Abstract
Located in the Eurasian heartland, Central Asia has played a major role in both the early spread of modern humans out of Africa and the more recent settlements of differentiated populations across Eurasia. A detailed knowledge of the peopling in this vast region would therefore greatly improve our understanding of range expansions, colonizations and recurrent migrations, including the impact of the historical expansion of eastern nomadic groups that occurred in Central Asia. However, despite its presumable importance, little is known about the level and the distribution of genetic variation in this region. We genotyped 26 Indo-Iranian- and Turkic-speaking populations, belonging to six different ethnic groups, at 27 autosomal microsatellite loci. The analysis of genetic variation reveals that Central Asian diversity is mainly shaped by linguistic affiliation, with Turkic-speaking populations forming a cluster more closely related to East-Asian populations and Indo-Iranian speakers forming a cluster closer to Western Eurasians. The scattered position of Uzbeks across Turkic- and Indo-Iranian-speaking populations may reflect their origins from the union of different tribes. We propose that the complex genetic landscape of Central Asian populations results from the movements of eastern, Turkic-speaking groups during historical times, into a long-lasting group of settled populations, which may be represented nowadays by Tajiks and Turkmen. Contrary to what is generally thought, our results suggest that the recurrent expansions of eastern nomadic groups did not result in the complete replacement of local populations, but rather into partial admixture.
Keywords: admixture, Central Asia, ethnic groups, genetic diversity, microsatellites, population genetics
INTRODUCTION
The evolutionary history of modern humans has been characterized by range expansions, colonizations and recurrent migrations over the last 100 000 years.1 Some regions of the world that have served as natural corridors between landmasses are of particular importance in the history of human migrations. Central Asia is probably at the crossroads of such migration routes.1, 2 Located in the Eurasian heartland, it encompasses a vast territory, limited to the east by the Pamir and Tien-Shan mountains, to the west by the Caspian Sea, to the north by the Russian taiga and to the south by the Iranian deserts and Afghan mountains. The role of Central Asia in both the early spread of modern humans out of Africa and the more recent settlement of differentiated populations3 is not precisely known.4, 5, 6 For example, it remains unclear as to whether this region harbored a Paleolithic ‘maturation phase' of modern humans before giving rise to waves of migration, resulting in colonization of the Eurasian continent6 or whether it has served as a meeting place for previously differentiated Asian and European populations following their initial expansions.3, 7
Central Asia entered the historical records about 1300 , when Aryan tribes invaded the Iranian territory from what is nowadays Turkmenistan and established the Persian Empire in the seventh century .8 A branch of those, the Scythians, described in ancient Chinese texts and in Herodotus' Histories, as having European morphological traits and speaking Indo-Iranian languages, expanded north into the steppes. Thereafter, Central Asia was faced with multiple waves of Turkic migrations, although it is difficult to know precisely when these westward expansions began. Between the second and the first century , Huns brought the East-Asian anthropological phenotype to Central Asia.8 At the same period, the Chinese established a trade route (the Silk Road), which connected the Mediterranean Basin and Eastern Asia for more than 16 centuries. In the thirteenth century the Turco-Mongol Empire lead by Genghis Khan became the largest of all time, from Mongolia to the Black Sea. All these movements of populations resulted in a considerable ethnic diversity in Central Asia, with Indo-Iranian speakers living as sedentary agriculturalists and Turkic speakers mainly living as traditionally nomadic herders.
Taken together with the ancient peopling of Central Asia, this intricate demographic history shaped patterns of genetic variability in a complex manner. Most previous studies, based on classical markers,1 mitochondrial DNA (mtDNA)3, 9, 10, 11, 12, 13 or the non-recombining portion of the Y-chromosome (NRY),6, 14, 15, 16 have shown that genetic diversity in Central Asia is among the highest in Eurasia.3, 6, 15 NRY studies suggest an early settlement of Central Asia by modern humans, followed by subsequent colonization waves in Eurasia,6 whereas some mtDNA studies point to an admixed origin from previously differentiated Eastern and Western Eurasian populations.11 Furthermore, a recent analysis of mtDNA data suggests east-to-west expansions waves across Eurasia.14 However, inferring more accurately the impact of population movements, including the expansion of eastern nomadic groups, requires additional, fast-evolving molecular markers. Here we report on the first multilocus autosomal genetic survey of Central Asian populations. Twenty-six populations from six ethnic groups were genotyped at 27 autosomal unlinked microsatellite markers. We aimed to shed light on the genetic origins of Central Asian populations, and to investigate how the recurrent westward expansions of eastern nomadic groups during historical times have shaped the Central Asian genetic landscape.
