus PrD must be due to Sup35M. Indeed, we previously observed that the Sup35M region of S. paradoxus is partly responsible for extreme mitotic instability of prions generated by intact S. paradoxus Sup35 in the S. cerevisiae cell (Chen et al., 2007). The non-PrD region of S. bayanus Sup35 also influences some patterns of cross-species interactions, as introduction of the chimeric protein with S. bayanus PrD into the strong S. cerevisiae [PSI+] strain results in a larger fraction of protein remaining in the supernatant (Fig. 2A) and a larger proportion of non-polymerized protein associated with aggregates (Fig. 2B), if compared to complete S. bayanus Sup35. Interestingly, the non-PrD region of S. bayanus acts “in favor” rather than “against” prionization. One possibility is that interactions with the host-specific cellular factor, such as chaperone Hsp104 (Chernoff et al., 1995; Rikhvanov et al., 2007), partly modulated by Sup35M (Liu et al., 2002), might influence physical stability of heteroaggregates and/or a freshly generated heterologous prion. If so, this may point to an additional level at which specificity of cross-species prion transmission could be controlled. Further experiments aimed at testing this hypothesis are currently underway. However it should be noted that in general, correlation between the mitotic instability (observable after 20-40 generations for weak prions in general and some heterologous prions in particular, see Supplement Table SIX) and species barrier has not been observed, as some constructs exhibiting instability (e.g. Sb-Sp-Sp) did not show a strong barrier.
Even in the case of a strong species barrier, a significant portion of heterologous protein associated with aggregates is converted into an SDS-insoluble polymer. For S. bayanus Sup35, that is distinguishable from S. cerevisiae Sup35 by size, it is obvious that in a heterologous combination, some of this protein is present in a polymerized fraction that can enter the gel only after boiling (see Supplement Fig. S8). It is possible that in some cases, heterologous protein may “poison” prion polymers and prevent further growth. We have previously observed such a “poisoning” effect in vitro (Chen et al., 2007), and our preliminary data (not shown) suggest that protein with S. bayanus PrD may poison weak S. cerevisiae prions in vivo. Parameters and mechanisms of this phenomenon are currently being investigated.
Prion variants and species barrier
Our results confirm previous findings in mammalian and yeast systems showing that variant-specific patterns of a prion affect cross-species prion transmission. In addition, we also demonstrate that different prion variants of Sup35 may influence prion transmission at a different level. Distribution of Sup35 protein between the soluble and precipitated fractions in the heterologous combinations indicated that weak S. cerevisiae prion (Fig. 2C) is either inefficient in promoting aggregation of a heterologous protein or incorporates it preferentially into small aggregates not precipitated at 39,000 g. It is possible that heteroaggregates of a weak prion are capped at a small size due to a “poisoning” effect of a heterologous protein. In contrast, a strong prion promotes heterologous aggregation (Fig. 2A) but is inefficient in converting aggregated heterologous protein into the SDS-resistant polymers (Fig. 2B). The stringency of the prion variant also influenced cross-species prion transmission to different orthologous proteins in different ways, so that a strong prion variant was transmitted to S. paradoxus PrD more efficiently than a weak variant, while for S. bayanus PrD, the ratio was the opposite (Fig. 3B and D). Interestingly, S. bayanus PrD usually drives a weak prion phenotype in S. cerevisiae (Chen et al., 2007), and this pattern is, at least in part, controlled by its module II region including ORs (see above).
Weak S. cerevisiae [PSI+] prion variants possess a larger portion of PrD that is “protected” from hydrogen exchange and is, therefore, likely to be included in the β-structured region (Toyama et al., 2007). Weak variants also require a larger PrD region for the faithful propagation of variant-specific patterns (Chang et al., 2008; Chernoff, 2008; Shkundina et al., 2006), as compared to the strong prion variants. It is possible that prions formed by S. bayanus PrD are weak because shorter β-structured regions are insufficient for keeping this protein in the amyloid-proficient state. Therefore, more efficient transmission of the weak S. cerevisiae prion variant to S. bayanus PrD could be due to a larger size of the β-structured region involved in such a conversion, while a shorter region generated in the case of a strong prion cannot be stably maintained by the S. bayanus PrD sequence.
