Abstract
Muscle strength is important in functional activities of daily living and the prevention of common pathologies. We describe the two-staged fine mapping of a previously identified linkage peak for knee strength on chr12q12-14. First, 209 tagSNPs in/around 74 prioritized genes were genotyped in 500 Caucasian brothers from the Leuven Genes for Muscular Strength study (LGfMS). Combined linkage and family-based association analyses identified activin receptor 1B (ACVR1B) and inhibin β C (INHBC), part of the transforming growth factor β pathway regulating myostatin – a negative regulator of muscle mass – signaling, for follow-up. Second, 33 SNPs, selected in these genes based on their likelihood to functionally affect gene expression/function, were genotyped in an extended sample of 536 LGfMS siblings. Strong associations between ACVR1B genotypes and knee muscle strength (P-values up to 0.00002) were present. Of particular interest was the association with rs2854464, located in a putative miR-24-binding site, as miR-24 was implicated in the inhibition of skeletal muscle differentiation. Rs2854464 AA individuals were ∼2% stronger than G-allele carriers. The strength increasing effect of the A-allele was also observed in an independent replication sample (n=266) selected from the Baltimore Longitudinal Study of Aging and a Flemish Policy Research Centre Sport, Physical Activity and Health study. However, no genotype-related difference in ACVR1B mRNA expression in quadriceps muscle was observed. In conclusion, we applied a two-stage fine mapping approach, and are the first to identify and partially replicate genetic variants in the ACVR1B gene that account for genetic variation in human muscle strength.
Keywords: complex trait, combined linkage and association analyses, family-based association, genotype/phenotype association
Introduction
Because of its beneficial health effects, skeletal muscle strength is an important trait, even in populations that live in modern societies and no longer rely heavily on physical labor. Indeed, both in the elderly and the general population, muscle strength is inversely associated with all-cause mortality,1 osteoporosis,2 cardiovascular diseases,3 metabolic syndrome prevalence4 and other pathological conditions and chronic diseases.5 Heritability estimates vary from 31 to 78% with large differences between muscle groups, contraction velocities or muscle lengths (reviewed in Peeters et al6). Apart from a genetic component, muscle strength is influenced by environmental factors such as nutrition, training or social status, and by the interaction between genes and environment, rendering it a complex multifactorial trait.
The ‘Human gene map for performance and health-related fitness phenotypes'7 indicates that only a limited number of genes has been implicated in muscle strength and even fewer associations have consistently been replicated. Genetic linkage studies regarding muscle strength are even scarcer. To date, only Tiainen et al8 and our research group9, 10, 11, 12, 13, 14 reported results on genetic linkage for skeletal muscle strength characteristics. We were the first to perform linkage analyses on a unique collection of young male Caucasian siblings drawn from the Leuven Genes for Muscular Strength study (LGfMS).9 Single and multipoint microsatellite marker-based linkage analyses revealed significant or suggestive linkage of chromosomal regions 12q12-14, 12q22-23 and 13q14.2 to knee muscle strength.9, 11 It can, however, not be excluded that genes other than the original myostatin pathway genes underlie these linkage findings and further fine mapping of the region surrounding these candidate genes is therefore warranted.
In this study, we further investigated the 12q12-14 region. We applied a two-staged genetic fine mapping approach, followed by a replication of the most promising associations and an evaluation of mRNA variation in relation to a ACVR1B SNP, in an independent study sample. We are the first to reveal evidence for variations in the ACVR1B gene to determine – at least in part – muscle strength properties. Our observations fit with available physiological data that demonstrate a potential role for ACVR1B in the regulation of muscle mass, as it is part of the transforming growth factor β (TGFβ) pathway regulating myostatin – a negative regulator of muscle mass – signaling.15, 16
Materials and methods
Study design
A two-staged approach was designed to fine map the chr12q12-14 linkage peak. The most significant findings were then replicated in an independent study sample and the influence of a specific SNP on mRNA expression levels in quadriceps muscle tissue was tested. A schematic representation of the study design can be found in Figure 1.
Figure 1.