MATERIALS AND METHODS
DNA samples
We sampled 767 men belonging to 26 populations from western Uzbekistan to eastern Kyrgyzstan (Table 1 and Figure 1) representative of the ethnological diversity in Central Asia: Tajiks, which are Indo-Iranian speakers (a branch of the Indo-European language family), and Kazakhs, Turkmen, Karakalpaks, Kyrgyz and Uzbeks, which are Turkic speakers (a branch of the Altaic language family). In two Uzbek populations from the Bukhara area (LUZa and LUZn), an extensive linguistic survey showed that individuals were bilingual, speaking both Tajik and Uzbek. As their home language was Tajik (an Indo-Iranian language), we further classified these two populations into the Indo-Iranian group for subsequent analyses. We collected individuals unrelated for at least two generations back in time. All individuals gave informed consent for their participation in this study. Total genomic DNA was isolated from blood samples by a standard salting out procedure17 followed by a phenol–chloroform extraction.18
Table 1. Description of the 26 Central Asian studied populations.
| Sampled populations (area) | Acronym | Location | Language family | Long. | Lat. | n |
|---|---|---|---|---|---|---|
| Tajiks (Samarkand) | TJA | Uzbekistan/Tajikistan border | Indo-Iranian | 39.54 | 66.89 | 31 |
| Tajiks (Samarkand) | TJU | Uzbekistan/Tajikistan border | Indo-Iranian | 39.50 | 67.27 | 29 |
| Tajiks (Ferghana) | TJR | Tajikistan/Kyrgyzstan border | Indo-Iranian | 40.36 | 71.28 | 29 |
| Tajiks (Ferghana) | TJK | Tajikistan/Kyrgyzstan border | Indo-Iranian | 40.25 | 71.87 | 26 |
| Tajiks (Gharm) | TJE | Northern Tajikistan | Indo-Iranian | 39.12 | 70.67 | 25 |
| Tajiks (Gharm) | TJN | Northern Tajikistan | Indo-Iranian | 38.09 | 68.81 | 24 |
| Tajiks (Gharm) | TJT | Northern Tajikistan | Indo-Iranian | 39.11 | 70.86 | 25 |
| Tajiks (Penjikent) | TDS | Uzbekistan/Tajikistan border | Indo-Iranian | 39.28 | 67.81 | 25 |
| Tajiks (Penjikent) | TDU | Uzbekistan/Tajikistan border | Indo-Iranian | 39.44 | 68.26 | 25 |
| Tajiks (Yagnobs from Dushanbe) | TJY | Western Tajikistan | Indo-Iranian | 38.57 | 68.78 | 25 |
| Uzbeks (Ferghana) | UZA | Uzbekistan/Kyrgyzstan border | Turkic | 40.77 | 72.31 | 25 |
| Uzbeks (Penjikent) | UZT | Northern Tajikistan | Turkic | 39.49 | 67.54 | 25 |
| Uzbeks (Bukhara) | LUZn | Central Uzbekistan | Indo-Iranian | 39.70 | 64.38 | 20 |
| Uzbeks (Bukhara) | LUZa | Central Uzbekistan | Indo-Iranian | 39.73 | 64.27 | 20 |
| Uzbeks (Karakalpakia) | UZB | Western Uzbekistan | Turkic | 43.04 | 58.84 | 35 |
| Karakalpaks (Qongrat from Karakalpakia) | KKK | Western Uzbekistan | Turkic | 43.77 | 59.02 | 45 |
| Karakalpaks (On Tört Uruw from Karakalpakia) | OTU | Western Uzbekistan | Turkic | 42.94 | 59.78 | 45 |
| Kazaks (Karakalpakia) | KAZ | Western Uzbekistan | Turkic | 43.04 | 58.84 | 49 |
| Kazaks (Bukhara) | LKZ | Central Uzbekistan | Turkic | 40.08 | 63.56 | 25 |
| Kyrgyz (Andijan) | KRA | Uzbekistan/Kyrgyzstan border | Turkic | 40.77 | 72.31 | 45 |
| Kyrgyz (Narin) | KRG | Eastern Kyrgyzstan | Turkic | 41.60 | 75.80 | 18 |
| Kyrgyz (Narin) | KRM | Eastern Kyrgyzstan | Turkic | 41.45 | 76.22 | 21 |
| Kyrgyz (Narin) | KRL | Eastern Kyrgyzstan | Turkic | 41.36 | 75.50 | 22 |
| Kyrgyz (Narin) | KRB | Eastern Kyrgyzstan | Turkic | 41.25 | 76.00 | 24 |
| Kyrgyz (Issyk Kul) | KRT | Eastern Kyrgyzstan | Turkic | 42.16 | 77.57 | 37 |
| Turkmen (Karakalpakia) | TUR | Western Uzbekistan | Turkic | 41.55 | 60.63 | 47 |
Abbreviations: Long., longitude; Lat., latitude; n, sample size.
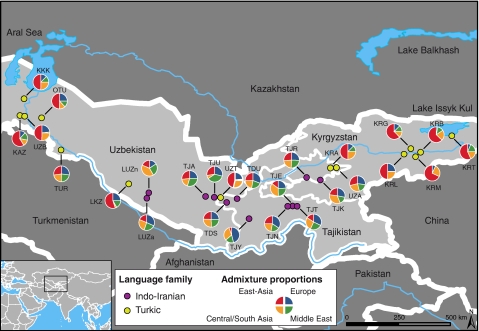
Figure 1.
Geographic location of the 26 Central Asian populations sampled. Linguistic affiliation as well as admixture proportions from putative parental origins (Central/South Asia, East Asia, Europe and Middle East) are also indicated. See Table 1 for acronyms.
Genotyping
We selected 27 microsatellite markers19 from the set of 377 markers used in the worldwide study by Rosenberg et al.20 The choice and description of markers, PCR and electrophoresis conditions are given in Ségurel et al.19 We further genotyped 20 individuals from the HGDP-CEPH Human Genome Diversity Cell Line Panel20, 21, 22 at the 27 microsatellite loci, in order to standardize the original Central Asian data presented here with the worldwide HGDP-CEPH data.