Fidelity of cross-species prion conversion
While transmission of the prion state from the S. cerevisiae protein to a protein with the S. paradoxus PrD, or transmission of the weak S. cerevisiae prion to a protein with the S. bayanus PrD resulted in phenotypically weakened prion variants, the patterns of the original S. cerevisiae prion were restored after reverse transmission back to S. cerevisiae protein (Fig. 4A and D). Possibly in these cases, sequence divergence led to the alteration of growth and/or fragmentation kinetics of prion polymers; however, the structural characteristics of prion units remained faithfully reproducible and were restored upon return back to the original sequence. In contrast, the variant patterns were switched irreversibly when strong S. cerevisiae prion was transmitted to the protein with S. bayanus PrD and then back to S. cerevisiae protein (Fig. 4A and C). Perhaps the prion state can be transmitted from the strong S. cerevisiae prion by the S. bayanus PrD only in exceptional situations when an extended β-structured region is occasionally formed in the heteroaggregate. Resulting S. bayanus prion represents a new (weaker) variant which in turn, generates weaker variants of the S. cerevisiae prion in the reverse shuffle. Appearance of the multiple prion variants reflects imprecise interactions between the divergent PrD regions. Such a mechanism could fit into the “conformational selection” model (Collinge & Clarke, 2007), with a clarification that formation of the new conformational variant is stimulated within a heteroaggregate when accurate transmission of the properties of a pre-existing conformer is impaired due to differences in the sequence. It is unlikely that new conformers pre-exist in the strong prion “population,” as intraspecies transmission of the strong prion does not produce weak variants at a detectable level. The variant switch apparently does not occur in the case of transmission of the weak S. cerevisiae prion variant via S. bayanus PrD (Fig. 4D) as the weak variant already contains a large β-structured region. Generation of multiple prion variants was previously reported in the case of promotion of prion formation by a highly divergent Sup35 protein (Vishveshwara & Liebman, 2009); however, this occurred with a much lower frequency than in the S. bayanus / S. cerevisiae reverse shuffle. Prion transmission between some artificially modified derivatives of Rnq1 protein with altered combinations of prionogenic regions also generated multiple prion variants (Kadnar et al., 2010).
Identity determinants of prion proteins
Previous work with highly divergent Sup35 proteins implicated the N-proximal QN-rich region of PrD, encompassing the first 40 aa residues, as a major determinant of the sequence-specificity in prion transmission (Chien & Weissman, 2001; Osherovich et al., 2004; Santoso et al., 2000). However, our data (Fig. 5) surprisingly show that in the S. cerevisiae / S. bayanus combination, specificity of transmission is primarily determined by module II of PrD encompassing residues 34-96. As the “tail” of the QN region located within module II (positions 34-40) is identical in both species, it is obvious that sequence elements located within the region of ORs contribute to transmission specificity. Indeed, mutational alteration at position 50 (Fig. 6E) and/or addition of the missing OR unit to S. bayanus PrD significantly increased cross-species prion conversion (Fig. 5B, C and D).
Moreover, even in the S. cerevisiae / S. paradoxus combination where the QN region is the primary determinant of the species barrier, it is not the overall sequence divergence of this region that is most important. Indeed, the QN region of S. paradoxus, which is responsible for the barrier in transmission of a weak prion from S. cerevisiae, is less divergent from S. cerevisiae than is the QN region of S. bayanus which does not show a barrier (Figs. 1D and 5C). A combination of the QN region of S. bayanus with the rest of the PrD sequence from S. paradoxus generates an artificial PrD that is highly susceptible to transmission of prion state from S. cerevisiae (Fig. 5), despite retaining only about 93% of sequence identity. This is less than in case of the complete S. paradoxus PrD (94%) which does not exhibit such promiscuity, at least for the weak prion variant. In contrast, the reciprocal chimeric combination (Sp-Sb-Sb) possesses a slightly higher identity to the S. cerevisiae PrD than does the complete S. bayanus PrD but exhibits an even stronger barrier. The only plausible explanation for these phenomena is that identity of the relatively short aa stretches located at different positions within the PrD is more important for determining conversion specificity than is overall conservation of PrD sequences. Indeed, if prion specificity is controlled by at least two short stretches, one of which is located within the QN region and is identical between S. cerevisiae and S. bayanus but different in S. paradoxus, while another stretch is located within ORs region and is identical for S. cerevisiae and S. paradoxus but different in S. bayanus, we would get the observed results. This model also agrees with the recent observations for Rnq1 prion, where multiple prion determinants control transmission specificity (Kadnar et al., 2010).
