Abstract
POLQ (polθ) is a large, multi-domain DNA polymerase0020encoded in higher eukaryotic genomes. It is important for maintaining genetic stability in cells and helping protect cells from DNA damage caused by ionizing radiation. POLQ contains an N-terminal helicase-like domain, a large central domain of indeterminate function, and a C-terminal polymerase domain with sequence similarity to the A-family of DNA polymerases. The enzyme has several unique properties, including low fidelity and the ability to insert and extend past abasic sites and thymine glycol lesions. It is not known whether the abasic site bypass activity is an intrinsic property of the polymerase domain, or whether helicase activity is also required. Three “insertion” sequence elements present in POLQ are not found in any other A-family DNA polymerase and it has been proposed that they may lend some unique properties to POLQ. In this work we analyzed the activity of the DNA polymerase in the absence of each sequence insertion. We find that the pol domain is capable of highly efficient bypass of abasic sites in the absence of the helicase-like or central domains. Insertion 1 increases the processivity of the polymerase but has little, if any, bearing on the translesion synthesis properties of the enzyme. However, removal of insertions 2 and 3 reduces activity on undamaged DNA and completely abrogates the ability of the enzyme to bypass abasic sites or thymine glycol lesions.
Keywords: replication, mutation, fidelity, enzyme kinetics
Introduction
DNA polymerase θ (POLQ) was originally described as sharing sequence homology with the A-family of DNA polymerases typified by Escherichia coli DNA polymerase I.1 Subsequent work showed POLQ to be a 290 kDa protein homologous to Drosophila Mus308, consisting of an N-terminal helicase-like domain and a C-terminal A-family polymerase domain connected by a large central domain.2 While the polymerase domain has been shown to perform template directed synthesis of DNA, no specific function has yet been assigned to the helicase domain or the central domain.
The role of POLQ in the cell is still a matter of debate and several lines of experiments have been performed to investigate its possible function in vivo. POLQ is important in defense against damage caused by ionizing radiation. Mouse bone marrow cell lines deleted for Polq are more sensitive than normal cells to ionizing radiation,3 and the ionizing radiation sensitivity of human tumor cells is increased by siRNA-mediated suppression of POLQ.4 Polq−/− mice develop normally, but have elevated frequencies of spontaneous and radiation-induced micronuclei,5–6 diagnostic of an increased frequency of chromosomal breakage. One possibility is that POLQ is involved in translesion synthesis opposite some lesion generated by ionizing radiation, helping to prevent generation of double strand breaks at stalled DNA replication forks. POLQ may also be involved in some aspect of double strand break repair. This is suggested by the partial impairment of synthesis-dependent microhomology-mediated end-joining described recently in Drosophila Mus308 mutants.7
A knockout of POLQ in the chicken DT40 B-cell line shows some increased sensitivity to hydrogen peroxide, indicating that polθ is involved in tolerance of damage caused by reactive oxygen species, perhaps by lesion bypass or as a backup DNA polymerase for BER.8 POLQ has intrinsic 5'-deoxyribose phosphate (5'-dRP) lyase activity that could function in BER.9 POLQ has also been suggested as a possible candidate for involvement in somatic hypermutation of immunoglobulin genes,10–13 but it appears to play a minor role, if any, in this process.14 POLQ gene expression is higher in tumor cell lines than in normal cell lines4,15 and is also elevated in human colon cancer tissues compared with surrounding normal tissues.16
The proposed functions of POLQ, and its effects upon under- or over-expression, are based upon several unusual features of this enzyme. Despite sharing sequence similarity with the A-family DNA polymerases, POLQ is a low fidelity enzyme with no editing function17 and exhibits error rates for single base insertions and deletions on par with the Y-family DNA polymerases κ and η.18 POLQ possesses the unique ability to bypass both abasic sites and thymine glycol lesions with a steady-state incorporation efficiency opposite an abasic site of only about 4-fold less than for incorporation of an A opposite T.19 POLQ also readily extends mismatched primer termini and, in concert with polymerase ι, is able to extend past a (6-4) photoproduct.20 Additionally, in the presence of single stranded DNA, POLQ exhibits ATPase activity,2 and shows no exonuclease activity on any substrate.2
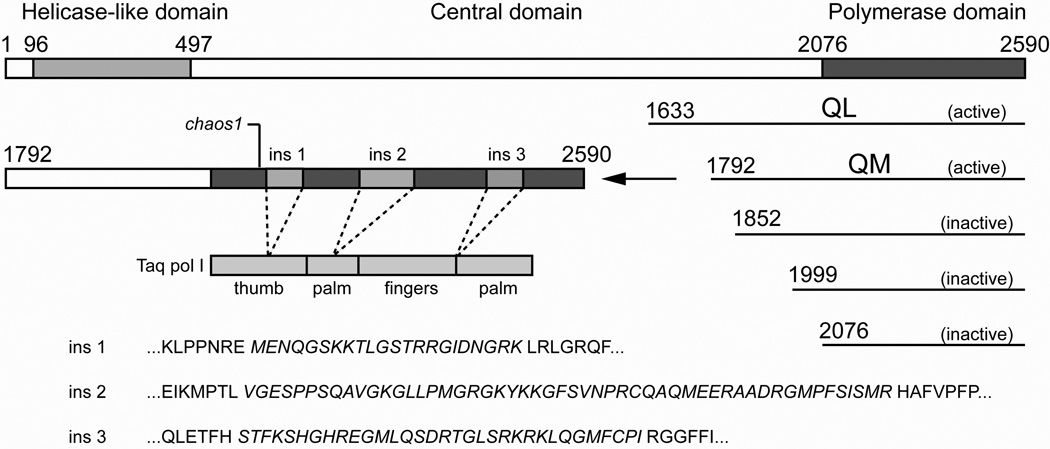
The unique ability of POLQ to bypass lesions such as abasic sites and thymine glycol and to extend past mismatches has been postulated to be associated with three sequence elements that are not present in prokaryotic members of the A-family of DNA polymerases or in invertebrate homologs such as Drosophila Mus308. Sequence alignment of human and mouse POLQ with other A-family polymerases show that insertion 1 likely resides in the thumb domain of the polymerase and insertions 2 and 3 are likely within the palm domain of the enzyme19 (Figure 1). A computationally derived model of the polymerase domain of POLQ (residues 2076–2590)19 based on the known structure of Taq DNA polymerase I21–22 shows the predicted location of these insertion loops with respect to duplex DNA and an incoming nucleoside triphosphate (Figure 2). In the current work, we have deleted each of the insertion elements in turn and examined the resulting polymerase’s ability to bypass an abasic site and a thymine glycol lesion. We show that deletion of insertion 1 has no significant effect on the polymerase’s ability to bypass lesions but instead reduces the enzyme’s processivity while deletion of insertion 2 or 3 significantly reduces polymerase activity on undamaged DNA and completely abolishes the polymerase’s ability to bypass abasic sites and thymine glycols.
