Abstract
The Philippines exhibits a rich diversity of people, languages, and culture, including so-called ‘Negrito' groups that have for long fascinated anthropologists, yet little is known about their genetic diversity. We report here, a survey of Y-chromosome variation in 390 individuals from 16 Filipino ethnolinguistic groups, including six Negrito groups, from across the archipelago. We find extreme diversity in the Y-chromosome lineages of Filipino groups with heterogeneity seen in both Negrito and non-Negrito groups, which does not support a simple dichotomy of Filipino groups as Negrito vs non-Negrito. Filipino non-recombining region of the human Y chromosome lineages reflect a chronology that extends from after the initial colonization of the Asia-Pacific region, to the time frame of the Austronesian expansion. Filipino groups appear to have diverse genetic affinities with different populations in the Asia-Pacific region. In particular, some Negrito groups are associated with indigenous Australians, with a potential time for the association ranging from the initial colonization of the region to more recent (after colonization) times. Overall, our results indicate extensive heterogeneity contributing to a complex genetic history for Filipino groups, with varying roles for migrations from outside the Philippines, genetic drift, and admixture among neighboring groups.
Keywords: Y-chromosome, Filipino, Negrito, heterogeneity, genetic affinity
Introduction
The Philippines figures prominently in two significant demographic events in the Asia-Pacific region. The first was the initial colonization of Sunda and Sahul around 40–50 thousand years ago (KYA)1 to which the human fossil remains from Tabon Cave, Palawan in the Philippines, with estimated ages ranging from 14 to 58 thousand years (KY),2 may be associated.3 The second was the Austronesian expansion, which is proposed to have originated in Taiwan around 5–6 KYA and spread first to the Philippines before continuing to Southeast Asia, Near and Remote Oceania, and Madagascar.4 The impact of the Austronesian expansion is evidenced by the fact that all Philippine languages today are Austronesian.5 In addition, a popularly accepted ‘waves of migration theory' suggests that the origins and diversity of people in the Philippines was the result of several migrations into the archipelago during the periods of low water levels, when there existed land bridges with the Sunda shelf via Palawan and the Sulu Archipelago, and with Celebes via Mindanao.6, 7
Currently, over 170 Filipino ethnolinguistic (FE) groups are recognized on the basis of language and culture.5 Of these FE groups, anthropologists have traditionally been most interested in those with short stature, frizzy hair, and dark skin color, historically defined as ‘Negrito' groups.8 There are more than 20 FE Negrito (FEN) groups in the Philippines.5 It has been proposed that Negrito groups represent a separate migration out of Africa.9 Although all FEN groups currently speak Austronesian languages, it has been proposed that they originally spoke non-Austronesian languages, which were subsequently replaced by Austronesian languages.10 This view, along with their physical features and hunter-gatherer mode of subsistence, has resulted in the historical depiction of the FEN groups as the aboriginal, first occupants of the archipelago, and as such, the popular distinction of FEN vs non-Negrito FE (FEnN) groups has been perpetuated.7
To date, no comprehensive genetic diversity study of FE groups has been presented. A mitochondrial DNA (mtDNA) study on Philippine populations showed further support for the ‘out of Taiwan' hypothesis for the Austronesian expansion, as well as identifying rare mtDNA haplotypes; however, samples were not from specific FE groups, but from urban populations, and from Filipinos living in Taiwan.11 Early studies have inferred genetic differences between some FEN groups (the Aeta of Bataan, Aeta of Zambales and the Mamanwa)12, 13 and possible affinities with populations, such as indigenous Australians and Papuan groups.12 A recent genome-wide study on Asian populations included six FEN groups, but only one other FEnN group, and three urban populations.14
We, therefore, report a survey of genetic variation in the non-recombining region of the human Y chromosome (NRY) of 16 FE groups (six FEN and 10 FEnN groups). Our results indicate that, a simple division of FEN vs FEnN groups is not supported by the NRY evidence. Instead, FE groups, in general, demonstrate extensive heterogeneity and diverse genetic affinities with their geographic neighbors, suggesting a complex history, reflecting varying roles of migration from outside the Philippines, genetic drift, and admixture among FE groups and with other Asia-Pacific groups.
Materials and methods
Population samples
DNA samples were collected from 390 unrelated males from 16 FE groups, representing eight Philippine geographical regions and three language groupings (Figure 1, Supplementary Text). Population sampling was conducted independently by the DNA Analysis Laboratory, Natural Sciences Research Institute, University of the Philippines (UP-NSRI-DAL) and the Max Planck Institute for Evolutionary Anthropology (MPI-EVA), and the sample collections of both institutions were combined to form the data set in this study (Supplementary Text). Human population sampling procedures followed the guidelines outlined by the Philippine National Bioethics Committee on Scientific Research, the National Commission on Indigenous Peoples (NCIP) and the MPI-EVA. Free, prior and informed consent for sample collection was obtained at the level of the community and the individual.
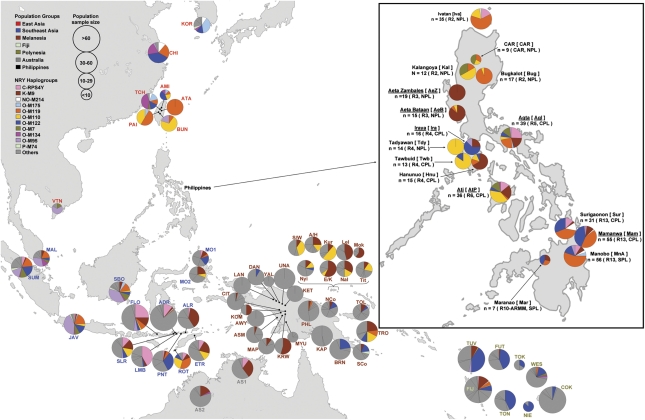
Figure 1.