Study design. The different stages of the study are displayed according to their chronological order from left to right. Sample description, genotyping details and statistical analyses are presented from top to bottom. LGfMS, Leuven Genes for Muscular Strength study; BLSA, Baltimore Longitudinal Study of Aging; SPAH, study conducted within the framework of the Flemish Policy Research Centre Sport, Physical Activity and Health.
Fine mapping stage 1
SNPs were selected under the 1-LOD confidence interval of the previously defined linkage peak,9, 11 using an empirical two-step strategy, in which candidate genes are prioritized using a bioinformatics approach, and the top genes are chosen for further SNP selection with a linkage disequilibrium (LD)-based method, as described in Supplementary file 1 (available online) and elsewhere.17 A list of the candidate genes and corresponding polymorphisms is available in Supplementary file 2 (available online) (Supplementary Table S1).
Joint linkage and association analyses, and family-based association analyses tested the association of these polymorphisms with knee strength measurements.
Fine mapping stage 2
Candidate genes were ranked on the basis of (1) their significance in (a) the joint linkage and association approach, (b) the family-based association approach and (c) both methods; (2) their significance over different strength measurements; (3) the knowledge about their function in muscle tissue (known, putative, general function in different tissues or no known/unclear function). The highest ranked gene ACVR1B was selected for further follow-up. INHBC was also included because it is part of the same TGFβ signaling pathway as ACVR1B, even though it had a lower ranking. An extended sample was genotyped for the stage 1 SNPs, complemented with additional tagSNPs and polymorphisms likely to have functional consequences, such as exonic SNPs, SNPs in intron/exon boundaries (possibility to influence the splicing process), in (putative) transcription factor-binding sites or in highly conserved regions (Table 1). Assessment of potential functionality was based on queries from SNPselector,18 SNPseek (http://snp.wustl.edu/cgi-bin/S NPseek/index.cgi) and publicly available genetic databases.
Table 1. Stage 2 genes and corresponding polymorphisms.
| SNP | No. | Coordinate | Minor allele | Minor allele frequency | Putative function |
|---|---|---|---|---|---|
| ACVR1B | |||||
| rs10431538 | 1 | 50605127 | A | 0.15 | Located in transcription factor-binding site (TFBS)/conserved |
| rs706824a | 2 | 50605518 | Located in TFBS | ||
| rs1979921 | 3 | 50606344 | T | 0.08 | Conserved |
| rs12230575a | 4 | 50608313 | Conserved | ||
| rs10783485 | 5 | 50622274 | A | 0.35 | Tagging SNP, genotyped stage 1 |
| rs746434 | 6 | 50628932 | A | 0.09 | Tagging SNP, genotyped stage 1 |
| rs808873a | 7 | 50634449 | C | 0 | Located in TFBS/conserved |
| rs11612312 | 8 | 50635355 | A | 0.22 | Conserved |
| rs12809597a | 9 | 50642590 | Conserved | ||
| rs10783486b | 10 | 50649053 | A | 0.27 | Tagging SNP, genotyped stage 1 |
| rs34488074c | 11 | 50656482 | C | 0 | Non-synonymous SNP (L/F) |
| rs2172603 | 12 | 50663079 | A | 0.02 | Located in TFBS |
| rs12296553a | 13 | 50663569 | Located in TFBS | ||
| rs2641516c | 14 | 50666953 | C | 0 | Synonymous SNP (G/G)/conserved/located in TFBS |
| rs928906a | 15 | 50666954 | Located in TFBS/non-synonymous (V/L) | ||
| rs34050429c | 16 | 50667165 | A | 0 | Non-synonymous SNP (H/R) |
| rs2854464 | 17 | 50675158 | G | 0.27 | Located in TFBS/miRNA-binding site |
| rs2714 | 18 | 50675551 | A | 0.10 | Conserved |
| rs11169974 | 19 | 50676563 | T | 0.04 | Located in TFBS |
| rs7955401a | 20 | 50685018 | Located in TFBS | ||
| INHBC | |||||
| rs7964492 | 1 | 56109852 | C | 0.25 | Tagging SNP/conserved |
| rs12826906 | 2 | 56114925 | G | 0.006 | 5′ UTR |