Figure 1.
POLQ constructs used in this study. Full length POLQ is shown on top as a 2590 amino acid sequence containing an N-terminal helicase-like domain, a large central domain and a C-terminal A-family polymerase domain. The N-terminal amino acid and level of polymerase activity are shown for each of the five truncation constructs. The truncation mutant QM is enlarged to show the relative location of the three insertion elements and their predicted location relative to the functional domains of Taq pol I. The positions of the insertions are exactly as previously described.19 The equivalent position of the chaos1 mutation is also indicated. The amino acid sequences of the loop deletions are shown in italics within their respective flanking sequences.
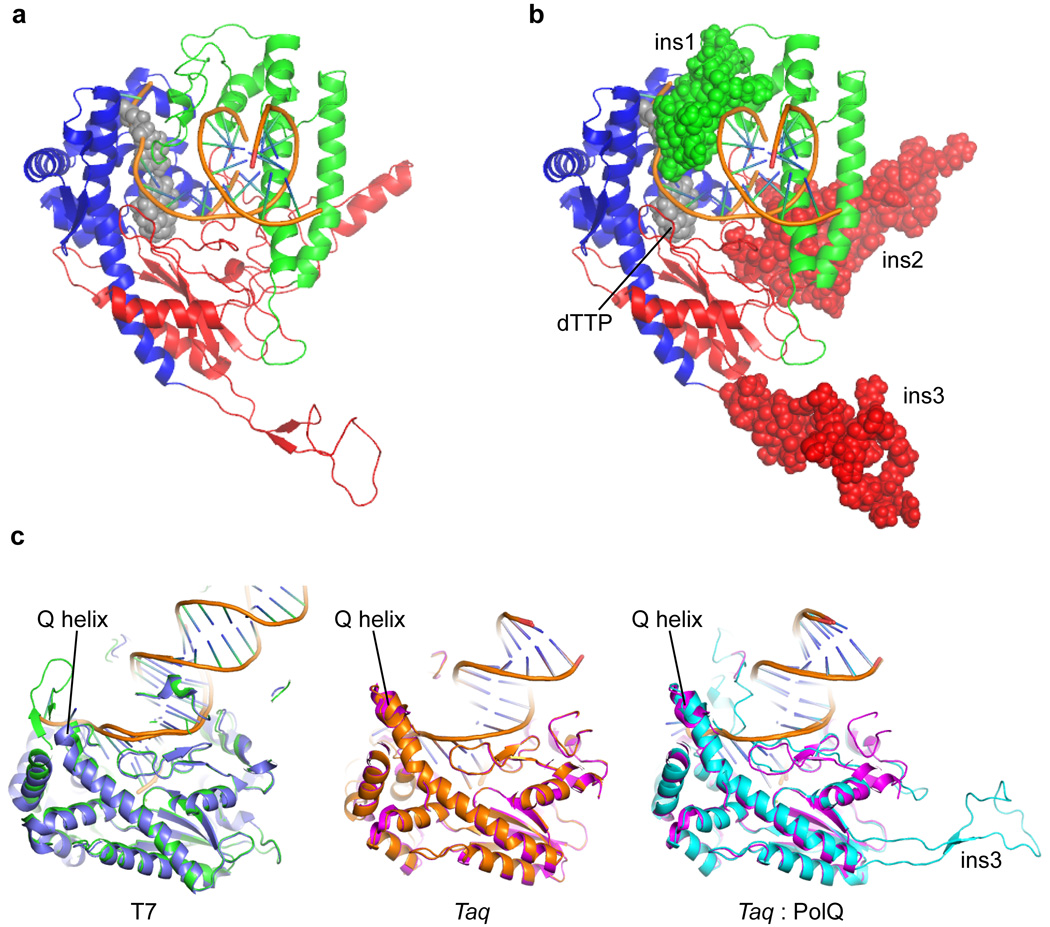
Figure 2.
a) The computationally derived model of the polymerase domain of POLQ as originally described.19 The thumb domain is shown in green, the palm domain in red, and the fingers domain in blue. The templating nucleotide and incoming dTTP are shown as space filling models (gray) and the duplex DNA as ribbons (orange). b) Insertion loops 1, 2 and 3 are shown as space filling models to highlight their relative orientation with respect to the duplex DNA and the nascent base pair. c) Alignments of the open (PDB ID: 1SL1)34 (blue) and closed (PDB ID: 1T8E)35 (green) forms of the DNA polymerase from bacteriophage T7, the open (PDB ID: 2KTQ)36 (orange) and closed (PDB ID: 1QTM)21 (magenta) forms of the DNA polymerase from Taq and the closed form of Taq DNA polymerase and the computationally derived model of POLQ (cyan). The Q helix of the fingers domain is labeled.