Distribution of Y-SNP haplogroups observed among Filipino groups and the reference data set. The figure shows the frequency and geographical distribution across the Asia-Pacific region of the 11 Y-SNP haplogroups observed among Filipino groups. Pie chart size represents population sample size. Colors distinguish haplogroups within pie charts, with pie chart sections representing haplogroup frequencies. Population groups (East Asian, Southeast Asia, Melanesian, Fiji, Polynesia, Australia, and the Philippines) each have a common color for their three-letter population codes. For the Philippines, labels include language group name, [population code], population sample size ‘n'. Labels in parenthesis indicate geographical region (R#, CAR – Cordillera Administrative Region, ARMM – Autonomous Region of Muslim Mindanao) and language classification: northern Philippine language (NPL), central Philippine language (CPL) or southern Philippine language (SPL) based on the Ethnologue.5 Underlined Filipino group names and codes indicate Negrito groups. Population codes for the reference data set are enumerated in Supplementary Text.
DNA typing
DNA samples were extracted from whole-blood or oral samples (buccal swabs and/or saliva) using commercially available (FTA Gene Guard system (Whatman Inc., Springfield Mill, Maidstone, Kent, UK), QIAamp DNA Blood Mini kit (QIAGEN Inc., Valencia, CA, USA)) and published protocols.15 DNA samples were typed for 48 NRY binary markers (Y-SNPs) (Supplementary Figure 1). A set of Y-SNP multiplexes previously described16, 17 were typed using the ABI Prism SNaPshot multiplex kit (Applied Biosystems, Foster, CA, USA) (Supplementary Text, Supplementary Table 1). Another set of multiplexes were typed using a single-base extension (SBE) assay described elsewhere18 (Supplementary Text, Supplementary Table 1). Y-chromosome microsatellite/short tandem repeat (Y-STR) markers were typed using the PowerPlex Y system (Promega Corporation, Madison, WI, USA) following manufacturer's instructions (Supplementary Text). For Y-SNP haplogroup nomenclature, the capital letter (major clade) – terminal mutation (NRY binary marker) naming system19 was adopted.
Data analyses
For population comparisons, a reference data set was assembled from previously published works,20, 21, 22, 23, 24 composed of 1756 males from 60 groups representing five Asia-Pacific regions (Figure 1). Reference data set population codes and measures to ensure compatibility with the Filipino data set are discussed in Supplementary Text.
The Arlequin software version 3.1125 (http://cmpg.unibe.ch/software/arlequin3) was used to calculate genetic diversity indices: Haplogroup diversity (HgD), Haplotype diversity (HtD), haplotype mean pairwise difference (MPD) and Analysis Of Molecular Variance (AMOVA). AMOVA analyses were performed using different groupings (geography, language classification, FEN vs FenN, and all groups collectively (Figure 1)). Mann–Whitney U tests and correspondence analyses (CA) were performed using the STATISTICA software package.26 Network analyses were performed using Network version 4.510 and Network Publisher version 1.1.0.6 (http://fluxus-engineering.com). The BATWING program27 (http://www.mas.ncl.ac.uk/~nijw/) was used to estimate haplogroup coalescent times (Time since the Most Recent Common Ancestor –(TMRCA)). The IM program28 (http://genfaculty.rutgers.edu/hey/software) was used to estimate divergence times and migration rates. Details for Network analyses, estimation of coalescent times, divergence times, and migration rates are given in Supplementary Text.
Results
NRY diversity and genetic relationships of Filipino ethnolinguistic groups
Eleven Y-SNP haplogroups were observed among 390 unrelated males from 16 FE groups (Figure 1). Five major haplogroups (C-RPS4Y, K-M9, O-M119, O-M110, and O-M122) had frequencies >5% and were present among a greater number of FE groups than minor haplogroups (NO-M214, O-M175, O-M7, O-M95, O-M134, and P-M74) with <5% frequency (Table 1, Figure 1). In general, there is a patchy distribution of all haplogroups (Figure 1), such that even the most frequent haplogroup O-M119, observed among northern, central, and southern FE groups, was present in only 12 out of 16 FE groups. This distribution may be the result of genetic drift, which is detectable with the NRY because of its low effective population size.
Table 1. Frequency of Y-SNP haplogroups observed among Filipino groups.
