Abstract
This study explores the relationship between sperm structural aberrations and age by using a multicolor multichromosome FISH strategy that provides information on the incidence of duplications and deletions on all the autosomes. ToTelvysion kit (Abbott Molecular, Abbott Park, IL, USA) with telomere-specific probes was used. We investigated the sperm of 10 male donors aged from 23 to 74 years old. The donors were divided into two groups according to age, a cohort of five individuals younger than 40 and a cohort of five individuals older than 60 years. The goal of this study was to determine (1) the relationship between donor age and frequency and type of chromosome structural abnormalities and (2) chromosomes more frequently involved in sperm structural aberrations. We found that the older patients had a higher rate of structural abnormalities (6.6%) compared with the younger cohort (4.9%). Although both duplications and deletions were seen more frequently in older men, our findings demonstrate the presence of an excess of duplications versus deletions in both groups at a ratio of 2 to 1. We demonstrate that the distribution of duplications and deletions was not linear along the chromosomes, although a trend toward a higher rate of abnormalities in larger chromosomes was observed. This work is the first study addressing the frequencies of sperm chromosome structural aberrations of all autosomes in a single assay thus making a contribution to the clarification of the amount and origin of damage present in human spermatozoa and in relation to age.
Keywords: paternal age, spermatozoa, chromosome duplications, FISH, structural chromosome abnormalities
Introduction
The causes of negative reproductive outcomes are poorly understood but chromosome abnormalities are known to be a very significant factor. In humans, it has been estimated that at least 8.1% of all clinically recognized pregnancies have a numerical (∼7%) or structural (∼1%) chromosome abnormality. Chromosome abnormalities are seen in 50% of spontaneous abortions, 6% still births and 1% live births.1 At birth, the incidence of aneuploidy (0.33%) is only slightly higher than structural abnormalities 0.25%.2 Most chromosomally abnormal conceptions are the result of an error in gametogenesis and, less frequently, the result of a post fertilization event.
In a study of the origin of de novo structural chromosome rearrangements using chromosome heteromorphisms, Olson and Magenis3 found that 80% of de novo reciprocal translocations and deletions and 100% of duplication and ring chromosomes were of paternal origin. The majority of 115 de novo unbalanced structural chromosome abnormalities detectable by light microscopy are of paternal origin4 varying from 84% of interstitial deletions to 58% of duplications and rings. Deletions of the long arm of chromosome 18 are disproportionately male5 as well as the deletions that cause Wolf–Hirschhorn syndrome6 and Cri-du-chat syndrome.7 More recently, the parental origin of de novo microdeletions associated with de novo reciprocal translocations and cases of complex chromosomes rearrangements was determined to be paternal in all cases.8 Similarly, all de novo deletions described by aCGH in carriers of balanced translocations and abnormal phenotype turned out to be of paternal origin.9 Moreover, studies on the origin of the recurrent de novo t(11;22) found that it was paternal in 100% of cases10, 11 and Thomas et al12 confirmed a paternal origin of non-recurrent de novo balanced reciprocal translocations in 96% of cases.
Despite the high paternal contribution of de novo rearrangements compared with the low frequency of paternally derived numerical abnormalities, the available information on structural aberrations in human sperm is more limited. In sperm karyotypes, obtained after in vitro penetration of hamster oocytes, structural chromosome abnormalities have been observed far more frequently than numerical aberrations.13, 14, 15 In a review, Templado et al16 reported a median percentage of 6.6% of structural aberrations and 1.8% of numerical abnormalities. Several authors have explored the relationship between age of the donor and sperm structural aberrations (reviewed by Buwe et al17). Martin and Rademaker18 found positive correlations with chromosomal breaks, and Sartorelli et al19 with acentric fragments and complex radial figures. Other authors noticed a trend toward elevated frequencies of breaks and/or acentric fragments with age but without reaching statistical significance.15, 20, 21 There have been further reports using multicolor FISH in decondensed human sperm heads but each study has been limited to a particular chromosome. Using this approach, a positive age effect on the frequencies of duplications and deletions for the centromeric and subtelomeric regions of chromosome 9 was reported by Bosch et al22 and, similarly, Sloter et al23 found that sperm of older men carried more breaks and segmental duplications and deletions of chromosome 1.
In Western societies, women's access to contraception and to the work force coupled with the availability of new reproductive technologies has resulted in couples delaying childbearing. Both the age of the father and that of the mother are thought to influence reproductive outcome. It has long been known that the age of the female increases the chances for meiotic errors in oogenesis resulting in offspring with aneuploid chromosome abnormalities. There seems to be a lesser impact for paternal than maternal age and the means by which advanced paternal age affects human reproduction are poorly understood.