Remarkably, our search for the altered prion identity determinant located within module I (QN region) of S. paradoxus led to the base substitution at position 12, that disrupts the only hexapeptide in module I satisfying the requirements for the amyloid stretch (Fig. 6D), a consensus sequence detected in most proteins that efficiently form amyloids in vitro (Lopez de la Paz & Serrano, 2004; Pastor et al., 2007). Even though position 12 is not solely responsible for the barrier, its alteration has a drastic effect on the efficiency of cross-species prion transmission (Fig. 6A and C). Notably, single aa substitutions with an anti-prion effect were previously generated in the region between positions 8 and 26 that surrounds and includes amyloid stretch I (De Pace et al., 1998). Amyloid stretch I also overlaps with the Sup35N peptide (7-13) shown to form amyloid-like microcrystals in vitro (Nelson et al., 2005), and is included in the region between positions 9 and 20, present in all peptides capable of efficiently immobilizing Sup35NM on the peptide array in vitro and promoting its conversion into an amyloid (Tessier & Lindquist, 2007).
There are two more amyloid stretches in the S. cerevisiae Sup35 PrD, that are located within modules II (ORs region) and III, respectively (Fig. 1B and D). Consensus of stretch II is broken by an aa substitution at position 50 in S. bayanus (Fig. 6D). As mentioned above, we have proven that this substitution has a drastic effect on the cross-species prion transmission (Fig. 6E). Interestingly, substitutions within amyloid stretches I or II weaken the phenotypic patterns of the S. cerevisiae prion (Fig. 6B and F). However, none of these substitutions alone is sufficient for the irreversible variant switch. Stretch III, located at positions 102-107, is conserved in S. paradoxus and conforms to consensus requirements despite two aa substitutions in S. bayanus (Fig. 6D). Interestingly, this stretch is located within the second (less stringent) region of intermolecular interactions uncovered by the peptide array analysis (Tessier & Lindquist, 2007). It remains to be seen if alterations within this region contribute to the species specificity of prion transmission.
In addition to specific sequences, the size of a PrD is apparently playing a role in the species barrier, as it could be seen in the case of addition of a missing OR unit to S. bayanus PrD (Fig. 5). Possibly, PrD size is important for the proper formation of the β-structured amyloid core and/or for correct alignment of the interacting sequences in different amyloid units. Experiments with Rnq1 protein also indicate that alterations of PrD size via removing certain prionogenic regions influence efficiency of prion transmission between the normal and altered protein (Kadnar et al., 2010).