| rs12831855c | 3 | 56114969 | T | 0 | Synonymous SNP (L/L)/conserved |
| rs12809642c | 4 | 56115002 | G | 0 | Synonymous SNP (A/A) |
| rs560048 | 5 | 56115396 | T | 0.45 | Regulatory region intron 1 |
| rs533975 | 6 | 56118251 | A | 0.45 | Tagging SNP/conserved |
| rs2943693 | 7 | 56122232 | A | 0.22 | Tagging SNP, genotyped stage 1 |
| rs34137943c | 8 | 56129647 | T | 0 | Synonymous SNP (L/L) |
| rs2229357 | 9 | 56129978 | A | 0.26 | Non-synonymous SNP (Q/R), genotyped stage 1 |
| rs3741414 | 10 | 56130316 | A | 0.26 | Tagging SNP, genotyped stage 1 |
| rs34435057c | 11 | 56130855 | T | 0 | 3′ UTR |
| rs11172223c | 12 | 56132314 | A | 0 | 3′ conserved SNP |
| rs7136620c | 13 | 56132489 | A | 0 | 3′ conserved SNP |
The physical location of each SNP is based on NCBI Build 36. The putative function was derived from SNPselector and SNPseek output. All SNP genotype distributions are in Hardy–Weinberg equilibrium (P>0.001).
Failed iPLEX assay.
Failed iPLEX assay but also genotyped in stage 1.
Monomorphic SNPs.
Replication
Genetic linkage and association analyses are prone to false-positive findings. Therefore, we applied a staged design, used complementary association methods and derived empirical P-values. Moreover, we replicated the associations with putatively functional polymorphisms, detected in the second stage of the fine mapping, in an independent sample.
Study samples
Two-staged fine mapping
Siblings analyzed in stage 1 and stage 2 of this study were selected from the LGfMS (748 men; 17–36 years), based on family size and DNA availability. The recruitment protocol and subject characteristics have been described elsewhere.9, 11 For stage 1, a total of 500 individuals were included (169 subjects overlap with the microsatellite-based linkage study11). For stage 2, a total of 536 subjects were included with an overlap of 464 subjects with stage 1. Subjects for whom genotyping failed in stage 1 were excluded from stage 2, and subjects for whom new DNA was collected between the two stages were included. Descriptive statistics of these samples can be found in Supplementary file 2 (Supplementary Table S2).
Replication sample
The replication sample consists of 193 Caucasian men (20–90 years) from the Baltimore Longitudinal Study of Aging (BLSA)19, 20, 21 and 74 healthy Caucasian men (60–78 years) from a study conducted within the framework of the Flemish Policy Research Centre Sport, Physical Activity and Health (SPAH).22, 23
The medical and ethical committee of the Katholieke Universiteit Leuven approved the LGfMS and SPAH studies. The BLSA experimental protocols were approved by the Institutional Review Boards for Human Subjects at Medstar Research Institute (Baltimore, MD, USA), Johns Hopkins Bayview Medical Center (Baltimore, MD, USA) and the University of Maryland (College Park, MD, USA). All subjects gave written informed consent before participating. Descriptive statistics of the replication subsamples can be found in Supplementary file 2 (Supplementary Tables S2 and S3).
Strength measurements
Isometric and dynamic strength of the knee extensors and flexors was measured using an isokinetic dynamometer as described in Supplementary file 1 and elsewhere (LGfMS,9, 11 BLSA20, 21 and SPAH23).
SNP genotyping
Detailed SNP genotyping methods can be found in Supplementary file 1. In short, DNA was extracted from whole blood (LGfMS, BLSA, SPAH) or from saliva (Oragene DNA Self-Collection Kits; Oragene, Ontario, Canada; LGfMS). Genotyping was performed on an Illumina Bead Array platform (Illumina Inc., San Diego, CA, USA; LGfMS stage 1), a Sequenom iPLEX Gold platform (Sequenom Inc., San Diego, CA, USA; LGfMS stage 2 and SPAH) or with Taqman allelic discrimination assays (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA; BLSA). Overall sample success rates, locus success rates and genotype call rates can be found in Supplementary file 1.