The purpose of this study is to explore the relationship between sperm structural aberrations and age by using a multicolor multichromosome FISH strategy (ToTelvysion multicolor FISH, Abbott Molecular, Abbott Park, IL, USA) that provides information on the incidence of duplications and deletions on all the autosomes. We investigated the sperm of 10 male donors aged from 23 to 74 years old. The donors were divided into two groups according to age, a cohort of five individuals younger than 40 (between 23 and 37 years old) and a cohort of five individuals older than 60 years of age (ages 60–74). We analyzed 1 000 spermatozoa per chromosome with a total of 15 000 spermatozoa per donor for a total of 150 000 cells. The multicolor multichromosome assay was used to explore (1) the nature of the relationship between donor age and frequency and type of chromosome structural abnormalities and (2) chromosomes more frequently involved in sperm structural aberrations.
Materials and methods
Sperm donors
We worked with spermatozoa from 10 healthy male donors ages from 23 to 74 years old from the Andrology Service of the Fundació Puigvert (Barcelona, Spain). Before the start of the research, all of the sperm donors signed their consent to the study. The study was approved by the Institutional Ethics Committee of the Universitat Autònoma of Barcelona, and the Ethics Committee of the Fundació Puigvert.
Our donors had no history of exposure to any known mutagens, clastogens, radiation or recreational drugs. Six of our donors, namely C1, C2, C6, C7, C9 and C10, had children. The same donors had been used in previous studies on the frequency of numerical abnormalities of chromosomes 6, 9, 21, X and Y22, 24 and the frequency of structural abnormalities of chromosoma 924 in human sperm. Sperm samples were obtained by masturbation after an abstinence period of at least 3 days.
FISH with subtelomeric DNA probes
Slide preparation and pretreatments before hybridization were performed following the protocols described elsewhere.24 Similarly, the supplier's protocol with slight modifications was followed for the hybridization procedure.24
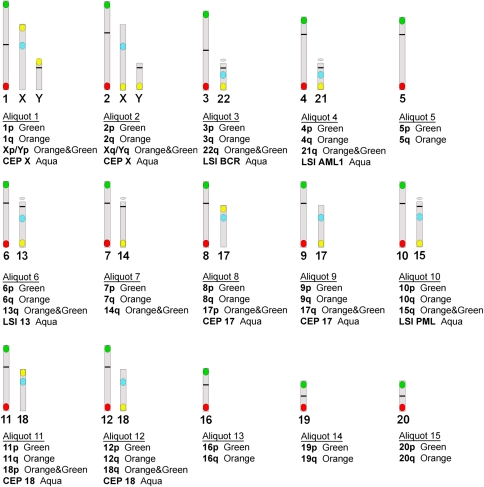
We used the panel of DNA probes ToTelvysion multicolor FISH (Abbott Molecular). Figure 1 depicts the chromosome location, color and aliquot distribution of the panel of probes used in this study. These include subtelomeric probes specific for the short and long arms of all chromosomes except the p arm of the acrocentrics (13, 14, 15, 21 and 22) and a number of centromeric (CEP) probes (CEP X, CEP 17, CEP 18) and locus specific probes (LSIs): LSI 13 (13q14), LSI PML, (15q22), LSI AML, (21q22), LSI BCR, (22q11). For the sex chromosomes, the probe mix used in this study (Figure 1) does not allow the discrimination between numerical and structural aberrations nor the determination of structural aberrations present in each of the sex chromosome, thus the sex chromosomes results have not been included in this study.
Figure 1.
The 15 aliquots of the FISH probe panel used showing both the chromosome location and the color of the probes (spectrum orange, spectrum green and spectrum aqua) from each aliquot.
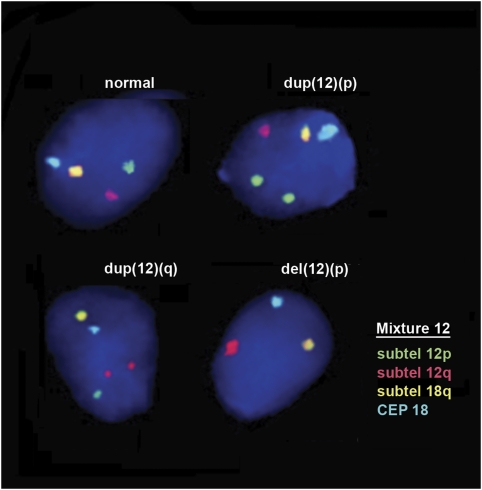
This FISH protocol will measure duplications and deletions that include the following chromosomal regions: (a) telomeres q of all autosomes, (b) telomeres p of all autosomes except acrocentric ones and (c) centromere and LSI specific for 6 out of 22 autosomes. The duplications and deletions detected are not necessarily limited to subtelomeric ones but could be whole arm or partial. They would correspond to chromosome structural aberrations, such as unbalanced reciprocal translocations, unbalanced Robertsonian translocations, unbalanced peri and paracentric inversions, isochromosomes, dicentric chromosomes, terminal deletions and duplications, segmental deletions and duplications, whole arm deletions and duplications, acentric fragments, and ring chromosomes. Interstitial deletions and/or duplications not including the targeted probes, most microdeletions and microduplications, and balanced structural aberrations would not be detected (Figure 2).