Specific mechanism of the action of amyloid stretches remains unclear, as structural models of Sup35 amyloids, based on different experimental approaches, disagree with each other (for example, see Krishnan & Lindquist, 2005; Shewmaker et al., 2006; Wickner et al., 2007a). It does not seem likely that amyloid stretches, deduced from in vitro experiments, are required for the prion formation in yeast per se, as some yeast PrDs (e. g. Ure2) do not appear to contain them (data not shown). Attempts were made recently to define compositional determinants of prion formation in yeast by approaches that are independent of amyloid stretch consensus (Toombs et al., 2010). However, it is possible that amyloid stretches mark some (although not necessarily all) regions of intermolecular interactions determining the specificity of transmission of the amyloid state to a newly immobilized protein molecule, rather than the initial amyloid formation. Due to a significant level of flexibility allowed by consensus requirements for an amyloid stretch, it can potentially be formed in various sequences of similar aa composition. Indeed, each of the “reshuffled” Sup35 PrDs retaining prion-forming properties (Ross et al., 2004; Ross et al., 2005) contains one or more amyloid stretches, however of different sequences and locations (data not shown), which may explain the generation of prion transmission barriers between these proteins. Previous data for both chimeric Candida-Saccharomyces prion (Chien & Weissman, 2001) and mammalian prions (Scott et al., 2005; Vanik et al., 2004) demonstrated that even single aa substitutions may generate transmission barriers, suggesting that short stretches rather than large regions are involved in control of prion specificity in these cases as well. Further experiments are needed to completely decipher the in vivo code of amyloid recognition.
Experimental Procedures
Yeast strains and plasmids
S. cerevisiae strains GT256-23C (strong [PSI+]) and GT988-1A (weak [PSI+]), as well as control [psi−] strain GT255-2A, were haploid derivatives of GT81 with the following genotype: MATα ade1-14 (UGA) his3 leu2 lys2 trp1 ura3 (see Chen et al., 2007 and references therein). They contained the sup35Δ::HIS3 transplacement on the chromosome (constructed as described previously, see Chernoff et al., 2000), and were maintained alive by the S. cerevisiae – E. coli shuttle plasmids bearing the SUP35 gene. The karyogamy-deficient recipient strains for cytoduction were constructed on the basis of the previously described (Chen et al., 2007) strain 1B-D910 (MATa ade1–14 his3 leu2 trp1 ura3 cyh R kar1–1sup35Δ::HIS3 [rho− psi− pin−]), by substituting the original plasmid bearing the S. cerevisiae SUP35 gene by plasmids with a LEU2 marker, each carrying one of the original or chimeric SUP35 constructs.
All plasmids used in this study were centromeric (low-copy) vectors with either URA3 or LEU2 markers. Plasmids containing the complete genes from S. cerevisiae, S. paradoxus or S. bayanus under control of the endogenous S. cerevisiae SUP35 (PSUP35) promoter were described previously (see Chen et al., 2007 and references therein). Major plasmids constructed in this work and primers used for plasmid construction and mutagenesis are listed in Supplement (Tables SI and SII, respectively).
For construction of the SUP35 genes with chimeric SUP35N domains, we employed recognition sites for restriction endonucleases HindIII (located between modules I and II) and PflMI (located between modules II and III) that are conserved among the three Saccharomyces species in this work. The PflMI site is unique while another HindIII site is present in SUP35M, close to the N/M boundary. The construction strategy is explained in detail in Supplement (Fig. S7). Due to the construction procedure, all chimeric proteins contained the insertion of two additional aa residues at Sup35N/M boundary. To make sure that this insertion does not influence prion transmission, the S. cerevisiae SUP35 gene with the same insertion was constructed and used as a control in all experiments; no differences from intact SUP35 were observed. The mutagenesis strategy for constructs with alterations within amyloid stretches is described in Supplement (Fig. S11). All chimeric SUP35N domains constructed as described here and further were verified by sequencing by Nevada Genomics Center and Eurofins MWG Operon.
Genetic and microbiological techniques
Standard yeast media and growth conditions, as well as standard techniques for yeast genetic analysis, transformation and phenotype characterization were employed (Kaiser et al., 1994). Yeast cultures were incubated at 30°C. [PSI+] detection, characterization, cytoduction, induction by transient overproduction and curing by GuHCl were performed according to routinely used procedures (see Chernoff et al., 2002).