Results
As part of an effort to produce large amounts of POLQ for structural and functional studies, a series of constructs were made that shortened the N-terminus of the protein. Our initial construct contained residues 1633–2590 and is shown as QL (long) in Figure 1. The shortest length with which we observed activity comprised residues 1792–2590, labeled QM (medium) in Figure 1. All constructs shorter than QM were either completely inactive or failed to produce soluble protein therefore construct QM was used as the starting point for making deletions of the three insertion loops: residues 2149–2170 (Δins1), residues 2264–2315 (Δins2) and residues 2497–2529 (Δins3) (Figure 2). All three deletion mutants yielded soluble protein but the yields were significantly decreased for the Δins2 and Δins3 variants. For example, we were consistently able to produce as much as 2 mg of soluble protein for every liter of cell culture for QM and Δins1 but this dropped to about 500 µg/L for Δins2 and to approximately 50 µg/L for Δins3.
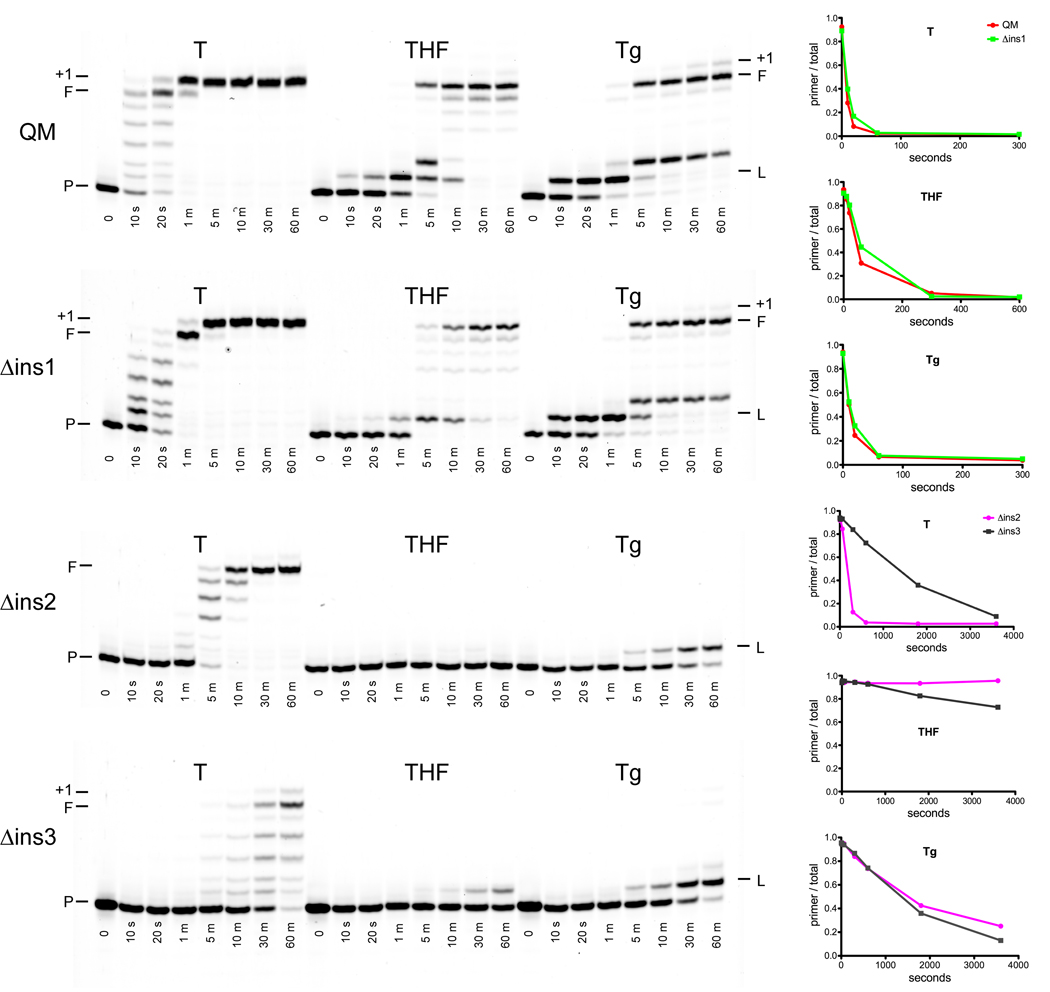
Primer extension assays with undamaged template DNA as well as with DNA containing an abasic site or a thymine glycol show that the truncated form of POLQ (QM) can readily copy undamaged as well as damaged DNA. As seen in Figure 3, QM can completely extend a primer past an undamaged template with no obvious pause sites. However, extension past an abasic site showed an initial pause at the point of incorporation opposite the abasic site followed by an accumulation of product one base past the abasic site and then complete extension of the primer. Extension past the thymine glycol lesion showed pausing at both the site of the lesion and one base past the lesion followed by full extension of the primers. These pause sites are in keeping with biochemical23–24 and structural25 studies with replicative polymerases, which demonstrated that the 5-methyl group of Tg disrupts the stacking of the 5’ base, thereby preventing extension of the Tg:A pair.
Figure 3.