Data analyses
Genetic diversity
In each population and for each locus, we calculated the allelic richness (AR) using the rarefaction method proposed by El Mousadik et al23 with the software package FSTAT.24 Unbiased estimates of expected heterozygosity (He)25 were computed in each population for each locus with GENETIX.26 Both AR and He estimates were averaged over the loci in each population. We tested heterogeneity in both AR and He among populations using the Kruskal–Wallis test, with locus-specific estimates taken as replicate observations. Locus-specific AR and expected heterozygosity were also estimated for populations pooled into Indo-Iranian- and Turkic-speaking groups, and averaged over loci within groups. We tested between-group differences in both AR and He using the Wilcoxon's signed-rank test, with locus-specific estimates taken as replicate observations. We further estimated AR and He for each locus over the pooled data from Central Asia and over the pooled data for Central/South Asia, East Asia, Europe and the Middle East from the HGDP-CEPH Panel, and calculated the averages over loci within groups. We tested heterogeneity in both AR and He across the five groups of Eurasian populations using the Kruskal–Wallis test, taking locus-specific estimates as replicate observations. When significant differences among groups were found, we ran the Tukey's range test to find which group statistics were significantly different from one another. All statistical analyses were performed with the software package – JMP5.1 (SAS Institute Inc.).27
Genetic structure
Population differentiation (FST) was calculated overall and between pairs of Central Asian populations with GENEPOP 4.0.28 Exact tests of differentiation were performed with FSTAT,24 adjusting P-values with Bonferroni correction for multiple tests. We performed a correspondence analysis (CA) based on tables of allele counts using GENETIX.26 The population structure was also inferred by means of a hierarchical analysis of molecular variance (AMOVA),29 with populations pooled into ethnic or linguistic groups. For ethnic grouping, populations were pooled as Tajiks (TJA, TDS, TJT, TJK, TJR, TJN, TDU, TJE, TJY and TJU), Karakalpaks (KKK and OTU), Kazakhs (KAZ and LKZ), Kyrgyz (KRA, KRG, KRL, KRB, KRT and KRM), Uzbeks (UZA, UZB, LUZa, LUZn and UZT) and Turkmen (TUR). For linguistic grouping, populations were pooled as Indo-Iranian speakers (Tajiks and the two Uzbek populations LUZa and LUZn) and Turkic speakers (all other populations). These analyses were performed with ARLEQUIN 3.11.30 Isolation-by-distance (IBD) was tested with GENEPOP 4.0.28 We used PATHMATRIX31 to compute the matrix of effective geographical distances, based on a least-cost path algorithm. The least-cost distances, which account for the cost of the movement through the slopes in the landscape, were calculated from the digital elevation model GTOPO30 of the Earth Resources Observation and Science Center.
Clustering analyses
We performed a clustering analysis with STRUCTURE32 on the Central Asian populations together with all the Eurasian and African populations from the HGDP-CEPH Panel H952 corrected data set.33, 34 We used the latest version of STRUCTURE35 (version 2.3), which allows structure to be detected at lower levels of divergence than the original model. Each Markov chain was run for 106 steps, after a 105-step burn-in period. In each case, the results were checked to ensure consistency over 40 independent runs. Potential distinct modes among the 40 runs were identified using the Greedy algorithm implemented in CLUMP.36 We varied the hypothetical number of clusters (K) from 1 to 8 for all analyses. All chains were run using the F model for correlations of allele frequencies across clusters.37
Admixture analyses
The Central Asian genetic pool may be more than just the result of admixture from Eurasian populations, but we were nonetheless interested in investigating the potential origins of Central Asian populations among all Eurasian populations. We used LEADMIX38 to calculate maximum likelihood estimates (MLE) of the admixture proportions for each Central Asian population. We ran the program independently for each of them, considering four putative parental groups from the HGDP-CEPH Panel: Central/South Asia, East Asia, Europe and Middle East. For the Central/South Asian group, we chose a pool of Balochi (n=25) and Makrani (n=25) individuals, both populations being non-significantly differentiated (FST=−0.002; exact test P=0.34). We chose the Han Chinese (n=44) for the East-Asian parental group, and we further considered a pool of French (n=28), Bergamo (n=13) and Tuscan (n=21) individuals for the European group, these three populations being non-significantly differentiated (FST <−0.006; P>0.42). Last, we chose the Palestinians (n=46) for the Middle Eastern group.39
RESULTS
Genetic diversity
Average AR and expected heterozygosity for each of the 26 Central Asian populations and across regions are given in Table 2. We found a significant difference in AR (Kruskal–Wallis test, χ2=105.29, d.f.=25, P<0.0001) and in expected heterozygosity (Kruskal–Wallis test, χ2=67.98, d.f.=25, P<0.0001) among populations. We found no significant difference in AR between Indo-Iranian (AR=13.8) and Turkic speakers (AR=13.7, Wilcoxon signed-rank test, Z=−0.69, P=0.49), although the expected heterozygosity was significantly higher in Indo-Iranian as compared with Turkic speakers (He=0.818 and 0.787, respectively, Wilcoxon signed-rank test, Z=−4.55, P<0.0001). We found a significant difference in AR across Central Asia, Europe, Central/South Asia, Middle East and East Asia (Kruskal–Wallis test, K=36.46, d.f.=4, P<0.0001), as well as in expected heterozygosity (Kruskal–Wallis test, K=52.94, d.f.=4, P<0.0001). Yet, these differences were rather owing to a lower heterozygosity in East Asia and also slightly higher AR in Middle East (Tukey's test, P<0.0001 for both AR and He). Central Asia therefore showed neither higher nor lower diversity than the rest of Eurasia.