Quantification of ACVR1B mRNA expression
Muscle tissue (right vastus lateralis) was obtained from 16 healthy male volunteers (18–28 years) from an independent study conducted in our research center.24 A detailed description of the mRNA extraction and real-time PCR protocol for ACVR1B mRNA expression is available in Supplementary file 1 and elsewhere.25
Statistical analyses
Pedstats v0.6.426 was used to check pedigrees for Mendelian consistency and to test Hardy–Weinberg equilibrium (HWE). Unlikely genotypes were identified and discarded using the error checking option of Merlin v1.0.1.27
QTDT v2.5.1 software28 was applied to compare a test for linkage between the SNPs and the strength measurements with a test for linkage while modeling association.29 When association between a SNP and the studied trait is present, the linkage signal should become nonsignificant when association is added to the model. Merlin v1.0.127 was used to calculate IBD probabilities. Genetic distances were derived from physical distances (NCBI build 35) using Rutgers Map Interpolator based on Rutgers first-generation combined linkage-physical map (Rutgers map v.1).30
Haploview31 was used to examine the LD structure across the region of interest and to calculate r2 between all pairs of SNPs within a candidate gene.
Complementary to the combined linkage and association analysis, we conducted family-based association tests (FBAT v1.7.3) under the null hypothesis of ‘linkage and no association'.32, 33 The haplotype version of this test (HBAT)34 was used to obtain empirical P-values by means of the Monte-Carlo permutation procedure (10 000 permutations). An additive model was tested and trait offsets were specified as the sample mean of the trait. As only empirical P-values are reported, we should note that these were derived under the null hypothesis of ‘no linkage and no association' since permutation tests are too computer-intensive under the alternative hypothesis. However, simulations show that tests under both null hypotheses render comparable results (data not shown).
SAS v9.1.3 (SAS Institute, Cary, NC, USA) was used to calculate descriptive statistics and to assess genotypic differences in muscle strength in the replication sample and in ACVR1B mRNA expression using analyses of (co)variance. Because the strength testing protocols between the replication samples differed (eg, in equipment, angle for the isometric test, measurement units), strength measurements were normalized to Z-scores (mean=0; standard deviation=1) per sample. Similar strength measurements were combined in the joined sample as described in Supplementary file 2, Supplementary Table S3. Because of the large age range of the replication individuals, age was included as a covariate in the analyses.
Probability levels <0.05 are considered significant. Instead of correcting P-values for multiple testing, we relied on the use of complementary association analysis methods, calculation of empirical P-values and a staged design including replication in an independent sample to minimize the possibility of false-positive findings.
Results
Fine mapping stage 1
In stage 1, candidate genes and SNPs in the 1-LOD region of the original chr12q12-14 linkage peak were selected using an empiric two-step approach. First, a bioinformatical prioritization approach was used to select 86 candidate genes out of the 454 genes in the region. Second, 181 tagging SNPs, determined on CEPH (Centre d'Etude du Polymorphisme Humain) genotypes downloaded from the Hapmap35 website, were selected in 74 candidate genes. For the remaining 12 genes, no polymorphisms meeting our criteria (Illumina designability rank=1 (ie, high success rate of assay design); MAF>0.05) were present. These polymorphisms were supplemented with 28 SNPs in the gaps in-between the genes, to enhance power for linkage.17 A list of the selected SNPs is available in Supplementary file 2 (Supplementary Table S1).
Of the 209 SNPs, 198 were successfully genotyped, of which 7 SNPs were discarded (2 were monomorphic and 5 departed from HWE at the P<0.001 level), yielding 191 SNPs for further analysis.