Figure 2.
FISH in human decondensed sperm nuclei using one of the 15 mixtures of the FISH panel (mixture 12) showing structural aberrations for chromosome 12: deletions for subtelomere 12p and duplications for subtelomeres 12p and 12q.
Data collection and scoring criteria
Only slides with a hybridization efficiency of at least 99% were scored. Only intact and non-overlapped decondensed sperm, identified because of the presence of a tail, were evaluated. For each probe, two signals were scored as such only if they had the same intensity, size and color and were separated by a distance equal to or higher than the probe signal diameter. If two signals were recorded for a subtelomeric, centromeric or LSI probe it was considered a duplication (partial or whole arm). When we observed the absence of one of these signals for a chromosome, it was recorded as deletion (partial or whole arm). The presence of two signals for subtelomere p and q regions and two signals for the corresponding centromeric or LSI probe was considered disomy and it was not scored as a structural duplication. In mixtures with only two subtelomeric probes, the presence of two signals for both subtelomeric probes was considered disomy. Slides were coded before the microscope scoring.
Statistical analysis
The propensity to duplications versus the propensity to deletions for each individual was assessed by both the paired Student's t-test and the non-parametric Wilcoxon signed-rank test. The χ2-test was used to analyze the propensity of structural abnormalities for each chromosome relative to a uniform distribution after including a correction for chromosome length. A generalized estimating equation (GEE)25 was used for assessing the differences between the two age groups for the different types of chromosomal instability. The GEE approach is an extension of generalized linear models designed to account for repeated within-individual measurements. The GEE statistical model has been proved instrumental for the comparison of chromosomal instability between two groups (see De la Chica et al26 for further statistical details). Statistical significance was set at P<0.05. Statistical analyses were carried out with SAS/STAT release 9.1 (SAS Institute Inc, Cary, NC, USA).
Results
We analyzed 1000 spermatozoa per chromosome (15 aliquots) with a total of 15 000 sperm for each donor for a total of 150 000 cells. The donors were divided into two groups according to age: a cohort of five individuals younger than 40 (between 23 and 37 years old) and a cohort of five individuals older than 60 years of age (ages 60–74).
Frequency of structural abnormalities
Table 1 shows the percentages of structural chromosome abnormalities (duplications and deletions) in each individual, and the mean and standard error of the mean (SEM) of response variables for each group age
Table 1. Percentages of structural chromosome abnormalities in spermatozoa from 10 healthy donors in each group of age.
| Donor | Age | % Duplications | % Deletions | % Total structural |
|---|---|---|---|---|
| C3 | 24 | 2.9 | 1.5 | 4.4 |
| C4 | 25 | 3.5 | 1.1 | 4.6 |
| C5 | 37 | 3.1 | 1.3 | 4.4 |
| C8 | 23 | 2.9 | 1.7 | 4.6 |
| C9 | 30 | 4.2 | 2.5 | 6.7 |
| Mean (SEM) | 28 | 3.3 (0.25) | 1.6 (0.2) | 4.9 (0.4) |
| C1 | 74 | 3.5 | 1.0 | 4.5 |
| C2 | 64 | 4.4 | 2.7 | 7.1 |
| C6 | 72 | 5.3 | 2.4 | 7.7 |
| C7 | 60 | 3.7 | 2.4 | 6,1 |
| C10 | 62 | 5.7 | 1.9 | 7.6 |
| Mean (SEM) | 66 | 4.5 (0.4) | 2.1 (0.3) | 6.6 (0.6) |
The frequency of structural abnormalities ranged from 4.4% (C3 and C5) to 6.7% (C9) in the younger group and from 4.5% (C1) to 7.7% (C6) in the older group. The SEM for the total of structural abnormalities was higher (0.60) in the older group than in the younger group (0.44) meaning that there was more heterogeneity or interindividual variability within the older group. Two outliers contribute significantly to the increase in variability within each group. Indeed, the frequency of deletions (2.5%), duplications (4.2%) and total structural abnormalities (6.7%) of donor C9 from the younger group was at or above the mean for older donors. Likewise, within the older group there was C1 whose percentages of deletions (1.0%), duplications (3.5%) and total structural (4.5%) were close to the mean of younger donors.