| Filipino groups | Sample size | C-RPS4Y | K-M9 | NO-M214 | O-M175 | O-M119 | O-M110 | O-M122 | O-M7 | O-M134 | O-M95 | P-M74 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negrito | 180 | 0.089 | 0.322 | 0.006 | 0.006 | 0.189 | 0.094 | 0.211 | 0.056 | 0.006 | 0.022 | |
| Aeta Z | 19 | 1.00 | ||||||||||
| Aeta B | 15 | 0.87 | 0.07 | 0.07 | ||||||||
| Agta | 39 | 0.26 | 0.21 | 0.03 | 0.23 | 0.05 | 0.08 | 0.13 | 0.03 | |||
| Iraya | 16 | 0.25 | 0.63 | 0.13 | ||||||||
| Ati | 36 | 0.14 | 0.22 | 0.03 | 0.39 | 0.06 | 0.14 | 0.03 | ||||
| Mamanwa | 55 | 0.02 | 0.11 | 0.02 | 0.42 | 0.42 | 0.02 | |||||
| Non-Negrito | 210 | 0.071 | 0.081 | 0.029 | 0.014 | 0.433 | 0.200 | 0.114 | 0.038 | 0.014 | 0.005 | |
| Ivatan | 35 | 0.17 | 0.63 | 0.17 | 0.03 | |||||||
| CAR | 9 | 0.33 | 0.22 | 0.44 | ||||||||
| Bugkalot | 17 | 0.94 | 0.06 | |||||||||
| Kalangoya | 12 | 0.17 | 0.50 | 0.33 | ||||||||
| Tadyawan | 14 | 1.00 | ||||||||||
| Tawbuid | 13 | 0.85 | 0.15 | |||||||||
| Hanunuo | 15 | 0.67 | 0.13 | 0.13 | 0.07 | |||||||
| Surigaonon | 32 | 0.13 | 0.03 | 0.13 | 0.41 | 0.31 | ||||||
| Manobo | 56 | 0.09 | 0.07 | 0.04 | 0.05 | 0.55 | 0.14 | 0.05 | ||||
| Maranao | 7 | 0.29 | 0.29 | 0.43 | ||||||||
| Philippines | 390 | 0.079 | 0.192 | 0.018 | 0.010 | 0.321 | 0.151 | 0.159 | 0.046 | 0.003 | 0.018 | 0.003 |
Abbreviation: CAR, Cordillera Administrative Region.
For genetic diversity indices (Table 2), HgD ranged from 0 (in the Aeta of Zambales and the Tadyawan, who were fixed for haplogroups K-M9 and O-M110, respectively) to 0.83. Y-STR diversity indices such as HtD also ranged widely, from 0.59 (in the Tadyawan) to 0.97 (Ivatan, CAR, and Surigaonon), and MPD ranged from 1.40 (again in the Tadyawan) to 4.63 (Agta). Extremely low HgD values were observed for some FEN (Aeta of Zambales and Bataan) and FEnN groups, (Bugkalot, Tadyawan, and Tawbuid) further suggesting genetic drift owing to small population sizes. The genetic diversity indices of FEN groups did not significantly differ from FEnN groups, based on Mann–Whitney U tests (HgD: Z=0.52, P=0.61; HtD: Z=−0.32, P=0.75; MPD: Z=1.10, P=0.27), implying no distinction between FEN and FEnN groups based on NRY diversity.
Table 2. Genetic diversity indices estimated for Filipino groups.
| Filipino groups | nPop | nHg | HgD | SE | nHt | HtD | SE | MPD | SE |
|---|---|---|---|---|---|---|---|---|---|
| Negrito | 180 | 10 | 0.80 | 0.01 | 70 | 0.98 | 0.00 | 4.34 | 2.16 |
| Aeta Z | 19 | 1 | 0.00 | 0.00 | 10 | 0.91 | 0.04 | 2.78 | 1.54 |
| Aeta B | 15 | 3 | 0.28 | 0.14 | 8 | 0.79 | 0.10 | 3.75 | 2.00 |
| Agta | 39 | 8 | 0.83 | 0.03 | 19 | 0.95 | 0.02 | 4.63 | 2.32 |
| Iraya | 16 | 3 | 0.57 | 0.11 | 5 | 0.65 | 0.11 | 2.34 | 1.35 |
| Ati | 36 | 7 | 0.78 | 0.04 | 12 | 0.83 | 0.05 | 3.61 | 1.87 |
| Mamanwa | 55 | 6 | 0.65 | 0.04 | 23 | 0.94 | 0.01 | 3.58 | 1.85 |
| Non-Negrito | 210 | 10 | 0.75 | 0.02 | 93 | 0.98 | 0.00 | 3.69 | 1.88 |
| Ivatan | 35 | 4 | 0.56 | 0.08 | 24 | 0.97 | 0.01 | 3.34 | 1.75 |
| CAR | 9 | 3 | 0.72 | 0.10 | 8 | 0.97 | 0.06 | 3.75 | 2.09 |
| Bugkalot | 17 | 2 | 0.12 | 0.10 | 8 | 0.82 | 0.08 | 2.08 | 1.22 |
| Kalangoya | 12 | 3 | 0.67 | 0.09 | 7 | 0.89 | 0.06 | 2.83 | 1.60 |
| Tadyawan | 14 | 1 | 0.00 | 0.00 | 5 | 0.59 | 0.14 | 1.40 | 0.91 |
| Tawbuid | 13 | 2 | 0.28 | 0.14 | 8 | 0.88 | 0.07 | 2.37 | 1.38 |
| Hanunuo | 15 | 4 | 0.55 | 0.14 | 9 | 0.90 | 0.05 | 2.80 | 1.57 |
| Surigaonon | 32 | 5 | 0.73 | 0.05 | 24 | 0.97 | 0.02 | 3.80 | 1.96 |
| Manobo | 56 | 7 | 0.66 | 0.06 | 24 | 0.93 | 0.02 | 3.58 | 1.85 |
| Maranao | 7 | 3 | 0.76 | 0.11 | 6 | 0.95 | 0.10 | 3.67 | 2.11 |
| Philippines | 390 | 11 | 0.81 | 0.01 | 143 | 0.98 | 0.00 | 4.06 | 2.03 |
Abbreviations: HgD, haplogroup diversity; HtD, haplotype diversity; MPD, mean pairwise haplotype difference; nHg, number of Y-SNP haplogroups; nHt, number of Y-STR haplotypes; nPop, population sample size.