Table 2. Genetic diversity in the studied populations and in Eurasia.
| World Area | Population | AR | He |
|---|---|---|---|
| Central Asia | KAZ | 7.9 | 0.784 |
| Central Asia | KKK | 7.8 | 0.782 |
| Central Asia | KRA | 7.5 | 0.769 |
| Central Asia | KRB | 7.3 | 0.757 |
| Central Asia | KRG | 7.7 | 0.779 |
| Central Asia | KRL | 7.8 | 0.778 |
| Central Asia | KRM | 7.6 | 0.752 |
| Central Asia | KRT | 7.7 | 0.761 |
| Central Asia | LKZ | 7.8 | 0.778 |
| Central Asia | LUZa | 8.3 | 0.817 |
| Central Asia | LUZn | 8.6 | 0.821 |
| Central Asia | OTU | 8.0 | 0.784 |
| Central Asia | TDS | 7.7 | 0.784 |
| Central Asia | TDU | 7.9 | 0.805 |
| Central Asia | TJA | 8.0 | 0.806 |
| Central Asia | TJE | 8.4 | 0.814 |
| Central Asia | TJK | 8.6 | 0.820 |
| Central Asia | TJN | 8.4 | 0.811 |
| Central Asia | TJR | 8.6 | 0.812 |
| Central Asia | TJT | 8.5 | 0.812 |
| Central Asia | TJU | 8.5 | 0.811 |
| Central Asia | TJY | 7.9 | 0.799 |
| Central Asia | TUR | 8.5 | 0.812 |
| Central Asia | UZA | 9.0 | 0.817 |
| Central Asia | UZB | 8.5 | 0.774 |
| Central Asia | UZT | 8.4 | 0.795 |
| Central Asia (pooled populations) | 12.58 | 0.803 | |
| Central/South Asia | 12.66 | 0.819 | |
| East Asia | 11.4 | 0.705 | |
| Europe | 11.83 | 0.808 | |
| Middle East | 13.17 | 0.827 |
Abbreviations: AR, allelic richness; He, expected heterozygosity.
AR was calculated using a common sample size of n=13 diploid individuals for the Central Asian samples, and a common samples size of n=123 diploid individuals for the regional samples. These sample sizes correspond to the smallest number of genes sampled at a locus, including missing data.
Population differentiation
The 26 Central Asian populations were slightly but significantly differentiated (FST=0.015, CI99%=[0.011–0.018], P<0.01). Pairwise FST estimates ranged from −0.004 to 0.056, with 205 out of 325 pairs of populations (ie, 63.1%) being significantly differentiated after Bonferroni correction for multiple tests (see Supplementary Table 1). These significant estimates mainly corresponded to pairwise comparisons between one Turkic and one Indo-Iranian population, as well as to comparisons between two Indo-Iranian populations. The apportionment of genetic variation among linguistic or ethnic groups of populations (Table 3) showed that more than 98% of the total variation lay within populations (P<0.0001). Yet, both ethnicity and linguistic affiliation accounted significantly for the observed variation (FCT=0.007, P<0.0001 and FCT=0.011, P<0.0001, respectively). We found no evidence of IBD within each of Turkic and Indo-Iranian group of populations (P=0.363 and 0.772, respectively).
Table 3. AMOVA of the 26 Central Asian studied populations.
| Grouping | Source of variation | Percentage of variation | FST | FSC | FCT |
|---|---|---|---|---|---|
| Linguistic affiliation | Among groups | 1.09 | 0.010*** | ||
| Among populations within groups | 0.91 | 0.009*** | |||
| Within populations | 98.0 | 0.020*** | |||
| Ethnicity | Among groups | 0.69 | 0.007*** | ||
| Among populations within groups | 0.91 | 0.009*** | |||
| Within populations | 98.39 | 0.016*** |
***P<0.0001.
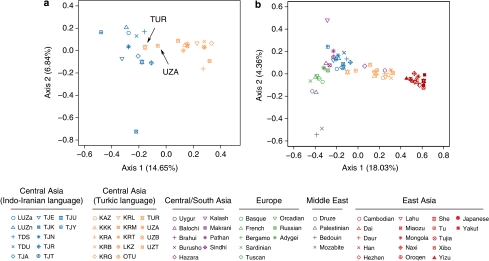
The CA based on the table of allele counts in Central Asia separated Turkic- and Indo-Iranian-speaking populations on the first axis (Figure 2a). The first two factorial components (FC) accounted for 20.5% of the total inertia. There were some exceptions though: two Turkic-speaking populations, TUR and UZA, were clearly clustered with Indo-Iranian-speaking populations. Interestingly, the Uzbek populations (LUZa, LUZn, UZA and UZT) showed a scattered pattern on the CA, which overlapped the Turkic-speaking and the Indo-Iranian-speaking groups of populations. The CA based on the table of allele counts in Eurasia placed Central Asian populations in an intermediate position between a group of European populations, a group of Middle Eastern populations, a group of Central/South Asian populations and a group of East-Asian populations (Figure 2b). The first two FC accounted for 22.4% of the total inertia. Turkic- and Indo-Iranian-speaking populations were separated on the first axis, with Turkic-speaking populations being closer to East-Asian populations and Indo-Iranian-speaking populations being closer to Central/South Asian, European and Middle Eastern populations. It is worth noting that Central Asian and Central/South Asian populations were more scattered than any other group of populations in Eurasia (Figure 2b). Interestingly, the Hazaras from Pakistan, who claim to be direct male-line descendants of Genghis Khan,40, 41 as well as the Uygurs, clustered together with the Turkic-speaking populations of Central Asia.