The mean frequency of duplications was statistically higher than that of deletions in both the older (4.5%) vs (2.1% P=0.0053) and the younger group (3.3%) vs (1.62% P=0.0012) when using the paired Student's t-test. However, under a more conservative approach (the non-parametric Wilcoxon signed-rank test) the P-values switch to P=0.0625 for both older and younger groups resulting in a lack of statistical relevance. All men from both groups had higher frequencies of duplications than deletions, regardless of age (see Table 1). The ratio of duplications to deletions is maintained throughout the age groups, 2.1 in the younger cohort and 2.2 in the older, thus, in each group of age, for each deletion observed we scored ∼2 duplications. Table 2 shows the frequencies of duplications and deletions per chromosome arm and the total of structural abnormalities per chromosome in all donors studied. The higher frequency of duplications is observed across the range of all chromosome arms with some exceptions, such as 2q, 3q, 4p, 7p, 8p and 19q.
Table 2. Percentages of structural abnormalities in each chromosome arm in spermatozoa from all of the 10 individuals studied.
| p arm | q arm | ||||||
|---|---|---|---|---|---|---|---|
| Chromosomea | % Duplication | % Deletion | % Total | % Duplication | % Deletion | % Total | % Total structural |
| 1 | 0.15 | 0.10 | 0.25 | 0.12 | 0.06 | 0.18 | 0.43 |
| 2 | 0.09 | 0.02 | 0.11 | 0.07b | 0.16b | 0.23 | 0.34 |
| 3 | 0.11 | 0.01 | 0.12 | 0.04b | 0.05b | 0.09 | 0.21 |
| 4 | 0.08b | 0.08b | 0.16 | 0.07 | 0.06 | 0.13 | 0.29 |
| 5 | 0.14 | 0.02 | 0.16 | 0.12 | 0.09 | 0.21 | 0.37 |
| 6 | 0.10 | 0.02 | 0.12 | 0.06 | 0.04 | 0.10 | 0.22 |
| 7 | 0.02b | 0.08b | 0.10 | 0.10 | 0.06 | 0.16 | 0.26 |
| 8 | 0.04b | 0.09b | 0.13 | 0.12 | 0.10 | 0.22 | 0.35 |
| 9 | 0.08 | 0.06 | 0.14 | 0.15 | 0.07 | 0.22 | 0.36 |
| 10 | 0.11 | 0.04 | 0.15 | 0.05 | 0.03 | 0.08 | 0.23 |
| 11 | 0.05 | 0.04 | 0.09 | 0.18 | 0.03 | 0.21 | 0.30 |
| 12 | 0.12 | 0.04 | 0.16 | 0.07 | 0.00 | 0.07 | 0.23 |
| 13 | ND | ND | ND | 0.22 | 0.04 | 0.26 | 0.26 |
| 14 | ND | ND | ND | 0.14 | 0.07 | 0.21 | 0.21 |
| 15 | ND | ND | ND | 0.15 | 0.03 | 0.18 | 0.18 |
| 16 | 0.08 | 0.05 | 0.13 | 0.06 | 0.03 | 0.09 | 0.22 |
| 17 | 0.01 | 0.00 | 0.01 | 0.16 | 0.0 | 0.16 | 0.17 |
| 18 | 0.09 | 0.00 | 0.09 | 0.09 | 0.0 | 0.09 | 0.18 |
| 19 | 0.18 | 0.04 | 0.22 | 0.03b | 0.07b | 0.10 | 0.32 |
| 20 | 0.12 | 0.09 | 0.21 | 0.11 | 0.03 | 0.14 | 0.35 |
| 21 | ND | ND | ND | 0.13 | 0.04 | 0.17 | 0.17 |
| 22 | ND | ND | ND | 0.11 | 0.01 | 0.12 | 0.12 |
Abbreviation: ND, not determined.
10 000 spermatozoa analyzed/chromosome.
Chromosome arms for which the frequency of duplications is not higher than that of deletions.
Structural abnormalities in the older and younger groups
We scored a total of 330 structural abnormalities in the older group and 247 in the younger group. The older cohort had a higher incidence of structural abnormalities across the board, including deletions, duplications and total structural anomalies.
The statistical analysis showed a significant difference in the incidence of total structural abnormalities between the older (6.6%) and younger groups (4.9%) (P=0.0499, GEE analysis). If donor C9 from the younger group who has outlier values for the group, was removed from the analysis, the P-value shifted to P=0.0190, increasing the significance between older and younger. A similar trend was obtained when removing instead the outlier in the older group (donor C1), the P-value shifted to P=0.0154.). The frequency of duplications was statistically different when comparing both groups of age (4.5 vs 3.3% P=0.0400 GEE analysis). As for deletions, although more were found in the older group (2.1%) than in the younger (1.6%), the difference was not statistically relevant.
Age effect on the distribution of structural abnormalities per chromosome
Pooling all chromosomes together, the mean frequency of structural abnormalities per chromosome was 0.31% in the older group and was higher (P=0.004) than that of the younger group (0.23%).