AMOVA analyses showed extensive heterogeneity among FE groups, among FEN groups, and among FEnN groups, with only about 70% of the variation observed within populations in each comparison (Table 3). Only a geographic grouping resulted in a slightly larger difference among groups than among populations within groups. Furthermore, the grouping of FEN vs FEnN also provided a poor description of genetic structure, consistent with the absence of a significant difference between FEN and FEnN diversity indices.
Table 3. Analysis of molecular variance (AMOVA) results.
| Grouping (number of groups) | Among groups | Among populations within groups | Within populations |
|---|---|---|---|
| Philippines (14) | — | 31.3 | 68.7 |
| Negrito (6) | — | 28.1 | 71.9 |
| Non-Negrito (8) | — | 32.2 | 67.9 |
| Negrito and non-Negrito (2) | 3.1 | 29.2 | 67.7 |
| Language (3) | 0.0 | 38.1 | 70.7 |
| Geography (6) | 17.1 | 16.0 | 67.0 |
| Reference data set | |||
| East Asia (6) | — | 24.7 | 75.3 |
| Southeast Asia (14) | — | 15.6 | 84.4 |
| Melanesia (24) | — | 29.6 | 70.4 |
| Polynesia (5) | — | 16.6 | 83.4 |
| Australia (2) | — | 2.2 | 97.8 |
All P-values were significant (P<0.05) except for the following estimates: Among groups-Philippines-Language (0.0; P=0.92); Among groups-Philippines-Negrito and non-Negrito (3.1; P=0.11) and Among populations-Australia (2.2; P=0.13).
Philippine and Reference data set groups are shown in Figure 1.
Philippine groupings are enumerated in Figure 1.
All groups with sample sizes <10 (Figure 1, Supplementary Text) were excluded from the analyses.
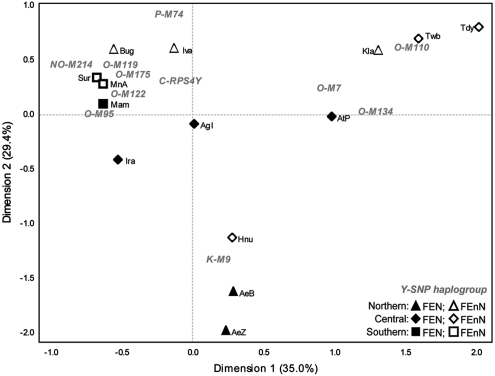
CA based on Y-SNP haplogroup frequencies were used to visualize the genetic relationships of 14 Filipino groups (with samples sizes >10) in association with the distribution of Y-SNP haplogroups. The CA plot (Figure 2) showed three clusters of FE groups: one consisting of the Aeta of Zambales, Aeta of Bataan, and Hanunuo, associated with haplogroup K-M9; one consisting of the Ati, Kalangoya, Tawbuid, and Tadyawan, associated with haplogroups O-M7, O-M110, and O-M134; and the third consisting of the remaining FE groups and haplogroups. There was no tendency for FEN groups to cluster together as they were distributed in all three clusters. There was also no tendency for the three clusters of FE groups to reflect geographic relationships, with the exception of the three southern groups (Surigaonon, Manobo, and Mamanwa), all from northern Mindanao, that cluster together.
Figure 2.
CA plot based on Y-SNP haplogroup frequencies of Filipino groups. Percentages in parentheses indicate the fraction of the variance explained by each dimension. Haplogroups are in gray text, while population codes are in black text. Population codes are the same as in Figure 1. All groups with sample sizes <10 (Figure 1) were excluded from the analyses.
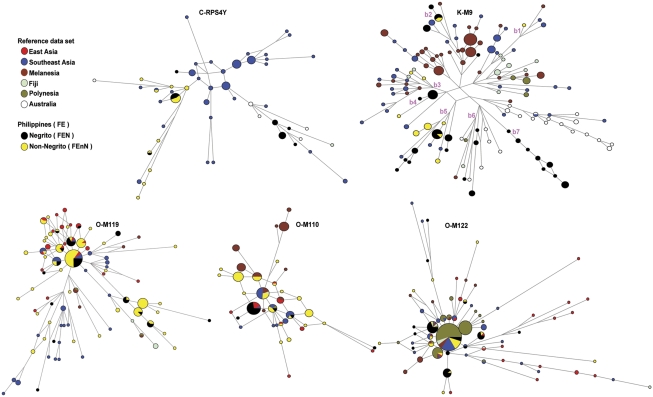
Network (Figure 3) and haplotype sharing analyses (Supplementary Figure 2) showed that haplotypes in major haplogroups O-M119, O-M110, and O-M122 were shared among 14 FE groups, regardless of geographic proximity. In contrast, the shared haplotypes in haplogroups C-RPS4Y and KM9 were distributed among nine FE groups, and for the most part tended to involve groups that are geographically close. These patterns seem to reflect different histories for these haplogroups and suggest that C-RPS4Y and K-M9 are likely to be older, while O-M119, O-M110, and O-M122 would have spread more recently. TMRCA estimates and their credible intervals (CI) were consistent with C-RPS4Y (13 245; CI: 7824–20 644) and K-M9 (13 608; CI: 9225–19 709) being older than O-M119 (7299; CI: 5046–10 204), O-M110 (4167; CI: 2837–5898), and O-M122 (6223; CI: 4151-8886).
Figure 3.
Networks for Y-SNP haplogroups C-RPS4Y, K-M9, O-M119, O-M110, and O-M122. Networks are based on seven haplogroup-associated Y-STR loci for Filipino groups and the reference data set. Circles represent Y-STR haplotypes, with the area of the circle proportional to the frequency of the haplotype. A circle with more than one color indicates a haplotype that is shared between different populations. Nodes without circles that connect branches are median vectors that represent inferred ancestral haplotypes. Branch lengths are proportional to the number of mutational steps. Marked K-M9 network branches (b1–b7) highlight the different associations of FE groups in the K-M9 network. For all networks, specific FE groups are stated in the main paper text.