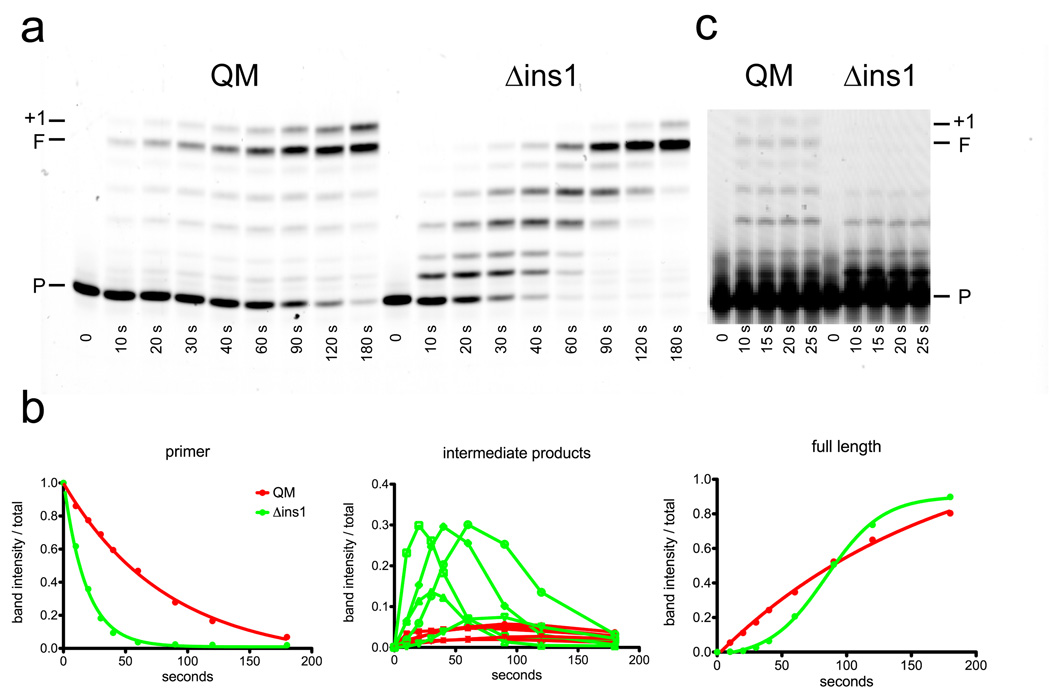
Primer extension assays. In these reactions, undamaged DNA is represented by a T in the templating position and lesion-containing DNA by a tetrahydrofuran (THF) or thymine glycol (Tg) in the templating position. The positions of the unextended primers are labeled with a P, the full extension products with an F, the untemplated extensions of the duplex DNA (if present) with a +1, and the positions of the lesions with an L. The graphs to the right of the figure represent the relative intensities of the remaining primer bands with respect to the intensities of all bands as a function of time. The decrease in the amount of unextended primer is shown for undamaged templates (T) and those containing a tetrahydrofuran (THF) or thymine glycol (Tg) at the templating position. The first three graphs compare the wild type polymerase (QM, red) to the insertion 1 deletion mutant (Δins1, green) and the last three graphs compare the insertion 2 deletion mutant (Δins2, magenta) to the insertion 3 deletion mutant (Δins3, grey).
Extension of the primers by Δins1 (Figure 3) was similar to that of QM with all three DNA templates as demonstrated by the plots of primer intensity over time. The translesion synthesis capabilities of POLQ are also retained in the Δins1 variant as full length products were formed when either an abasic site or thymine glycol was present at the templating position. Δins2 and Δins3, however, were significantly reduced in their abilities to extend primers with all three DNA templates (Figure 3). Based on the plots of primer utilization with an undamaged template, all primers had been extended by about 60 seconds for the QM and Δins1 polymerases but a similar level of extension was not attained until about 600 seconds for the Δins2 polymerase and 3600 seconds for the Δins3 polymerase. The Δins2 mutant was completely devoid of any activity when an abasic site was in the templating position while the Δins3 mutant still retained the ability to incorporate a nucleotide opposite the lesion, albeit at a significantly reduced rate and with no detectable ability to extend beyond the lesion. Both the Δins2 and Δins3 mutants were able to incorporate a base opposite the thymine glycol lesion at similar rates but were significantly slower than the QM and Δins1 mutant polymerases. After about one minute, most primers had been extended by the QM and Δins1 polymerases but similar amounts of primer utilization by the Δins2 and Δins3 mutants were not achieved until about one hour. Neither the Δins2 nor Δins3 mutants were able to extend past the thymine glycol lesion to any appreciable extent (only a very faint band was observed for a single incorporation after the lesion at one hour for the Δins3 mutant). Also in contrast to the wild type QM polymerase and Δins1 mutant protein, no extension of the primer to full length was observed.