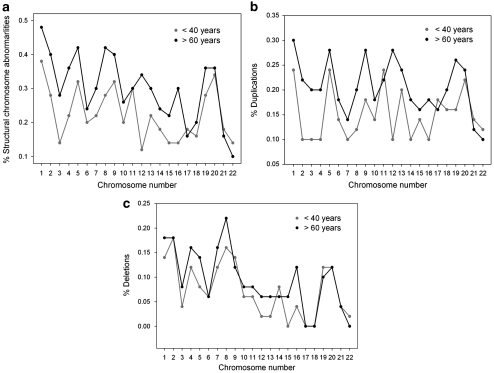
The percentage of structural abnormalities for each chromosome (Figure 3) follows the same trajectory in both groups with the exception of chromosome 12 where there is a statistical divergence between older and younger (0.34 and 0.12%, respectively; P=0.01, GEE analysis). For chromosome 16 the frequency of structural abnormalities also shows a divergent trajectory but it is not statistically significant (Figure 3a). The percentage of duplications is statistically different for chromosome 12 (0.28 older vs 0.1 younger; P=0.0286), and for chromosome 19 (0.26 older vs 0.16 younger) in which there is a clear divergence between the curves, (P=0.0239; Figure 3b). The incidence of deletions follows a similar trajectory for both groups with absence of statistical significance in any of the chromosomes (Figure 3c).
Figure 3.
(a) Percentage of chromosome structural abnormalities per chromosome in each group of age. (b) Percentage of duplications per chromosome.(c) Percentage of deletions per chromosome.
When we pooled all donors together (Table 2), we observed a percentage of structural abnormalities ranging from 0.12 for chromosome 22 to 0.43 for chromosome 1. The second chromosome in number of structural abnormalities was chromosome 5 (0.37%) followed by chromosome 9 (0.36%), which was third. A tendency to a higher rate of abnormalities in larger chromosomes was observed, however, a direct proportionality between the number of abnormalities and chromosome length was not maintained. Indeed, the rate of abnormalities in chromosomes 19 and 20 (0.32 and 0.35, respectively) were on the same order as larger chromosomes, namely chromosome 2 (0.34), 5 (0.37), 8 (0.35) and 9 (0.36). In those donors younger than 40 chromosome 3 had a lower (0.14%) than expected (0.34%) percentage of duplications and deletions, whereas chromosomes 19 (0.28%), 20 (0.34%) and 21 (0.18%) had higher than expected percentages (0.13, 0.13, 0.09%, respectively; P<0.05). In those 60 or older, chromosomes 19 and 20 demonstrated a higher percentage of structural abnormalities than expected (0.36 vs 0.18%, respectively) and (0.36 vs 0.17%, respectively; P<0.05).
Discussion
The multicolor multichromosome assay provides a measure of the number of duplications and deletions (terminal, partial including the subtelomere or other targeted areas, or whole arm) present in the spermatozoa of an ejaculate. These duplications and deletions result from presumably de novo structural chromosome rearrangements that may arise during the spermatogonial divisions or in meiosis. The specific type of structural aberration that lead to the duplications and deletions observed by our assay could not be determined because the multicolor multichromosome FISH assay does not discriminate among these aberrations.
The overall mean of structural abnormalities was 5.8% which is in line with what has been reported with the hamster oocyte assay for normal donors (6.6%)15 and reports of FISH studies of structural abnormalities of chromosome 1 extrapolated to the whole genome (4.4–6.1%).27 Although these numbers are comparatively similar it is important to keep in mind that our study includes only unbalanced rearrangements and direct chromosome studies with the hamster test included both balanced and unbalanced aberrations.
The older patients had a higher rate of structural abnormalities (6.6%) compared with the younger cohort (4.9%). Along these lines, a positive age effect on the frequencies of duplications and deletions for chromosome 922 and for chromosome 123 was reported in FISH studies of spermatozoa. We demonstrate that distribution of duplications and deletions was not linear along the chromosomes. A tendency toward a higher rate of abnormalities in larger chromosomes was observed, however, direct proportionality between the number of abnormalities and chromosome length was not detected. In both younger and older patients, chromosomes 19 and 20 had an excess of abnormalities, whereas chromosomes 3 and 6 had less than expected. Non-random distribution of breaks on chromosome 9 has been reported in sperm karyotypes14 and in FISH studies an age dependent linear trend on chromosome 9 breaks was reported.22. Our observations show a propensity of chromosome 9 to break higher than expected yet without statistical significance. Moreover, previous reports of chromosome 4 having a lower than expected number of breaks in sperm karyotypes15 were confirmed in this study but without reaching statistical significance.