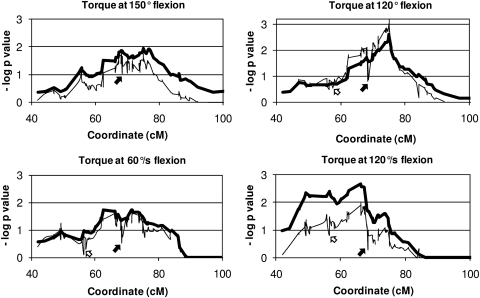
These SNPs were successfully genotyped in a sample of 499 young male siblings. Association between a SNP and the studied muscle strength characteristics was evaluated using a combined linkage and association approach.29 Consistent drops in –log P-values after modeling a joint test for linkage and association – indicating the presence of association between SNP and trait – were present for rs10784948 in contactin 1 (CNTN1) and rs7960176 in YY1 associated factor 2 (YAF2; Figure 2, open arrow) and for rs10783485 and rs10783486 in activin receptor 1B (ACVR1B; Figure 2, black arrow).
Figure 2.
Combined linkage and association analyses on selected strength measurements. A drop in –log P-value for the combined linkage and association analysis (thin line) compared to the linkage analysis (thick line) indicates evidence for association. Associated SNPs rs10783485 and rs10783486 in ACVR1B are marked with a black arrow and associated SNPs rs10784948 (CNTN1) and rs7960176 (YAF2) are marked with an open arrow.
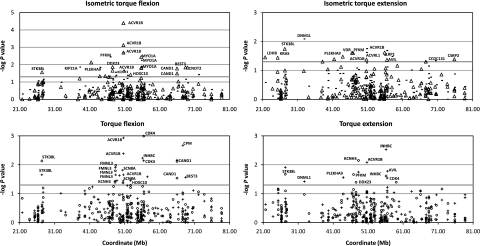
In addition, family-based association analyses were performed. Figure 3 shows the empirical P-values for an additive model for the 191 SNPs. SNPs in five genes (ACVR1B, cyclin-dependent kinase 4 (CDK4), cullin-associated and neddylation-dissociated 1 (CAND1), inhibin β C (INHBC) and myosin IA (MYO1A)) were repeatedly highly suggestively associated (P<0.01 or −log P>2) with different strength measurements.
Figure 3.
Family-based association results for selected strength measurements using single SNP markers. Empirical P-values from HBAT analyses were calculated after 10 000 permutations. Strength measurements at 150° (bold line), at 120° (triangle), at 60°/s (cross) and at 120°/s (circle) are shown. Most suggestive significant results (P<0.05, −log P-value >1.3) were marked with associated genes.
Stage 2 candidate genes were ranked on the basis of significance in the different association tests, significance over different strength measurements and (putative) functionality in muscle tissue. ACVR1B got the highest ranking because of its high significance in both the combined linkage and association approach (Figure 2) and the family-based association analyses (Figure 3), and because the ACVR1B protein has a role in the molecular TGFβ pathway regulating the signaling of myostatin, a negative regulator of skeletal muscle mass.15, 16 Even though it ranked lower, INHBC was also included for follow-up because it is a part of the same superfamily of proteins as ACVR1B.
Fine mapping stage 2
In stage 2, 20 and 13 polymorphisms were selected within ACVR1B and INHBC, respectively, (Table 1 and Supplementary Figures S1 and S2; available in Supplementary file 2) based on their probability to influence ACVR1B and INHBC gene function or expression. Of these 33 SNPs, 7 failed during analyses (including rs10783486, a stage 1 tagSNP), and 10 were 100% homozygous. The resulting 16 informative SNPs were genotyped in 536 siblings. Genotype distributions for all successfully genotyped polymorphisms were in HWE (P>0.001).
ACVR1B
Family-based association results for the ACVR1B SNPs and knee muscle strength can be found in Table 2. For rs10783485 and rs746434 (two tagSNPs), the association results from stage 1 were confirmed in the extended sample (stage 2: N=536 versus stage 1: N=499; overlap of 464 individuals) with significant associations for most of the knee flexion strength and some of the extension strength parameters (0.00002<P<0.039). No additional subjects were genotyped for rs10783486, so stage 1 results are presented in Table 2. The conserved SNP rs11612312 and SNP rs2854464, located in a putative miRNA-binding site, both showed an association with the dynamic knee flexion and extension measurements at 60°/s. Additionally, the polymorphisms show (a trend toward) significance for isometric strength.