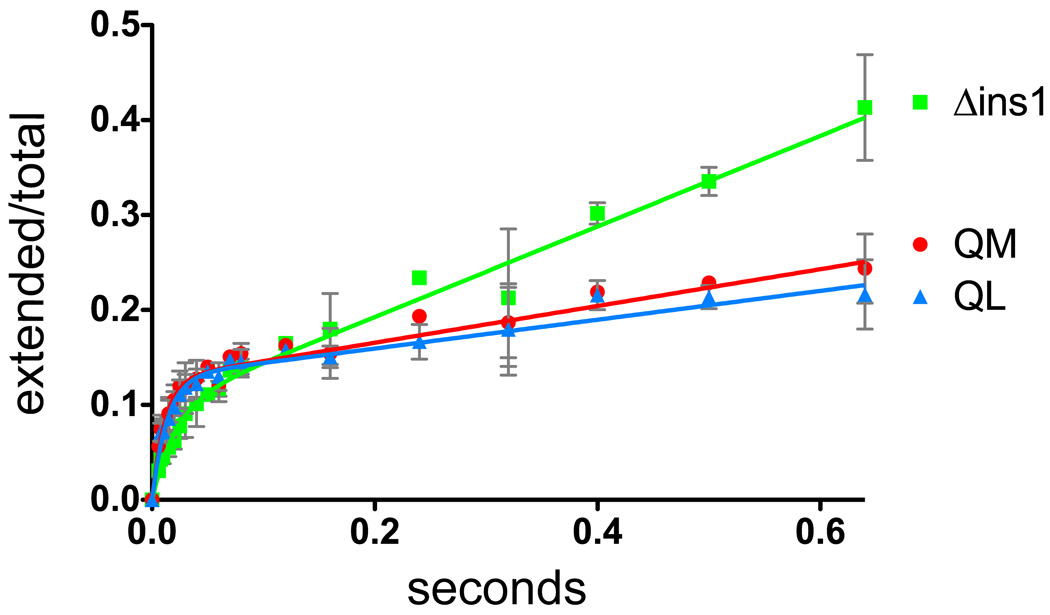
Due to the absence of a qualitative defect in bypassing lesions for Δins1, we examined the effect of this variant on the pre-steady state and steady state kinetic rates of polymerization by POLQ. Figure 4 shows the curves generated by measuring the incorporation of dAMP opposite a templating T by POLQ at very early time points. As a control, the longer version of POLQ (QL) was included in the analyses and all three enzymes exhibited an initial burst of activity. The burst amplitude of Δins1 was reduced by about 25% from the wild type enzymes (Table 1). The burst rate of Δins1 was also reduced when compared to the two constructs in which insertion 1 is retained. The steady-state rate of incorporation after the initial burst phase, however, was increased for Δins1 by more than two-fold (Table 1). These data suggest that Δins1 is less tightly bound to the DNA (it has a lower burst amplitude) with a reduced activity for incorporating nucleotides (it has a lower burst rate) but that it can dissociate and re-associate with DNA more rapidly (it has a higher steady state polymerization rate). To test this idea we examined the processivity of the Δins1 enzyme by comparing it to QM under conditions of significantly reduced enzyme and nucleotide concentrations. As can be seen in Figure 5a, under multiple hit conditions Δins1 initially extended the primer at a faster rate than the wild type enzymes but intermediate length products accumulated and full length product appeared more slowly than for the wild type polymerases (Figure 5b). Single hit conditions in the presence of a DNA trap (Figure 5c) confirm that in a single binding event the wild type enzyme is able to extend the primer further than the Δins1 polymerase.
Figure 4.
Pre-steady state burst curves for incorporation of A opposite T by the QL and QM truncations of POLQ and the deletion mutant of insertion 1 (Δins1). The curves are fit to the equation P=A(1-e−kt)+ksst as described in the Materials and Methods. The curve for Δins1 is shown in green, that for QM is shown in red and that for QL is shown in blue. The error bars represent one standard deviation based on at least three separate experiments and are shown in grey.
Table 1.
Pre-steady state burst kinetics of primer extension by POLQ.
| Burst amplitudea | Burst rate (s−1) | Steady-state rate (s−1) | |
|---|---|---|---|
| QM | 0.13 ± 0.005 | 86.5 ± 12 | 0.19 ± 0.02 |
| Δins1 | 0.10 ± 0.010 | 48.4 ± 12 | 0.48 ± 0.03 |
| QL | 0.13 ± 0.006 | 74.4 ± 10 | 0.15 ± 0.02 |
The values for the burst amplitudes are defined as the ratio of the intensity of the extended primer bands to total primer.
Figure 5.
Processivity experiments. These reactions compare the ability of Δins1 to synthesize undamaged DNA in a processive manner to that of the polymerase constructs containing all three insertions (QM). a) Multiple turnover experiments. The unextended primer is labeled with a P, the full length extension with an F, and untemplated extension of duplex DNA with +1. b) Quantification of bands resulting from the multiple turnover experiments. The first graph illustrates the decrease in the amount of starting primer (P). The second graph shows the formation of fragments of intermediate length between primer and full length. The third graph shows the formation of full length product (F). For all three graphs, the green plots represent the data for the Δins1 polymerase and the red plots represent the QM polymerase. All three graphs are plotted as the ratio of band intensity to the total intensity for all bands in the lane as a function of time in seconds. c) Single hit experiments showing the products formed by the Δins1 and QM polymerases in the presence of an unlabeled DNA trap.
Discussion
Of all the DNA polymerases studied to date, POLQ appears to be unique in that it shares sequence similarity with high fidelity replicative polymerases yet exhibits the low fidelity and lesion bypass ability of translesion synthesis polymerases. Thus it was important to explore how these properties may be related to the presence of three insertion elements that are not found in the A-family polymerases of prokaryotes or invertebrates.19 POLQ also possesses an N-terminal helicase domain and a large central domain, both of indeterminate function.
We first set out to determine the smallest construct of POLQ that retained activity. The five constructs were designed based on secondary structure predictions to avoid truncating the polymerase within any putative helix or β-strand (Figure 1). We found that the polymerase construct comprising residues 1792 through 2590 is close to the minimum size required for polymerase activity as truncation of a further 60 amino acids from the N-terminus results in protein lacking any observable activity. While this work was underway, Prasad et al published some findings relevant to required polymerase length.9 Their smallest active fragment consisted of residues 1712–2590. However, this minimal construct was unable to processively extend DNA. Polymerization rates for their minimal construct were similar to those of the full-length polymerase but their enzyme was only able to extend a primer by two nucleotides. The minimal construct in the current work readily extends a primer by six nucleotides and then incorporates one more untemplated base off of the resulting blunt-ended duplex DNA, similar to that observed previously for several other DNA polymerases.26 It is possible that the fragment chosen by Prasad et al is inherently non-processive although its length is intermediate between QM and QL (Figure 1), both of which are fully processive (Figure 5). We note that our purification scheme is significantly different and that the DNA substrates and reaction conditions also differ. These differences may account for the lack of observed processivity. Moreover, the previous work with truncated constructs did not test the lesion bypass ability of the resulting polymerase.9
One major finding of this work is that that bypass of abasic sites and thymine glycol lesions is an intrinsic function of the truncated form of the polymerase (Figure 3) and is not dependent on the helicase-like domain or most of the central domain present in the full-length enzyme. It is clear, however, that approximately 300 amino acids of the central domain are required for activity as the polymerase domain alone (residues 2076–2590) does not yield a functional enzyme. These central domain residues may play a largely structural as opposed to functional role since soluble polymerase was not readily obtained from the shorter truncations despite the use of several different N-terminal fusion tags and expression protocols (data not shown). It is interesting that the chaos1 mutation of Polq in the mouse produces a phenotype of the same severity as a full gene disruption.5–6 This point mutation is located at a conserved serine in the central domain (Ser1977 in the human protein, Fig. 1), and so this region may be functionally important for additional reasons.