Both duplications and deletions were seen more frequently in the older men. The increase in duplications with age was statistically significant as opposed to the increase in the frequency of deletions, which, although higher in older men, was not significant. The ratio of duplications to deletions was maintained around 2 in both groups, meaning that for each deletion observed we scored 2 duplications (2.1 in the younger cohort and 2.2 in the older). The higher frequency of duplications was observed in all donors, younger and older, and across the range of all chromosome arms with a few exceptions (2q, 3q, 4p, 7p, 8p and 19q). Sloter et al23 observed a slight trend toward higher frequencies of sperm with segmental duplications vs deletions in each man, regardless of age. Likewise, several authors28, 29, 30 showed that duplications of 1p were more frequent than deletions in human spermatozoa. The excess of duplications observed in sperm nuclei could be a reflection of the excess in acentric fragments described in sperm karyotypes.15, 18, 19, 20, 21 and metaphase I spermatocytes.31 Acentric fragments can either acquire a neocentromere and be rescued, or be lost.32 This could explain the excess of duplications detected in spermatozoa with respect to the rate of subtelomeric duplications in subjects with a clinical phenotype
The higher rate of duplications could be explained by a mitotic origin rather than a meiotic origin for some of these de novo abnormalities. Translocations, inversions, insertions, isochromosomes and small deletions and duplications are considered stable rearrangements and it has been postulated that they would originate in spermatogenic mitotic divisions or during meiosis.15
The number of spermatogonial divisions accumulates with increasing age. In males, germ cells divide continuously. At the spermatogonial phase a man of 50 years could accumulate 840 replications each increasing the chances for DNA damage and errors in transcription. Thomas et al12 report a paternal origin in 96% of de novo non-recurrent balanced reciprocal translocations. They all appeared to be unique events not mediated by sequence homology (unlike meiotic rearrangements) and showed a strong paternal age effect suggesting a mitotic spermatogonial origin. On the contrary, the frequency of recurrent de novo t(11;22) in sperm has been found to be independent of the age of the donors33 and it might be homology mediated and of meiotic origin.
Environmental insults and age
Environmental insults and exposures over a lifetime may increase DNA damage in sperm of older men and contribute to the incidence of childhood diseases that increase with paternal age.34 Men over 50 have a higher pregnancy loss and decreased live birth rate.35 There is increased DNA fragmentation with age, a consequence of defective apoptosis, oxidative stress, meiotic and spermiogenic abnormalities.36 Singh et al37 report that the amount of DNA damage in sperm of men aged 35–57 is three times that of men <35 years. The accumulation of unrepaired DNA damage in male germ cells, plus a progressively deteriorating testicular environment–host factors, could manifest as an age-related increase in chromosomal structural abnormalities that could be passed onto the zygote in the form of de novo structural rearrangements.
Clinical significance
The sperm carrying de novo structural rearrangements may father offspring with clinically significant de novo chromosome aberrations (microscopic and submicroscopic) or de novo submicroscopic clinical variants.
Contrary to that observed in sperm nucli, studies carried out in selected populations of patients, have found that de novo deletions are more frequent than duplications. Ravnan et al38 in a FISH study limited to subtelomeric regions of a highly selected population of patients with developmental disabilities and normal karyotypes, detected more de novo clinically significant subtelomeric deletions than subtelomeric duplications, and more subtelomeric deletion variants than subtelomeric duplication variants. A contributing factor to explain the differences between spermatozoa and clinical studies could be that duplications are associated with milder phenotypes and thus individuals with duplications are less likely to be included in highly selected populations, such as patients with developmental disabilities. Moreover, the FISH subtelomeric probes used in this study can potentially detect benign polymorphic variants39, 40 that do not result in pathologic phenotype. This could be another contributing factor to explain the differences between spermatozoa and clinical studies.
Conclusion
Our work shows an increase in structural abnormalities (duplications and deletions) in the spermatozoa of older men. Our findings demonstrate the presence of an excess of duplications versus deletions in both groups at a ratio of 2 to 1 which can only be explained by mechanisms other than meiotic recombination as their source or by a negative selection of germinal cells carrying deletions during spermatogenesis. Partial or whole arm duplications exceeded deletions across the board for most of the chromosome arms. Chromosome 12 rates of structural abnormalities differ significantly in the two groups of age. This work is the first study addressing the frequencies of sperm chromosome structural aberrations of all autosomes in a single assay thus contributing to clarify the amount of damage present in human spermatozoa and in relation to age. The next challenge resides in the elucidation of the specific types of chromosome abnormalities that are produced at each stage of germ cell production, the relative contributions of each mechanism involved in the production of sperm with structural chromosome abnormalities, and how both processes are affected by age.
Acknowledgments
This research was supported by the Ministerio de Ciencia e Innovación, Spain (SAF2007-65913) and the Generalitat de Catalunya, Spain (2005FI00399, 2009SGR-01107).
The authors declare no conflict of interest.