Genetic affinities of Filipino ethnolinguistic groups
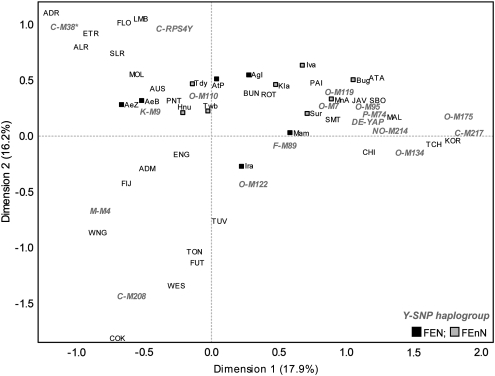
The FE group data set was compared with a reference data set composed of 1756 males from 60 groups across the Asia-Pacific region.20, 21, 22, 23, 24 Figure 1 shows the distribution of shared Y-SNP haplogroups among FE groups and the reference data set. AMOVA results (Table 3) show that the heterogeneity of FE groups were comparable to or exceeded the heterogeneity of the other regions in the reference data set. A CA plot (Figure 4) shows that FE groups have different affinities. The major O haplogroups appear to influence the association of the majority of FE groups with East Asian and Southeast Asian groups and, to a lesser extent, with Melanesian and Polynesian groups. Haplogroups C-RPS4Y and K-M9, drive the association of several FEN groups (Aetas, Ati, and Agta) and the Hanunuo with Southeast Asian and Australian groups. For the major O haplogroups, O-M119, O-M110, and O-M122, Y-STR diversity indices (HtD and MPD) (Supplementary Table 3) were similar for the FE, East, and Southeast Asian groups. Network (Figure 3) and haplotype sharing (Supplementary Table 4) analyses for these three haplogroups showed haplotype sharing between FE groups and all Asia-Pacific groups, with the exception of haplogroup O-M119, for which FE groups shared haplotypes only with East Asia and Southeast Asia. For haplogroup C-RPS4Y, network analysis (Figure 3) showed that FEnN (Ivatan, Manobo, and Surigaonon) and FEN (Ati and Mamanwa) haplotypes were associated with Southeast Asian haplotypes on one branch; while on another, FEN haplotypes (one Ati and all Agta C-RPS4Y haplotypes) were associated with indigenous Australians from Arnhem Land, one Fijian, and several Southeast Asians. This implies two different origins of haplogroup C-RPS4Y in FE groups, one associated with Southeast Asians, and another providing a possible signal of a genetic link between FEN groups (in particular, the Agta, who have the highest frequency of C-RPS4Y among FE groups) and indigenous Australians. It should be noted that the indigenous Australian groups in the reference data set possess, at high frequency, a unique DYS390.1 deletion on the background of haplogroup C-RPS4Y,20 as such these samples were excluded from the C-RPS4Y analysis. The DYS390.1 deletion has been associated with DYS390 allele lengths ranging from 18 to 22 repeats and so far is specific to Australians.20, 24, 29 Several FE groups (Ivatan, Agta, Ati, Surigaonon, Mamanwa, and Manobo) have C-RPS4Y/DYS390 allele lengths within this range (Supplementary Table 2). Although sequence analysis would be required to definitely rule out the DYS390.1 deletion in these individuals, network analysis does not show any association between these FE group haplotypes and the Australian DYS390.1 haplotypes (Supplementary Figure 3). With only three C-RPS4Y haplotypes shared (two among FE groups and one between FE and Southeast Asians) (Figure 3 and Supplementary Table 4), these NRY links between FE, Southeast Asian, and Australian groups are likely to be old. Haplogroup K-M9 was shared between FE groups and different Asia-Pacific regions; but within K-M9, different affinities of FE groups were observed, as seen on different branches of the K-M9 network (Figure 3, b1-b7). FEN (Iraya and Mamanwa) and FEnN (Maranao and Manobo) haplotypes clustered with Southeast Asian and Melanesian haplotypes on b1 and b2. FEnN (Surigaonon and Manobo) haplotypes clustered with Southeast Asian, Melanesian, and Fijian haplotypes on b3. All Ati (FEN) haplotypes were clustered with one Hanunuo (FEnN) haplotype on b4. FEN (two Aeta groups and the Agta) haplotypes were associated with Hanunuo (FEnN) and Southeast Asian haplotypes on b5. FEN (two Aeta groups) haplotypes were associated with one Polynesian, one Fijian, one Melanesian, and six Australian haplotypes on b6. Only FEN (two Aeta groups and the Agta) haplotypes were clustered on b7. Overall, results of FE group-reference data set comparisons for the major O haplogroups, C-RPS4Y, and K-M9 showed the diverse genetic affinities of FE groups. These comparisons also showed support for the observed genetic differences (based on genetic diversity, AMOVA, and CA) among all FE groups.
Figure 4.
CA plot based on haplogroup frequencies of Filipino groups and the reference data set. Percentages under each dimension label indicate the fraction of the variance explained by each dimension. All groups with sample sizes <10 (Figure 1, Supplementary Text) were excluded from the analyses. Haplogroups are in gray text, while population codes are in black text. Population codes are the same as in Figure 1 and enumerated in Supplementary Text.
Negrito – indigenous Australian association: ancient divergence or recent contact?