Figure 2.
Correspondence analysis (CA) based on the table of allele counts in Central Asia (a). The first two factorial components (FC) are represented, and their relative contribution to the total inertia are indicated. Colors indicate language affiliation; blue: Indo-Iranian speakers; orange: Turkic speakers. CA based on the table of allele counts in Eurasian populations (b). Colors represent major geographic regions; green: Europe; gray: Middle East; purple: Central/South Asia; red: East Asia.
Cluster analyses
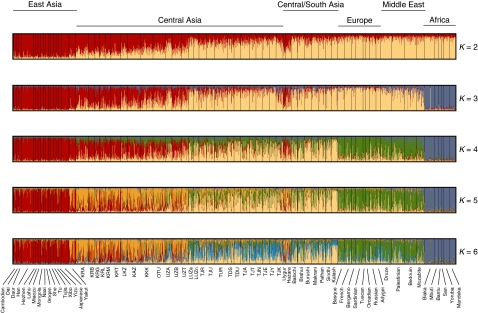
Analyzing the Eurasian plus the African populations altogether, we found that the highest average posterior probability of the data (D), across 40 runs, was obtained for K=7 putative clusters, with Log[P(K=7∣D)]=−167 565.4 (SD=22.8), although the average posterior probability for K=6 was only slightly lower, with Log[P(K=6∣D)]=−167 653.8 (SD=10.6). The symmetric similarity coefficients computed with CLUMPP across independent runs were all larger than 0.99 for K varying from 2 to 5, and larger than 0.87 for K=6, which suggests the absence of genuine multimodality across runs. As seen in Figure 3, at K=2, we observed a clear east–west cline. Central Asia seemed to be intermediate between one cluster made of European, Middle Eastern, Central/South Asian and African populations on the one hand and one cluster of East-Asian populations on the other hand, which is consistent with the CA (Figure 2b). There was no individual assigned exclusively to one cluster, with Turkic-speaking individuals having a higher membership coefficient in the East-Asian cluster and Indo-Iranian-speaking individuals having a higher membership coefficient in the cluster made of Europe, Middle East, Central/South Asia and Africa. At K=3, the six African populations clustered together. At K=4, the European and Middle Eastern populations clustered together, with Central/South Asian and Central Asian populations (mostly Indo-Iranian speakers) showing a small contribution from this European/Middle Eastern cluster (represented in green in Figure 3). At K=5, the Turkic-speaking populations from Central Asia showed a large contribution from a fifth cluster (in orange in Figure 3). At K=6, the Indo-Iranian-speaking populations from Central Asia show a large contribution from a sixth cluster (in light blue in Figure 3). The two latter clusters were found almost exclusively in Central Asian populations. Most Turkic-speaking populations showed a contribution from the East-Asian cluster (in red), and most Indo-Iranian populations showed a contribution from Europe and Middle East (in green). It is worth noting that Uygur and Hazara populations showed the same pattern as the Turkic-speaking populations from Central Asia. At K=7, all Eurasian populations (but mostly Turkic-speaking populations) had a variable proportion of the new component. Yet, no run at K=7 resulted in a new cluster of populations, as compared with K=6, which is the reason why the output for K=7 is not represented in Figure 3.
Figure 3.
Population structure inferred from microsatellite data using the software package STRUCTURE. K represents the number of putative clusters. Each individual is represented by a vertical line, divided into up to K colored segments, each of which represents the individual's estimated membership fraction to that cluster. Each output represents the matrix of membership coefficients averaged over 40 independent runs with CLUMPP. The data consisted in 767 individuals from 26 Central Asian populations genotyped at 27 microsatellite loci, plus 869 individuals from 44 African and Eurasian populations from the HGDP-CEPH Human Genome Diversity Cell Line Panel. See Table 1 for acronyms.