References
- Jacobs PA. The chromosome complement of human gametes. Oxf Rev Reprod Biol. 1992;14:47–72. [PubMed] [Google Scholar]
- Hassold TJ. Nondisjunction in the human male. Curr Top Dev Biol. 1998;37:383–406. doi: 10.1016/s0070-2153(08)60181-7. [DOI] [PubMed] [Google Scholar]
- Olson SD, Magenis RE.Preferential paternal origin of de novo structural rearrangementsIn: Daniel A (ed): The Cytogenetics of Mammalian Autosomal Rearrangements New York: Alan R Liss; 1988583–599. [Google Scholar]
- Thomas NS, Durkie M, Van Zyl B, et al. Parental and chromosomal origin of unbalanced de novo structural chromosome abnormalities in man. Hum Genet. 2006;119:444–450. doi: 10.1007/s00439-006-0157-6. [DOI] [PubMed] [Google Scholar]
- Cody JD, Perce JF, Brkanac Z, et al. Preferential loss of the paternal alleles in the 18q- syndrome. Am J Hum Genet. 1997;69:280–286. doi: 10.1002/(sici)1096-8628(19970331)69:3<280::aid-ajmg12>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Dallapiccola B, Mandich P, Bellone E, et al. Parental origin of chromosome 4p deletions in Wolf-Hirschhorn syndrome. Am J Med Genet. 1993;47:921–924. doi: 10.1002/ajmg.1320470622. [DOI] [PubMed] [Google Scholar]
- Overhauser J, McMahon J, Oberlender S, et al. Parental origin of chromosome 5 deletions in the Cri-Du-Chat syndrome. Am J Med Genet. 1990;37:83–88. doi: 10.1002/ajmg.1320370119. [DOI] [PubMed] [Google Scholar]
- De Gregori M, Ciccone R, Magini P, et al. Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet. 2007;44:750–762. doi: 10.1136/jmg.2007.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, et al. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Bolor H, Kato T, et al. Recent advance in our understanding of the molecular nature of chromosomal abnormalities. J Hum Genet. 2009;54:253–260. doi: 10.1038/jhg.2009.35. [DOI] [PubMed] [Google Scholar]
- Ohye T, Inagaki H, Kogo H, et al. Paternal origin of the de novo constitutional t(11;22)(q23;q11) Eur J Hum Genet. 2010;18:783–787. doi: 10.1038/ejhg.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NS, Morris JK, Baptista J, Ng BL, Crolla JA, Jacobs PA. De novo apparently balanced translocations in man are predominantly paternal in origin and associated with a significant increase in paternal age. J Med Genet. 2010;47:112–115. doi: 10.1136/jmg.2009.069716. [DOI] [PubMed] [Google Scholar]
- Martin RH, Rademaker AW, Hildebrand K, Long-Simpson L, Peterson D, Yamamoto J. Variation in the frequency and type of sperm chromosomal abnormalities among normal men. Hum Genet. 1987;77:108–114. doi: 10.1007/BF00272374. [DOI] [PubMed] [Google Scholar]
- Brandriff BF, Gordon LA, Moore D, II, Carrano AV. An analysis of structural aberrations in human sperm chromosomes. Cytogenet Cell Genet. 1988;47:29–36. doi: 10.1159/000132500. [DOI] [PubMed] [Google Scholar]
- Estop AM, Marquez C, Munne S, et al. An analysis of human sperm breakpoints. Am J Hum Genet. 1995;56:452–460. [PMC free article] [PubMed] [Google Scholar]
- Templado C, Bosch M, Benet J. Frequency and distribution of chromosome abnormalities in human spermatozoa. Cytogenet Genome Res. 2005;111:199–205. doi: 10.1159/000086890. [DOI] [PubMed] [Google Scholar]
- Buwe A, Guttenbach M, Schmid M. Effect of paternal age on the frequency of cytogenetic abnormalities in human spermatozoa. Cytogenet Genome Res. 2005;111:213–228. doi: 10.1159/000086892. [DOI] [PubMed] [Google Scholar]
- Martin RH, Rademaker AW. The effect of age on the frequency of sperm chromosome abnormalities in normal men. Am J Hum Genet. 1987;41:484–449. [PMC free article] [PubMed] [Google Scholar]
- Sartorelli EM, Mazzucatto LF, de Pina-Neto JM. Effect of paternal age on human sperm chromosomes. Fertil Steril. 2001;76:1119–1123. doi: 10.1016/s0015-0282(01)02894-1. [DOI] [PubMed] [Google Scholar]
- Brandriff B, Gordon L, Ashworth L, et al. Chromosomes of human sperm: variability among normal individuals. Hum Genet. 1885;70:18–24. doi: 10.1007/BF00389451. [DOI] [PubMed] [Google Scholar]
- Rosenbusch B, Strehler E, Abt M, Sterzik K. Correlation between cytogenetic anomalies of human spermatozoa and sperm morphology as well as age of patients studied. Zentralbl Gynakol. 1993;115:113–116. [PubMed] [Google Scholar]