As there seem to be signals of genetic links between several FEN groups (Aeta-Bataan, Aeta-Zambales, and Agta) and indigenous Australians (Arnhem Land and Great Sandy Desert) (Figure 3: C-RPS4Y and K-M9; Figure 4), divergence times and migration rates between these groups were estimated through pairwise, simulation-based analyses using the IM program.28 Divergence time estimates ranged from 14 549 to 19 799 Y (CI: 4939–75 059), although gene flow was detected from the Arnhem Land Australians to the Aeta of Bataan and the Agta, and from the Great Sandy Desert Australians to the Agta (Table 4 and Supplementary Figure 4). Point estimates indicate divergence times of about 15–20 KYA, after the initial colonization of the Asia-Pacific region around 40–50 KYA,1 though CI values overlap with the initial colonization time period. Time estimates, therefore, do not distinguish between ancient vs more recent divergence between the ancestors of FEN and indigenous Australian groups.
Table 4. Scaled estimates of divergence times (T) and migration rates (M1and M2).
| Philippines | Australia | Australia |
|---|---|---|
| Negrito | Arnhem Land | Great Sandy Desert |
| Aeta B | ||
| T | 19 710 (6540–74 258) | 15 350 (6184–73 101) |
| M1 | 0.072 (0.055–7.267) | 0.072 (0.042–9.587) |
| M2 | 1.690 (0.009–15.348) | 0.047 (0.009–12.852) |
| Aeta Z | ||
| T | 15 528 (4939–65 360) | 14 549 (5740–68 029) |
| M1 | 0.072 (0.054–8.275) | 0.073 (0.043–8.101) |
| M2 | 0.007 (0.003–7.729) | 0.008 (0.003–7.913) |
| Agta | ||
| T | 19 799 (9210–75 059) | 15 083 (6451–69 453) |
| M1 | 0.072 (0.056–6.548) | 0.074 (0.044–9.290) |
| M2 | 3.611 (0.327–18.094) | 1.897 (0.018–13.359) |
Scaled estimates of divergence times (T) and migration rates (M1 and M2) for Negrito groups and indigenous Australians with observed genetic associations. Analyses were based on Y-STR haplotype data (7 Y-STRs) using the IM program.28 Divergence times are in units of years while population migration rates (M1: from Negrito groups to Australian groups and M2: from Australian groups to Negrito groups) indicate the effective rate at which genes come into a population per generation. Credible intervals are in parenthesis. Posterior distributions for these parameter estimates (t, m1 and m2) are shown in Supplementary Figure 4.
Discussion
Our analyses revealed several attributes of FE group NRY lineages, namely: extensive heterogeneity, signatures of old and recent periods, and diverse affinities. Heterogeneity is seen in FEN groups, who show an old substrate (haplogroups C-RPS4Y and K-M9) suggesting ancient shared ancestry. Similarities in physical features and mode of subsistence of FEN groups have been the main, if not the only (anthropological), support for the inference of common ancestry.30 Haplogroups K-M9 and C-RPS4Y in this study provide genetic support for the common ancestry of FEN groups. FEN groups also share haplogroups with FEnN groups (ie, Mamanwa with Manobos and Surigaonons; Aeta with Hanunuo), but rather than ancient shared ancestry, gene flow between FEN and FEnN groups has been inferred by early anthropological studies.30 Heterogeneity among FEnN groups is best exemplified by the Hanunuo, Tadyawan, Tawbuid, and Iraya. Historically grouped and called ‘Mangyan' (with the Iraya also considered a FEN group because of physical features), these groups showed distinct NRY lineages despite their geographical proximity on the island of Mindoro; this may reflect differing cultural practices and language barriers between these groups.30 Owing to readily distinguishable physical features that may be the result of adaptation to their environment,31 confounded by historical and social circumstances, a general grouping of FE groups into FEN and FEnN has been perpetuated.7 However, we find that from a NRY perspective, the extensive heterogeneity of FE groups, influenced by drift and admixture, does not support a simple distinction between FEN and FEnN groups.
Summarized recently, haplogroups C-RPS4Y and K-M9, represent founding lineages in the Asia-Pacific region, while O-M119, O-M110, and O-M122 are associated with the Austronesian expansion.32 The phylogeographic analyses of C-RPS4Y and K-M9 in FE groups suggest that these are old NRY lineages. TMRCA estimates for C-RPS4Y (13 245; CI: 7824–20 644) and K-M9 (13 608; CI: 9225–19 709) also reflect a late Paleolithic timeline and overlap with the ages (14–58 KY) of upper Pleistocene human remains from Tabon Cave, Palawan, Philippines.2 TMRCA estimates for O-M119 (7299; CI: 5046–10 204), O-M110 (4167; CI: 2837–5898), and O-M122 (6223; CI: 4151–8886) in FE groups do overlap with the Austronesian expansion time (5–6 KY).4 In association with the Austronesian expansion, these haplogroups are presumed to have reached the Philippines via Taiwan. However, restricted to the reference data set in this study, only O-M110 has a clear Taiwan-specific origin.23 An alternative scenario was suggested for O-M122, involving expansion from East Asia to Southeast Asia and on to Melanesia.33 O-M122 was found only among central and southern FE groups with significant frequency differences among northern, central, and southern groups (χ2 test; P=0.000029). O-M122 is one of several NRY haplogroups whose diversity has been affected by drift,34 which could explain the absence of O-M122 in northern FE groups. However, a southern entry point into the Philippines could also explain our results. The FE group data for the major O-haplogroups simply suggests that they expanded into the Philippines, either from Taiwan or elsewhere in East Asia, in association with the Austronesian expansion. Overall, this study presents genetic evidence for the settlement of NRY lineages in the Philippines within a time frame after the initial colonization (after 40–50 KYA) of the Asia-Pacific region, extending into the time frame of the Austronesian expansion.