Plasmid shuffle
Plasmid shuffle was performed as described previously (Chen et al., 2007) and shown above (Fig. 1E). In brief, the S. cerevisiae [PSI+] sup35Δ strain with the S. cerevisiae SUP35 gene on a LEU2 plasmid was transformed by a URA3 plasmid bearing the homologous, heterologous, chimeric or mutated SUP35 construct. Transformants were obtained on the medium lacking uracil and leucine (-Ura, Leu) that is selective for both plasmids, and checked for suppression of the ade1-14 reporter on both medium lacking only adenine (-Ade) and medium lacking uracil, leucine and adenine (-Ura, Leu, Ade). The former medium enabled us to identify and exclude from further analysis colonies that have lost [PSI+] prior to or in the process of transformation, while the latter medium was used to determine whether newly introduced Sup35 protein is converted into a non-functional form or remains functional. Transformation-associated [PSI+] loss was almost negligible for strong [PSI+] but significant for weak [PSI+]. In parallel, transformants were streaked out on –Ura medium and velveteened to –Leu medium, in order to identify the Ura+ Leu− colonies that have lost the original LEU2 plasmid. Only one Ura+ Leu− colony was analyzed from each individual [PSI+] transformant, to ensure independence of all colonies from each other. Reverse shuffle was performed in a similar way, except that medium with 5-fluoroorotic acid (5-FOA) was used to cure transformants of the URA3 plasmid.
Cytoduction
Cytoduction experiments were performed as described previously (Chen et al., 2007). Donor strains were mated to the respective derivatives of the strain 1B-D910 by mixing them on YPD medium. After overnight incubation, mixtures were streaked onto –Leu medium with 5 ug/ml cycloheximide, containing 2% ethanol and 2% glycerol instead of glucose. This medium is selective for cytoductants getting the cytoplasm with mitochondrial DNA from the donor. Selected colonies were tested on the medium lacking adenine, and for the transfer of the donor plasmid on –Ura medium. Rare colonies getting URA3 plasmid from the donor were excluded.
Protein isolation and analysis
Proteins were isolated from yeast and fractionated by centrifugation at 39,000 g as described previously (Chen et al., 2007). “Boiled gel” analysis was performed as described (Kushnirov et al., 2006). In brief, protein samples containing 2% SDS were loaded onto the SDS-PAGE gel and run for a while, followed by interruption of electrophoresis and addition of the new portion of acrylamide to the wells. Once newly added polyacrylamide was solidified, gel was incubated in the boiling water bath for about 10 min, cooled, and electrophoresis was resumed. SDS-resistant polymers initially accumulated in the wells were now destroyed by boiling and capable of entering the gel.
Supplementary Material
Acknowledgements
We thank E. Afanasieva, M. Borodovsky, P. Burns, V. Kushnirov, S. Lindquist and P. Tessier for helpful discussion, and D. Bedwell for antibodies. This work was supported by grants MCB-0614772 from NSF and R01GM58763 from NIH to YOC. BC was a recipient of the Suddath Award from Institute for Bioengineering and Bioscience. SG was a recipient of the Undergraduate Research Scholarship from Georgia Tech – Emory Cell and Tissue Engineering Center.