Admixture analyses
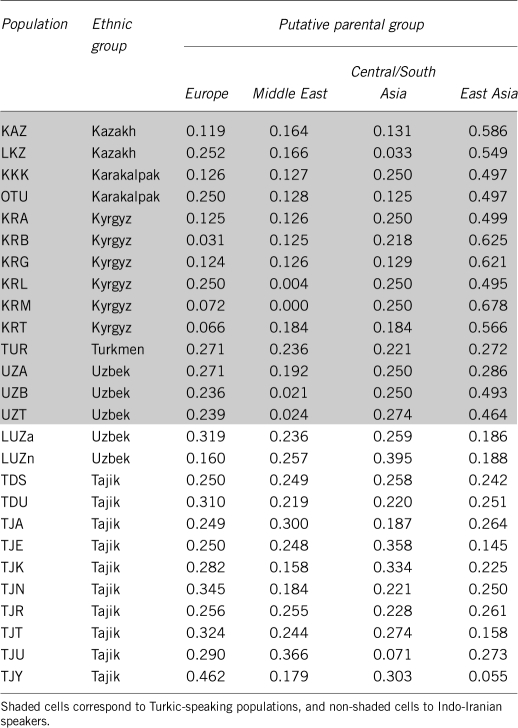
The MLE of admixture proportions obtained with LEADMIX for each Central Asian population are given in Figure 1 and Table 4. Most Turkic-speaking populations had a large East-Asian ancestral contribution, which represented in general 46.4%, or more, of the total contribution. There were two notable exceptions, although, with the Turkmen (TUR) and one Uzbek population (UZA) showing a lower contribution from East-Asian populations (27.2 and 28.6%, respectively). Indo-Iranian-speaking populations had a large western Eurasian contribution (Central/South Asia, Europe and Middle East), which represented 72.7–94.5% of the total contribution, although the relative contributions from these three parental groups differ across Indo-Iranian-speaking populations. It is worth noting that, in general, many geographically close populations that speak different languages showed contrasted admixture proportions (see, eg, UZT and TJU in Table 4), which supports the idea that language is a major determinant of population differentiation in Central Asia.
Table 4. Maximum-likelihood estimates of admixture proportions in the 26 Central Asian populations.
DISCUSSION
Central Asia in the heartland of Eurasia
We found a high level of autosomal genetic diversity in Central Asia, consistent with previous observations,3, 16 and similar in extent to other major regions in Eurasia (Table 2). Population differentiation among Central Asian populations was similar, or even stronger, than that measured among populations within other regions in Eurasia: the pairwise FST estimates ranged from −0.004 to 0.056 in Central Asia, a range which should be compared with that found in the European group (−0.011; 0.015), the Middle Eastern group (0.008; 0.021), the Central/South Asian group (−0.002; 0.062) and in the East-Asian group (−0.011; 0.046), based on the same set of 27 microsatellite loci as we used in our study. This pattern is also apparent in the CA (Figure 2b), where Central Asian and Central/South Asian populations were more scattered than each of the East-Asian, European and Middle Eastern groups, which suggests a higher diversification within Central Asia and Central/South Asia. Most importantly, the observed diversity was mainly owing to the differentiation into two main groups of populations (Figure 3): on the one hand, Indo-Iranian-speaking populations (which include Tajiks and three Uzbek populations) that are genetically closer to populations from Western Eurasia, and on the other hand, Turkic-speaking populations (which include Karakalpaks, Kazakhs, Kyrgyz and two other Uzbek populations) that are closer to Eastern Asian populations (with the exception of the Turkmen). This pattern was also apparent in the CA (Figure 2b), and consistent with the significant differentiation of almost all pairwise comparisons between an Indo-Iranian- and a Turkic-speaking population (Supplementary Table 1).
Although several studies have shown that geography is, in general, a better predictor of genetic differentiation than ethnicity and linguistics,42, 43 language affiliation appears as the most important factor explaining the distribution of genetic diversity in Central Asia (Table 3). We found indeed that, although most (98%) of the variation lay within Central Asian populations (P<0.0001), a significant part of the total variation (1.09% P<0.0001) lay among linguistic groups, which provides an estimate of differentiation among groups equal to FCT=0.011. For comparison purpose, the differentiation among Central/South Asia, East Asia, Europe and Middle East was found to be FCT=0.044, with 94.1% of the total variation found within populations (P< 0.0001) and 4.4% found among groups, based on the same set of 27 microsatellite loci as we used in our study. We found no evidence of a correlation between geography and genetics within each of the Indo-Iranian or Turkic groups of Central Asian populations. For the Turkic-speaking populations, this may be explained by their recent arrival in the region and/or their nomadic lifestyle. However, more striking is the fact that no geographic pattern of genetic variation was found among sedentary Indo-Iranian speakers either.
Putative origins of Indo-Iranian- and Turkic-speaking populations
The clustering analysis showed that most individuals from the Indo-Iranian-speaking populations had large membership coefficients into two clusters (light blue and beige in Figure 3) that were found mostly in these populations. Altogether, the significant pairwise FST estimates between almost all pairs of Indo-Iranian-speaking populations (Supplementary Table 1), the high level of diversity across Indo-Iranian populations (Table 2) and the variable level of admixture from the putative parental populations (Table 4) seem consistent with the premise that Indo-Iranian speakers are long-term settled populations in the area. This latter hypothesis is strongly supported by archeological evidence.44 Conversely, we found a lower genetic differentiation among Turkic-speaking populations despite their wide geographic distribution (Figure 1), which suggests a more recent common origin of these populations as compared with Indo-Iranian-speaking populations, in consistence with historical records.
Our study further shed some light on the origins of the Turkic-speaking populations in Central Asia. The clustering analyses indeed showed that most individuals from the Turkic-speaking populations had large membership coefficients into one Central Asian cluster (in orange in Figure 3) and smaller membership coefficients into the East-Asian cluster (in red in Figure 3), thus confirming the result of Li et al45 based on a small central Asian cluster for Uygur, Kazakh and Khanty. This pattern likely reflects the existence of an ancestral group of Turkic speakers (orange cluster in Figure 3), which popular Turkic culture considers as originating from the Altai region. The East-Asian ancestry of Turkic-speaking populations (red cluster in Figure 3) may then correspond to the westward expansions of nomadic groups form East Asia during historical times.