Recently, analysis of about 50 000 SNPs in a wide variety of Asian populations, including several FEN groups, found no clear-cut genetic distinction between FEN and FEnN groups, and concluded that, FEN and FEnN groups were part of the same wave of migration.14 This conclusion seems at odds with both the NRY data, which indicate an earlier presence of haplogroups C-RPS4Y and K-M9 in the Philippines that is associated with FEN groups, and with mtDNA data indicating novel and ancient mtDNA lineages in a FEN group (E Gunnarsdottir and M Stoneking, unpublished data). A possible reconciliation of the genome-wide SNP data with the uniparental marker data is, if the ancestors of FEN groups were isolated from the ancestors of FEnN groups (either as a separate migration or as part of the same migration), but then in recent times FEN groups have experienced substantial, primarily male-mediated admixture with FEnN groups.35 Such substantial admixture, documented in the NRY data as discussed above, could account for the genome-wide SNP data.14
Another interesting finding was the link between C-RPS4Y and K-M9 haplotypes of some FEN groups and indigenous Australians. An early study on FEN groups, using immunoglobulin G genetic markers, inferred a possible ancient link with northeastern indigenous Australians.12 With information only from the NRY, our demographic inference yielded broad divergence time estimates, but with inferred gene flow that seem to run counter to the commonly-held view that there was little contact between Australia and the rest of the world after initial colonization.36 However, given the broad divergence time estimates, as well as the ancient associations inferred from network and haplotype sharing analyses, the inferred gene flow between FEN groups and indigenous Australians need not necessarily imply direct contact between these groups. An alternative possibility is that, gene flow from Australia to the Philippines was only via neighboring populations, in a ‘stepping-stone' manner. Data from additional loci would be needed to confirm this apparent signal of more recent gene flow involving FEN and indigenous Australian groups.
Our study provides a number of important insights into the genetic history of Filipinos. First, there is extensive diversity in the NRY lineages of FE groups. In general, a simple dichotomy of Negrito vs non-Negrito is not supported by Filipino NRY data. Second, Filipino NRY lineages reflect a chronology and thus, a history that extends from after the initial colonization of the Asia-Pacific region, into the time frame of the Austronesian expansion. Third, the diverse NRY genetic affinities of Filipinos can also be associated with the Austronesian expansion, with older affinities reflecting demographic scenarios after the initial colonization of the Asia-Pacific region, including the possible divergence and subsequent gene flow between some Negrito groups and indigenous Australians. Although the Y-chromosome is a powerful tool for studying demographic events and population history, and has yielded some important new insights in the present study, it is still a single genetic locus. To obtain more accurate estimates of demographic parameters, to verify genetic associations, and to further characterize Filipino genetic diversity, genetic studies from other loci are needed.
Acknowledgments
We gratefully acknowledge all participants for kindly providing samples for this study and all individuals, institutions and organizations for their assistance in the population sampling process, including all funding organizations that supported this study (see Supplementary Text). We acknowledge the support provided by Miriam Dalet, Minerva Sagum, Michelle Music Tan, Nota Magno, Laarni Carpina, Liza Faustino and Alma Ferrer (UP-NSRI-DAL), Dr Victor Paz (UP Archeological Studies Program) Dr Francisco Datar (UP Anthropology), Dr Sabino Padilla (UP Anthropology) and Maria Concepcion Sakai (NCIP, Head Office). For assistance with the MPI-EVA sample collection in northern Mindanao, we are especially grateful to Fernando A Almeda Junior, Dr Irinetta C Montinola, Dr Wilfredo Sinco (all from the Surigaonon Heritage Center), Girlie Patagan (NCIP, Surigao), Elizabeth S Larase and .Juliet P Erazo (Office of Non-Formal Education), and the Rotary Club of Surigao. We also thank Peter Ahnert and Holger Kirsten (Department of Biochemistry, University of Leipzig) for support with the MALDI-TOF MS assays and Jody Hey for support with the IM program. We thank the MPI-EVA Molecular Anthropology Group and the Research Group on Comparative Population Linguistics for helpful discussions, in particular Cesare de Filippo and Cristoph Theunert for additional support in computer program implementations.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- O'Connell JF, Allen J. Dating the colonization of Sahul (Pleistocene Australia–New Guinea): a review of recent research. J Arch Sci. 2004;31:835–853. [Google Scholar]
- Détroit F, Dizon E, Falguères C, Hameau S, Ronquillo W, Sémah F. Upper Pleistocene Homo sapiens from the Tabon cave (Palawan, The Philippines): description and dating of new discoveries. C R Palevol. 2004;3:705–712. [Google Scholar]
- Barker G, Barton H, Bird M, et al. The ‘human revolution' in lowland tropical Southeast Asia: the antiquity and behavior of anatomically modern humans at Niah Cave (Sarawak, Borneo) J Hum Evol. 2007;52:243–261. doi: 10.1016/j.jhevol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Gray RD, Drummond AJ, Greenhill SJ. Language Phylogenies Reveal Expansion Pulses and Pauses in Pacific Settlement. Science. 2009;323:479–483. doi: 10.1126/science.1166858. [DOI] [PubMed] [Google Scholar]