References
- Baudin-Baillieu A, Fernandez-Bellot E, Reine F, Coissac E, Cullin C. Conservation of the prion properties of Ure2p through evolution. Mol Biol Cell. 2003;14:3449–3458. doi: 10.1091/mbc.E03-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce M, Chree A, McConnell I, Foster J, Pearson G, Fraser H. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos Trans R Soc Lond B Biol Sci. 1994;343:405–411. doi: 10.1098/rstb.1994.0036. [DOI] [PubMed] [Google Scholar]
- Chang H-Y, Lin J-Y, Lee H-C, Wang H-L, King C-Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc Natl Acad Sci U S A. 2008;105:13345–13350. doi: 10.1073/pnas.0802215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci U S A. 2007;104:2791–2796. doi: 10.1073/pnas.0611158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO. Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition? Current Opinion in Chemical Biology. 2004a;8:665–671. doi: 10.1016/j.cbpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Chernoff YO. Do amyloids remember their origin? New insights into the prion species barrier. Molecular Cell. 2004b;14:147–148. doi: 10.1016/s1097-2765(04)00208-4. [DOI] [PubMed] [Google Scholar]
- Chernoff YO. Replication vehicles of protein-based inheritance. Trends in Biotechnology. 2004c;22:549–552. doi: 10.1016/j.tibtech.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Chernoff YO. Identity determinants of infectious proteins. Proc Natl Acad Sci U S A. 2008;105:13191–13192. doi: 10.1073/pnas.0806234105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Uptain SM, Lindquist SL. Analysis of prion factors in yeast. Methods in Enzymology. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]
- Chien P, Weissman JS. Conformational diversity in a yeast prion dictates its seeding specificity. Nature. 2001;410:223–227. doi: 10.1038/35065632. [DOI] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- De Pace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009;181:1159–1167. doi: 10.1534/genetics.108.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Wickner RB. Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full-length protein. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16384–16391. doi: 10.1073/pnas.162349599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DA, True HL. New insights into prion structure and toxicity. Neuron. 2006;50:353–357. doi: 10.1016/j.neuron.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Inge-Vechtomov SG, Zhouravleva GA, Chernoff YO. Biological roles of prion domains. Prion. 2007;1:228–235. doi: 10.4161/pri.1.4.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, True HL, Chernoff YO, Lindquist S. Molecular population genetics and evolution of a prion-like protein in Saccharomyces cerevisiae. Genetics. 2001;159:527–535. doi: 10.1093/genetics/159.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadnar ML, Articov G, Derkatch IL. Distinct type of transmission barrier revealed by study of multiple prion determinants of Rnq1. PLoS Genet. 2010;6:e1000824. doi: 10.1371/journal.pgen.1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Alexandrov IM, Mitkevich OV, Shkundina IS, Ter-Avanesyan MD. Purification and analysis of prion and amyloid aggregates. Methods. 2006;39:50–55. doi: 10.1016/j.ymeth.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Kochneva-Pervukhova NV, Chechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury PT, Jr., Caughey B. The chemistry of scrapie infection: implications of the ‘ice 9’ metaphor. Chemistry & Biology. 1995;2:1–5. doi: 10.1016/1074-5521(95)90074-8. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de la Paz M, Serrano L. Sequence determinants of amyloid fibril formation. Proc Natl Acad Sci U S A. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Lee CI, Ostapchenko VG, Baskakov IV. Highly promiscuous nature of prion polymerization. The Journal of Biological Chemistry. 2007;282:36704–36713. doi: 10.1074/jbc.M704926200. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biology. 2004;2:E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Pastor MT, Esteras-Chopo A, Serrano L. Hacking the code of amyloid formation: the amyloid stretch hypothesis. Prion. 2007;1:9–14. doi: 10.4161/pri.1.1.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone effects on prion and nonprion aggregates. Prion. 2007;1:217–222. doi: 10.4161/pri.1.4.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- Scott MR, Peretz D, Nguyen HO, Dearmond SJ, Prusiner SB. Transmission barriers for bovine, ovine, and human prions in transgenic mice. J Virol. 2005;79:5259–5271. doi: 10.1128/JVI.79.9.5259-5271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci U S A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkundina IS, Kushnirov VV, Tuite MF, Ter-Avanesyan MD. The role of the N-terminal oligopeptide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics. 2006;172:827–835. doi: 10.1534/genetics.105.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- Vanik DL, Surewicz KA, Surewicz WK. Molecular basis of barriers for interspecies transmissibility of mammalian prions. Mol Cell. 2004;14:139–145. doi: 10.1016/s1097-2765(04)00155-8. [DOI] [PubMed] [Google Scholar]
- Vishveshwara N, Liebman SW. Heterologous cross-seeding mimics cross-species prion conversion in a yeast model. BMC Biol. 2009;7:26. doi: 10.1186/1741-7007-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C. The state of the prion. Nature Reviews Microbiology. 2004;2:861–871. doi: 10.1038/nrmicro1025. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nature Reviews. 2007a;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T, Engel A, McCann L, Kryndushkin D. Yeast prions: evolution of the prion concept. Prion. 2007b;1:94–100. doi: 10.4161/pri.1.2.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.