The Westernized view of westward invasions usually emphasizes the extreme violence and cruelty of the hordes led by Attila the Hun ( 406–453), or that from the Mongolian empire led by Genghis Khan. However, our results somehow challenge this view and rather suggest that these more recent expansions did not lead to the massacre and complete replacement of the locally settled populations, but rather to partial admixture. We found almost no eastern ancestry in Indo-Iranian-speaking populations (see Figure 3), which suggests that the group of people from which the current-day Tajik and Turkmen populations would be the descendants did not suffer from the westward expansions of eastern nomadic groups. This is consistent with Zerjal et al's study,16 which showed the absence of the ‘Genghis Khan lineage' in the Tajik and Turkmen populations they studied. Furthermore, the present finding that the partial admixture with eastern nomadic groups concerned almost exclusively the Turkic-speaking populations is consistent with the fact that Turks and Mongols share cultural traditions and lifestyle, which may have facilitated inter-groups marriages.
Our study also contradicts the claim that these westward invasions resulted in founder effects.16 The high level of autosomal diversity observed in all Turkic-speaking populations (Table 2) contrasts indeed with the low level of Y-chromosome diversity found in some populations of the region.10, 16 Our recent studies based on the analysis of uni-parental markers in Central Asia already showed that the low-level Y-chromosome diversity is only found in the Turkic-speaking group,46 which may therefore be explained by the social organization of Turkic-speaking populations, which is based on patrilineal descent groups.10, 18
Overall, our results are partly consistent with Comas et al's hypothesis11 that Central Asia has been a contact zone between two differentiated groups. Our study suggests that one of these groups is a long-lasting group of settled populations, now represented by Tajiks and Turkmen, although the origin of this group is difficult to infer; the second of these groups is likely to have a more recent origin, resulting from the movements of eastern nomadic Turkic-speaking groups. Interestingly, we found almost no African ancestry in the genetic pool of Central Asian population from clustering analyses (Figure 3). Yet, with the same level of clustering, we found no African ancestry either in Europe or in East-Asia. Further work is therefore required to infer the more ancient peopling of Central Asia, after the spread of modern humans out of Africa.
We found that the Uzbek populations were scattered across Turkic- and Indo-Iranian-speaking populations (Figure 2b). Some Uzbek populations (LUZa, LUZn, UZA) were closer to Indo-Iranian-speaking populations, whereas other populations (UZB, UZT) clearly clustered with Turkic-speaking populations. This is consistent with the fact that Uzbek populations include the seventeenth century Uzbeks, which were nomadic herders before they sedentarized around the sixteenth century,10 and the former Chagatai Turk groups who were already settled in Uzbekistan.47 Uzbeks therefore result from the union of different tribes, some of recent origin clustering with Turkic-speaking populations and some tracing back to Chagatai Turks who were strongly admixed with Iranian dwellers of Central Asia.
Evidence for linguistic replacements
We found two presumable cases of linguistic replacements in Central Asia. The Turkic-speaking populations, TUR (Turkmen) and UZA (Uzbek) were found to cluster together with Indo-Iranian-speaking populations (Figure 2). The Uzbek population UZA, a currently Turkic-speaking population, is indeed genetically more similar to Indo-Iranian speakers, which suggests a linguistic shift in this population. Concerning the Turkmen, their genetic similarity with Tajiks (see also Table 4) is consistent with the hypothesis that they may be the present-day descendants of populations established over long periods of time. The indigenous cultural history of the Turkmen in Turkmenistan can indeed be dated back to 10 000 years and similarities between the cultures and technologies found in the archaeological record suggest that this region has been continually occupied since 6000 . A recent linguistic replacement in the TUR population would then explain the observed pattern of a Turkic-speaking population clustering with Indo-Iranian speakers.
A Central Asian origin of the Hazaras?
Our study confirms the results of Li et al's study48 that cluster the Hazara population with Central Asian populations, rather than Mongolian populations, which is consistent with ethnological studies.49 Our results further extend these findings, as we show that the Hazaras are closer to Turkic-speaking populations from Central Asia than to East-Asian or Indo-Iranian populations.