- Bosch M, Rajmil O, Egozcue J, Templado C. Linear increase of structural and numerical chromosome 9 abnormalities in human sperm regarding age. Eur J Hum Genet. 2003;11:754–759. doi: 10.1038/sj.ejhg.5201049. [DOI] [PubMed] [Google Scholar]
- Sloter ED, Marchetti F, Eskenazi B, et al. Frequency of human sperm carrying structural aberrations of chromosome 1 increases with advancing age. Fertil Steril. 2007;87:1077–1086. doi: 10.1016/j.fertnstert.2006.08.112. [DOI] [PubMed] [Google Scholar]
- Bosch M, Rajmil O, Egozcue J, Templado C. Linear increase of diploidy in human sperm with age: a four-colour FISH study. Eur J Hum Genet. 2001;9:533–538. doi: 10.1038/sj.ejhg.5200659. [DOI] [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- De la Chica RA, Ribas I, Giraldo J, Egozcue J, Fuster C. Chromosomal instability in amniocytes from fetuses of mothers who smoke. JAMA. 2005;293:1212–1222. doi: 10.1001/jama.293.10.1212. [DOI] [PubMed] [Google Scholar]
- Schmid TE, Brinkworth MH, Hill F, et al. Detection of structural and numerical chromosomal abnormalities by ACM-FISH analysis in sperm of oligozoospermic infertility patients. Hum Reprod. 2004;19:1395–1400. doi: 10.1093/humrep/deh278. [DOI] [PubMed] [Google Scholar]
- Van Hummelen P, Lowe XR, Wyrobek AJ. Simultaneous detection of structural and chromosome abnormalities in sperm of healthy men by multicolor fluorescence in situ hybridization. Hum Genet. 1996;98:608–615. doi: 10.1007/s004390050268. [DOI] [PubMed] [Google Scholar]
- Baumgartner A, Van Hummelen P, Lowe XR, Adler ID, Wyrobek AJ. Numerical and structural chromosome abnormalities detected in human sperm with a combination of multicolor FISH assays. Environ Mol Mutagen. 1999;33:49–58. doi: 10.1002/(sici)1098-2280(1999)33:1<49::aid-em6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Sloter E, Lowe X, Moore DI, Nath J, Wyrobek AJ. Multicolor FISH analysis of chromosomal breaks, duplications, deletions, and numerical abnormalities in the sperm of healthy men. Am J Hum Genet. 2000;67:862–872. doi: 10.1086/303088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroz L, Rajmil O, Templado C.Meiotic chromosome abnormalities in fertile men: Are they increasing Steril Fertil 2010. doi: 10.1016/j.fertnster2010.06.042 [DOI] [PubMed]
- Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12 (6:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- Kato T, Yamada K, Inagaki H, et al. Age has no effect on the de novo constitutional t(11;22) translocation frequency in sperm. Fertil Steril. 2007;88:1446–1448. doi: 10.1016/j.fertnstert.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken RJ, Koopman P, Lewis SEM. Seeds of concern. Nature. 2004;432:48–52. doi: 10.1038/432048a. [DOI] [PubMed] [Google Scholar]
- Fratarelli JL, Miller KA, Miller BT, Elkin-Hirsch J, Scott RT., Jr Male age negatively impacts embryo development and reproductive outcome in donor oocyte assisted reproductive technology cycles. Fertil Steril. 2008;90:97–103. doi: 10.1016/j.fertnstert.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Tarozzi N, Bizarro D, Flamigni C, Borini A. Clinical relevance of sperm DNA damage in assisted reproduction. Reprod Biomed Online. 2007;14:746–757. doi: 10.1016/s1472-6483(10)60678-5. [DOI] [PubMed] [Google Scholar]
- Singh NP, Muller CH, Berger RE. Effects of age on DNA double-strand breaks and apoptosis in human sperm. Fertil Steril. 2003;80:1420–1430. doi: 10.1016/j.fertnstert.2003.04.002. [DOI] [PubMed] [Google Scholar]
- Ravnan JB, Tepperberg JH, Papenhausen P, et al. Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J Med Genet. 2006;43:478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Regan R, Nicod A, et al. Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet. 1999;354 (9191:1676–1681. doi: 10.1016/S0140-6736(99)03070-6. [DOI] [PubMed] [Google Scholar]
- Martin CL, Waggoner DJ, Wong A, et al. Molecular rulers for calibrating phenotypic effects of telomere imbalance. J Med Genet. 2002;39:734–740. doi: 10.1136/jmg.39.10.734. [DOI] [PMC free article] [PubMed] [Google Scholar]