Having established that the QM fragment of POLQ was active, we sequentially deleted insertions 1, 2 and 3 to qualitatively analyze the phenotypes of such mutations. In primer extension assays (Figure 3), when insertion loop 1 was deleted there was little difference compared to wild type in the ability of the polymerase to extend primers using either undamaged DNA or DNA containing an abasic site or thymine glycol lesion in the templating position since primer utilization was similar for both enzymes. Based on sequence alignment to the structure of Taq DNA polymerase, insertion loop 1 was predicted to be found within the thumb domain of the polymerase and may act to hold the polymerase onto the duplex DNA as polymerization occurs. Our results suggest that indeed this is the case. The increased steady state rate, despite a decrease in pre-steady state burst amplitude and burst rate (Table 1), suggested that the Δins1 polymerase more readily dissociates from the DNA substrate as steady state rates of DNA polymerase activity have been generally shown to be measuring the release of DNA from the polymerase.27 A decrease in association between the polymerase and duplex DNA is evidenced by the reduction in processivity of Δins1 as shown in Figure 5. Under multiple turnover conditions (Figure 5a and 5b), primer is used up more rapidly by the Δins1 mutant than the wild type protein. At any given time point, intermediate length products are observed for the Δins1 polymerase at higher concentrations than for wild type polymerase QM. We also observe a slower rate of formation of full length product by the Δins1 mutant. These results support the conclusion drawn from the burst experiments that the Δins1 is more readily able to dissociate from a primer/template and rebind to another but, during any particular binding event, is unable to extend the primer to similar lengths as the wild type polymerase. Single hit reactions in the presence of an unlabeled DNA trap (Figure 5c) show that, during a single binding event, the Δins1 can only extend the primer by about three nucleotides while the wild type polymerase can extend the primer at least twice as far in that it reaches the end of the template and can even add a single un-templated nucleotide to the end of the DNA molecule. These results for Δins1 confirm that this portion of the polymerase is functionally homologous to the tip of the thumb domain in other A-family DNA polymerases that influences processivity by interacting with the duplex DNA during replication.28–29 This loss of processivity, however, does not appear to have a significant effect on the ability of the enzyme to bypass lesions (Figure 3).
In contrast, removal of insertions 2 and 3 resulted in a considerable reduction in the ability of POLQ to synthesize DNA using normal, undamaged templates (Figure 3). Based on the plots of primer utilization (Figure 3), Δins2 appears to extend opposite a templating thymine about 10 times more slowly than wild type while Δins3 appears to be about 60 times slower than wild type. Δins2 was also completely unable to incorporate even a single nucleotide opposite an abasic site and this variant was also deficient in its ability to extend past a thymine glycol. While Δins3 showed an even greater reduction in its ability to synthesize DNA than Δins2, it was still able, albeit weakly, to incorporate a nucleotide opposite an abasic site but was unable to bypass either lesion. The lack of lesion bypass in this assay is likely to be the result of the insertion loop deletions and not just a consequence of a reduced amount of active enzyme. Sufficient Δins2 polymerase was added to allow full extension of the undamaged template by 10 minutes, yet no incorporation opposite furan is observed even after a further 50 minute incubation. Likewise, as shown in the graphs of primer utilization in Figure 3, the differences observed between Δins2 and Δins3 are not due to the differences in amounts of relative active enzyme. Δins3 showed significant reduction in its utilization of an undamaged template but still showed some incorporation opposite furan, something that is completely lacking in Δins2. We also observe no apparent difference in the two mutants’ ability to incorporate nucleotides opposite thymine glycol as their utilization of primer is similar. Both enzymes do, however, incorporate nucleotides opposite thymine glycol about 60 times less efficiently than wild type and the Δins1 mutant (Figure 3).
A computationally derived model of POLQ19 (Figure 2) predicts that insertion 2 lies on the primer side of the DNA duplex, with only minimal interactions with the DNA. Based on the alignment with Taq pol I that places this loop within the palm domain of the polymerase, we predict that removal of this loop disrupts proper protein folding. In addition, our observation that soluble protein yields were significantly reduced by removal of insertions 2 and 3 suggests that the inserts are necessary for the proper folding of the enzyme that would allow for bypass of DNA lesions. This proper folding could possibly involve altering the volume of the polymerase active site. Polymerases of the Y-family are notable for their ability to bypass lesions such as abasic sites, as well as bulky lesions, and these enzymes tend to have larger, more “open” polymerase active sites than the high-fidelity replicative DNA polymerases.30 The computationally derived model19 (Figure 2) also predicts that insertion 3 may lie at the junction between the palm domain and the Q helix of the fingers domain of the enzyme. While comparisons between open and closed forms of A-family DNA polymerases show that the Q helix is not a mobile element (Figure 2C), it is possible that removal of insertion 3 alters the relative orientation of the fingers domain with respect to the palm domain. We must stress that the results presented in this work are qualitative in nature. While we observe clear effects on the processivity and translesion synthesis capability of POLQ upon deletion of the three insertion loops, we cannot state whether these defects are related to catalytic events or substrate binding affinities. We can only speculate, therefore, as to a possible explanation for the observed effects of insertion loop deletion in POLQ. It must also be noted that the computationally derived model only encompasses the polymerase domain of POLQ, a fragment that we have observed to be completely insoluble. Thus the portion of the central domain that is essential for enzyme solubility and activity may well alter the relative positions of these insertion loops and regulate their activity in a way that is not yet observable.