Acknowledgments
We are indebted to everyone who volunteered to participate to this study. We also thank R Leblois and P Verdu for insightful discussions on previous versions of this paper, H Cann for providing CEPH samples, the Service de Systématique Moléculaire (SSM) at the Museum national d'Histoire naturelle (MNHN) for making facilities available and J.A. Godoy for technical assistance. We are very grateful to CESGA (Supercomputational Centre of Galicia) and to the Computational Biology Service Unit from the Museum national d'Histoire naturelle (MNHN – CNRS UMS 2700) where the computational analyses were performed. This work was supported by the Centre National de la Recherche Scientifique (CNRS) ATIP program (to EH), by the CNRS interdisciplinary program ‘Origines de l'Homme du Langage et des Langues' (OHLL), the European Science Foundation (ESF) EUROCORES program ‘The Origin of Man, Language and Languages' (OMLL) and the ANR Grant ‘NUTGENEVOL' (07-BLAN-0064).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, NJ: Princeton University Press; 1994. [Google Scholar]
- Nei M, Roychoudhury AK. Evolutionary relationships of human populations on a global scale. Mol Biol Evol. 1993;10:927–943. doi: 10.1093/oxfordjournals.molbev.a040059. [DOI] [PubMed] [Google Scholar]
- Comas D, Calafell F, Mateu E, et al. Trading genes along the silk road: mtDNA sequences and the origin of central Asian populations. Am J Hum Genet. 1998;63:1824–1838. doi: 10.1086/302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Deepa E, Vishwanathan H, Stoneking M. Genetic evidence for the demic diffusion of agriculture to India. Science. 2004;304:1125. doi: 10.1126/science.1095819. [DOI] [PubMed] [Google Scholar]
- Karafet T, Xu LP, Du RF, et al. Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet. 2001;69:615–628. doi: 10.1086/323299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian Heartland: a continental perspective on Y-chromosome diversity. Natl Acad Sci USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles G. The peoples of Asia. Weidenfeld and Nicolson: London; 1977. [Google Scholar]
- Gumilyov LN. Ancient Turks. Moscow: Institute of Ethnology and Anthropology of the Academy of Sciences of USSR; 1967. [Google Scholar]
- Chaix R, Austerlitz F, Khegay T, et al. The genetic or mythical ancestry of descent groups: lessons from the Y chromosome. Am J Hum Genet. 2004;75:1113–1116. doi: 10.1086/425938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix R, Quintana-Murci L, Hegay T, et al. From social to genetic structures in Central Asia. Curr Biol. 2007;17:43–48. doi: 10.1016/j.cub.2006.10.058. [DOI] [PubMed] [Google Scholar]
- Comas D, Plaza S, Wells RS, et al. Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages. Eur J Hum Genet. 2004;12:495–504. doi: 10.1038/sj.ejhg.5201160. [DOI] [PubMed] [Google Scholar]
- Lalueza-Fox C, Sampietro ML, Gilbert MTP, et al. Unravelling migrations in the steppe: mitochondrial DNA sequences from ancient Central Asians. Proc R Soc LondSer B. 2004;271:941–947. doi: 10.1098/rspb.2004.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lezaun A, Calafell F, Comas D, et al. Sex-specific migration patterns in central Asian populations, revealed by analysis of Y-chromosome short tandem repeats and mtDNA. Am J Hum Genet. 1999;65:208–219. doi: 10.1086/302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix R, Austerlitz F, Hegay T, Quintana-Murci L, Heyer E. Genetic traces of east-to-west human expansion waves in Eurasia. Am J Phys Anthropol. 2008;136:309–317. doi: 10.1002/ajpa.20813. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Karafet TM, Redd AJ, et al. Hierarchical patterns of global human Y-chromosome diversity. Mol Biol Evol. 2001;18:1189–1203. doi: 10.1093/oxfordjournals.molbev.a003906. [DOI] [PubMed] [Google Scholar]
- Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C. A genetic landscape reshaped by recent events: Y-chromosomal insights into Central Asia. Am J Hum Genet. 2002;71:466–482. doi: 10.1086/342096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, et al. Current Protocols in Molecular Biology. John Wiley & Sons: New York; 2001. [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. A Laboratory Manual. New York: Cold Spring Harbor; 1982. [Google Scholar]
- Ségurel L, Martinez-Cruz B, Quintana-Murci L, et al. Sex-specific genetic structure and social organization in Central Asia: insights from a multi-locus study. PLoS Genet. 2008;4:e1000200. doi: 10.1371/journal.pgen.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElMousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree Argania spinosa (L) Skeels endemic to Morocco. Theoret Appl Genet. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. 1996.
- JMP Statistics and Graphics Guide, Version 5.1. Cary, NC: SAS Institute Inc.; 2003. [Google Scholar]
- Rousset F. GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Res. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes – application to human mitochondrial-DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval LG, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Ray N. PATHMATRIX: a geographical information system tool to compute effective distances among samples. Mol Ecol Notes. 2005;5:177–180. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH human genome diversity cell line panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Gonzalez-Quevedo C, et al. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2006;2:2052–2061. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Res. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL. Maximum-likelihood estimation of admixture proportions from genetic data. Genetics. 2003;164:747–765. doi: 10.1093/genetics/164.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle EMS, Landry PA, Barbujani G. Origins and evolution of the Europeans' genome: evidence from multiple microsatellite loci. Proc R Soc Ser B. 2006;273:1595–1602. doi: 10.1098/rspb.2006.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar R, Ayub Q, Mohyuddin A, et al. Y-chromosomal DNA variation in Pakistan. Am J Hum Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Xue YL, Bertorelle G, et al. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Gonzalez-Neira A, et al. Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- Manica A, Prugnolle F, Balloux F. Geography is a better determinant of human genetic differentiation than ethnicity. Hum Genet. 2005;118:366–371. doi: 10.1007/s00439-005-0039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet F. La Néolithisation en Asie Centrale: un état de la question. Paléorient. 1999;24:27–48. [Google Scholar]
- Li H, Cho K, Kidd JR, Kidd KK. Genetic landscape of Eurasia and ‘admixture' in Uyghurs. Am J Hum Genet. 2009;85:934–937. doi: 10.1016/j.ajhg.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Balaresque P, Jobling MA, et al. Genetic diversity and the emergence of ethnic groups in Central Asia. BMC Genet. 2009;10:8. doi: 10.1186/1471-2156-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek S. A History of Inner Asia. Cambridge, MA: Cambridge University Press; 2000. [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Dupaigne B.L'artisanat Hazârain: Paysage du centre de l'Afghanistan – Paysages naturels, paysages culturels. CEREDAF: Paris; 2010212–222. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.