- Gordon RG, Jr. (ed).: Ethnologue: Languages of the World15th edn.Dallas, Tex: SIL International; Online version: http://www.ethnologue.com/ , 2005 [Google Scholar]
- Scott WH. Prehispanic Source Materials For the Study of Philippine History. Quezon City, Philippines: New Day Publishers; 1984. [Google Scholar]
- Jocano FL. Anthropology of the Filipino People I: Philippine Prehistory, Rediscovering Precolonial Heritage. Diliman, Quezon City, Philippines: PUNLAD Research House, Inc.; 1998. [Google Scholar]
- Bean RB. Types of Negritos in the Philippines. Am Anthro. 1910;12:220–236. [Google Scholar]
- Kashyap VK, Sitalaximi T, Sarkar BN, Trivedi R. Molecular relatedness of the aboriginal groups of Andaman and Nicobar Islands with similar ethnic populations. Int J Hum Genet. 2003;3:5–11. [Google Scholar]
- Reid LA.Unravelling the linguistic histories of Philippine Negritosin Dutton T, Tyron DT (eds): Language Contact and Change in the Austronesian World Berlin, Germany: Mouton de Gruyter; 1994443–475. [Google Scholar]
- Tabbada KA, Trejaut J, Loo J-H, et al. Philippine mitochondrial DNA diversity: A populated viaduct between Taiwan and Indonesia. Mol Biol Evol. 2010;27:21–31. doi: 10.1093/molbev/msp215. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Miyazaki T, Omoto K, et al. Population Genetic Studies of the Philippine Negritos II Gm and Km Allotypes of Three Population Groups. Am J Hum Genet. 1979;31:70–76. [PMC free article] [PubMed] [Google Scholar]
- Omoto K, Ueda S, Goriki K, Takahashi N, Misawa S, Pagaran IG. Population Genetic Studies of the Philippine Negritos. III. Identification of the Carbonic Anhydrase-1 Variant with CA1 Guam. Am J Hum Genet. 1981;33:105–111. [PMC free article] [PubMed] [Google Scholar]
- The HUGO Pan-Asian SNP Consortium Mapping Human Genetic Diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- Salvador JM, De Ungria MCA. Isolation of DNA from saliva of betel quid chewers using treated cards. J Forensic Sci. 2003;48:794–797. [PubMed] [Google Scholar]
- Paracchini S, Arredi B, Chalk R, Tyler-Smith C.Hierarchical high-throughput SNP genotyping of the human Y-chromosome using MALDI-TOF mass spectrometry Nucleic Acids Res 200230e2721–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Sykes BC, Jobling MA, Forster P. The dual origin of the Malagasy in island Southeast Asia and East Africa: Evidence from maternal and paternal lineages. Am J Hum Genet. 2005;76:894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DA, Tang K, Strotmann R, et al. Parallel Selection on TRPV6 in Human Populations PLoS ONE 20083e16861681–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, Schiefenhövel W, Underhill PA, Stoneking M. Independent Histories of Human Y Chromosomes from Melanesia and Australia. Am J Hum Genet. 2001;68:173–190. doi: 10.1086/316949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, et al. Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am J Hum Genet. 2003;72:281–302. doi: 10.1086/346065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Cordaux R, et al. Melanesian and Asian Origins of Polynesians: mtDNA and Y Chromosome Gradients Across the Pacific. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- Kayser M, Choi Y, van Oven M, et al. The Impact of the Austronesian Expansion: Evidence from mtDNA and Y Chromosome Diversity in the Admiralty Islands of Melanesia. Mol Biol Evol. 2008;25:1362–1374. doi: 10.1093/molbev/msn078. [DOI] [PubMed] [Google Scholar]
- Mona S, Grunz KE, Brauer S, et al. Genetic Admixture History of Eastern Indonesia as Revealed by Y-Chromosome and Mitochondrial DNA Analysis. Mol Biol Evol. 2009;26:1865–1877. doi: 10.1093/molbev/msp097. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- StatSoft, Inc STATISTICA (data analysis software system), version 8.0 2008 , www.statsoft.com .
- Wilson IJ, Weale ME, Balding DJ. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J R Stat Soc Ser A. 2003;166:155–201. [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. perimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Kayser M, Meyer E, et al. Phylogenetic Resolution of Complex Mutational Features at Y-STR DYS390 in Aboriginal Australians and Papuans. Mol Biol Evol. 1998;15:1108–1114. doi: 10.1093/oxfordjournals.molbev.a026018. [DOI] [PubMed] [Google Scholar]
- Lebar FM.(ed.): Ethnic Groups of Insular Southeast Asia Volume 2: Philippines and Formosa New Haven, USA: Human Relations Area Files Press; 1975 [Google Scholar]
- Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24:220–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Hallmark B, Cox MP, et al. Major East-West Division Underlies Y Chromosome Stratification Across Indonesia MBE Advance Access published on March 5, 2010doi: 10.1093/molbev/msq063 [DOI] [PubMed]
- Kayser M, Brauer S, Weiss G, et al. Melanesian origin of Polynesian Y chromosomes. Curr Biol. 2000;10:1237–1246. doi: 10.1016/s0960-9822(00)00734-x. [DOI] [PubMed] [Google Scholar]
- Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. PNAS. 2009;106:20174–20179. doi: 10.1073/pnas.0910803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M, Delfin F. The Human Genetic History of East Asia: Weaving a Complex Tapestry. Curr Biol. 2010;20:R188–R193. doi: 10.1016/j.cub.2009.11.052. [DOI] [PubMed] [Google Scholar]
- Pardoe C.Becoming Australian: evolutionary processes and biological variation from ancient to modern times Before Farming [online version] 2006. 2006/1 article41–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.