In summary, removing the helicase domain and most of the N-terminal portion of the central domain does not eradicate the lesion bypass activity of POLQ, although the last 300 amino acids of the central domain must be retained for the enzyme to remain soluble and active. Removal of insertion 1 results in an enzyme that exhibits reduced processivity, likely due to decreased affinity for duplex DNA, but shows no significant loss of lesion bypass ability. Removal of insertions 2 or 3 yields an enzyme with a markedly reduced ability to synthesize DNA and completely unable to bypass abasic sites or thymine glycol lesions. Recently, expression profiles for the human nuclear DNA polymerase genes were examined in patients with previously untreated primary breast cancers. Interestingly, POLQ was the only DNA polymerase gene significantly up-regulated in breast cancer compared with normal breast tissues.31 Further, POLQ expression correlated with poor clinical outcome, independently of Cyclin E expression or the number of positive nodes, which are currently considered as markers for poor outcome. These observations underline the importance of biochemical studies designed to explain the unique properties of POLQ.
Materials and Methods
Materials
All chemicals and reagents were purchased from Sigma (St. Louis, MO) or Fisher (Waltham, MA) and were of the highest purity. Nucleoside triphosphates were purchased from New England Biolabs (Ipswich, MA). The oligonucleotides were synthesized by the Midland Certified Reagent Company (Midland, TX) and were purified on 16% polyacrylamide gels and desalted on Sep-Pak C18 cartridges (Waters Corp., Milford, MA). The sequence of the primer was 5’-GCGGCTGTCATAAG-3’ and the 5’ end of the primer strand was labeled with tetrachlorofluorescein (TET) for subsequent visualization. The template sequence was 5’-GACCAXCTTATGACAGCCGCG-3’ where X denotes a thymine (T), thymine glycol (Tg) or tetrahydrofuran (THF), an abasic site analog that is resistant to cleavage.32 Primer and template oligonucleotides were annealed in 10 mM Tris-HCl pH 7.5, 50 mM NaCl and 1 mM EDTA with a 20% excess of template strand by heating to 70°C and cooling. All duplex oligonucleotides were tested to ensure that at least 90% of the primers were extendable.
Generation of mutants
The open reading frame for full length POLQ cloned into plasmid pFastBacHTc2 was used in polymerase chain reactions wth Pfu DNA polymerase to isolate the C-terminal part of the protein encoding amino acid residues 1633 (beginning with amino acid sequence GASFDL) to 2590. This was cloned into pGEX6P-1 (G.E. Healthcare) and propagated in E. coli STBL2 host cells (Invitrogen). This subcloning step was carried out by Dr. Luis Brieba in Dr. Tom Ellenberger’s laboratory at Harvard Medical School. Correct cloning was verified by DNA sequencing.
To generate deletion mutants, primers annealing just outside of the target region to be deleted were used, with a SapI recognition sequence attached to the 5’ end of each primer. This allowed amplification of the entire plasmid by PCR, except for the deleted region. The PCR products were digested with SapI and purified with QIAquick PCR Purification Kit (Qiagen). The plasmid was recircularized by incubation with T4 DNA ligase. After subcloning and verifying correct deletion of the inserts by DNA sequencing, residues 1792 to 2590 were subcloned by PCR into the pSUMO3 vector (LifeSensors).
The primers used to delete the amino acids in inserts 1, 2, or 3 as shown in Figure 1 were as follows:
Primer1 (Insert1): aaggttggccatgctcttccGAGCTAAGGCTGGGAAGACA
G Primer2 (Insert1): aaggttggccatgctcttccCTCTCTATTTGGGGGCAAC
Primer1 (Insert2): aaggttggccatgctcttcCCTACATGCCTTTGTGCCTTTCCCAGGTGG
Primer2 (Insert2): aaggttggccatgctcttccTAGTGTTGGCATTTTGA
Primer1 (Insert3): aaggttggccatgctcttccCACAGAGGAGGCTTCTTCATCC
Primer2 (Insert3): aaggttggccatgctcttccGTGGAAGGTCTCTAATTGC
Protein purification
pSUMO3 vectors containing the wild type and mutant polymerase genes were transformed into Rosetta2(DE3)/pLysS cells (Stratagene). Five freshly grown colonies were picked from a plate and resuspended in 20 ml LB broth. 1 ml of resuspended cells was added to 1 L of autoinduction medium (1X Terrific Broth (USB Corporation), 0.5% w/v glycerol, 0.05% w/v dextrose, 0.2% w/v alpha-lactose, 100 µg/ml ampicillin and 34 µg/ml chloramphenicol) in a 2.8 L Fernbach flask.33 The flasks were shaken at 20°C for 60 hours. For each construct, 6 L of culture were grown and resulting E. coli pellets were stored at −80°C.
Frozen pellets were thawed on ice and resuspended in buffer containing 50 mM HEPES pH 8, 300 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole pH 8, 5 mM CaCl2, 1.5 % (v/v) NP-40 substitute (Fluka), 5 mM 2-mercaptoethanol (BME), 10 mM PMSF, 100 mM benzamidine and 500 mg DNase I at a volume of 5 ml of buffer per gram of cell pellet. The resuspended cells were sonicated on ice with constant stirring and then spun down twice at 27,000 g. The clarified cell lysate was loaded onto a 5 ml His-Trap column (GE Lifesciences) and washed with buffer A (50 mM HEPES pH 8, 300 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole pH 8, 5 mM BME and 0.005% v/v NP-40 substitute). Bound fractions were then eluted with buffer B (50 mM HEPES pH 8, 300 mM NaCl, 10% (v/v) glycerol, 0.005% (v/v) NP-40 substitute, 5 mM BME and 125 mM imidazole pH 8). The eluted fractions were loaded onto a 15 ml column packed with type-II ceramic hydroxyapatite (Bio-Rad) and washed with buffer C (50 mM HEPES pH 8, 300 mM NaCl, 10% (v/v) glycerol, 0.005% NP-40 substitute and 5 mM BME). Bound fractions were eluted with a shallow gradient to 10% buffer D (500 mM K2HPO4/KH2PO4 pH 8, 300 mM NaCl, 10% (v/v) glycerol, 0.005% NP-40 substitute and 5 mM BME). Eluted fractions were loaded onto a 5 ml Heparin Hi-Trap column (GE Lifesciences) and washed with buffer C. Bound fractions were eluted with a gradient to buffer E (50 mM HEPES pH 8, 2 M NaCl, 10% (v/v) glycerol, 0.005% (v/v) NP-40 substitute and 5 mM BME). Fractions containing POLQ were pooled and incubated with 5 units of SUMO protease 2 (LifeSensors) for 2 hours. The digested fractions were then loaded onto a 5 ml His-Trap column and washed with buffer C. Cleaved POLQ was separated from uncleaved POLQ and the protease by applying a gradient to buffer B. POLQ fractions were concentrated to 0.5 ml and run through a 25 ml Superdex GS-200 column (GE Lifesciences) pre-equilibrated with buffer C. Fractions containing POLQ were concentrated, frozen in 5 µl aliquots by plunging into liquid nitrogen and stored at −80°C. All steps in the purification process were carried out at 4°C.
Primer extensions
For reactions measuring extension past undamaged DNA, tetrahydrofuran or thymine glycol, 100 nM enzyme and 250 nM DNA were pre-incubated at room temperature in a reaction buffer containing 20 mM Tris-HCl pH 8.8, 4% (v/v) glycerol, 80 µg/ml BSA, 0.1 mM EDTA and 5 mM BME. Reactions were initiated by adding a mixture of dNTP and MgCl2 to final concentrations of 500 µM and 10 mM, respectively. At the indicated time points, the reactions were quenched in a gel loading buffer consisting of 95% formamide and 20 mM EDTA. For pre-steady state burst reactions, the above conditions were used (except that only dATP was included) and reactions were mixed in an RQF-3 rapid quench device (KinTek Corp., State College, PA) and quenched with 500 mM EDTA. Reaction products were separated on 16% denaturing polyacrylamide gels and scanned at 532 nm in a Molecular Imager FX (Bio-Rad) to excite the 5’ TET fluorophore. Band intensities were measured with the Quantity-One software package (Bio-Rad) and the amount of product formed was calculated as the ratio of intensities for extended primers over the intensities of both extended and unextended primers. This was then fit to the equation P=A(1-e−kt) + ksst where P is the ratio of extended primers to total primers, A is the amplitude of the burst phase, k is the rate constant of the burst phase, kss is the steady state rate and t is time. Processivity reactions were carried under similar reaction conditions except that only 10 nM enzyme was used and the concentration of dNTPs was reduced to 100 µM. Single hit processivity experiments were carried in the same manner except that a 75-fold excess of unlabeled oligonucleotides were added to the mixture containing dNTPs and magnesium ions.
Acknowledgments
We thank Drs. Luis Brieba and Tom Ellenberger for assistance with some POLQ constructs and April Averill for advice regarding protein expression. Work in the laboratories of SD and SSW is supported by NIH RO1 CA52040 awarded by the National Cancer Institute. DNA sequencing was performed by the DNA Analysis Facility supported by the Vermont Cancer Center. Work in RDW’s laboratory is supported by grant NIH P30-ES007784 from the National Institute of Environmental Health Sciences, and NIH Cancer Center Support Grants P30-CA016672 (University of Texas M. D. Anderson Cancer Center).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- 2.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, Wittschieben J, Wood RD, Greenberger JS. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat. Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ, Taylor S, Yoshimura M, Hickson ID, Bernhard EJ, McKenna WG. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–2993. doi: 10.1158/0008-5472.CAN-09-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, J OW. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J. Biol. Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- 11.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, J OW. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, J OW. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl. Acad. Sci. U S A. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger Iii AJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, J OW. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 16.Pillaire MJ, Selves J, Gordien K, Gourraud PA, Gentil C, Danjoux M, Do C, Negre V, Bieth A, Guimbaud R, Trouche D, Pasero P, Mechali M, Hoffmann JS, Cazaux C. A 'DNA replication' signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 17.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 19.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Mitaxov V, Waksman G. Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc. Natl. Acad. Sci. U S A. 1999;96:9491–9496. doi: 10.1073/pnas.96.17.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y, Eom SH, Wang J, Lee DS, Suh SW, Steitz TA. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 23.Clark JM, Beardsley GP. Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ide H, Kow YW, Wallace SS. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aller P, Rould MA, Hogg M, Wallace SS, Doublié S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. U S A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark JM. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedford E, Tabor S, Richardson CC. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. U S A. 1997;94:479–484. doi: 10.1073/pnas.94.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 30.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U S A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemée F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire M-J, Bieth A, Gentil C, Baker L, Martin A-L, Leduc C, Lam E, Magdeleine E, Filleron T, Oumouhou N, Jordan L, Kaina B, Seki M, Grimal F, Lacroix-Triki M, Thompson A, Roche H, Bourdon J-C, Wood RD, Hoffmann J-S, Cazaux C. POLQ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication and promotes genetic instability Proc. Natl. Acad. Sci. U S A. 2010 doi: 10.1073/pnas.0910759107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 33.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Dutta S, Doublié S, Bdour HM, Taylor JS, Ellenberger T. Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7. Nat. Struct. Mol. Biol. 2004;11:784–790. doi: 10.1038/nsmb792. [DOI] [PubMed] [Google Scholar]
- 35.Brieba LG, Eichman BF, Kokoska RJ, Doublié S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]