Table 2. Empirical P-values for association of ACVR1B polymorphisms and selected strength measurements.
| Marker | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs10431538 | rs1979921 | rs10783485 | rs746434 | rs11612312 | rs10783486 | rs2172603 | rs2854464 | rs2714 | rs11169974 | |
| Strength increasing allele | C | G | T | G | A | |||||
| Knee flexion | ||||||||||
| Isometric at 150°, Nm | 0.84 | 0.63 | 0.028 | 0.067 | 0.034 | 0.0017 | 0.50 | 0.083 | 0.94 | 0.95 |
| Isometric at 120°, Nm | 0.26 | 0.15 | 0.0035 | 0.0038 | 0.00084 | 0.00002 | 0.90 | 0.0083 | 0.67 | 0.82 |
| Torque at 60°/s, Nm | 0.15 | 0.39 | 0.0069 | 0.023 | 0.0038 | 0.00066 | 0.58 | 0.029 | 0.50 | 0.91 |
| Torque at 120°/s, Nm | 0.67 | 0.35 | 0.053 | 0.13 | 0.076 | 0.073 | 0.60 | 0.23 | 0.57 | 0.76 |
| Knee extension | ||||||||||
| Isometric at 150°, Nm | 0.25 | 0.88 | 0.54 | 0.025 | 0.10 | 0.034 | 0.85 | 0.045 | 0.46 | 0.51 |
| Isometric at 120°, Nm | 0.84 | 0.76 | 0.30 | 0.19 | 0.052 | 0.039 | 0.88 | 0.033 | 0.37 | 0.88 |
| Torque at 60°/s, Nm | 0.34 | 0.098 | 0.10 | 0.18 | 0.0076 | 0.0075 | 0.43 | 0.011 | 0.89 | 0.68 |
| Torque at 120°/s, Nm | 0.52 | 0.47 | 0.12 | 0.057 | 0.13 | 0.074 | 0.29 | 0.14 | 0.88 | 0.92 |
Significant P-values are marked in bold.
INHBC
Only two of four selected tagSNPs showed limited levels of association with muscle strength (Table 3). Marker rs533975 was associated with torque flexion at 60°/s and 120°/s (P<0.049) and rs2943693 with torque flexion at 60°/s, and torque extension at both 60°/s and 120°/s (P<0.021). None of the other SNPs showed association with isometric or dynamic knee torque.
Table 3. Empirical P-values for association of INHBC polymorphisms and selected strength measurements.
| Marker | |||||||
|---|---|---|---|---|---|---|---|
| rs7964492a | rs12826906 | rs560048 | rs533975a | rs2943693a | rs2229357 | rs3741414a | |
| Strength increasing allele | T | A | A | ||||
| Knee flexion | |||||||
| Isometric at 150°, Nm | 0.52 | 0.68 | 0.17 | 0.27 | 0.43 | 0.35 | 0.32 |
| Isometric at 120°, Nm | 0.60 | 0.63 | 0.40 | 0.49 | 0.57 | 0.40 | 0.39 |
| Torque at 60°/s, Nm | 0.70 | 0.14 | 0.068 | 0.047 | 0.040 | 0.72 | 0.61 |
| Torque at 120°/s, Nm | 0.70 | 0.86 | 0.056 | 0.048 | 0.11 | 0.73 | 0.76 |
| Knee extension | |||||||
| Isometric at 150°, Nm | 0.095 | 0.68 | 0.092 | 0.066 | 0.57 | 0.25 | 0.22 |
| Isometric at 120°, Nm | 0.23 | 0.60 | 0.096 | 0.16 | 0.58 | 0.12 | 0.14 |
| Torque at 60°/s, Nm | 0.66 | 0.47 | 0.19 | 0.15 | 0.0039 | 0.43 | 0.49 |
| Torque at 120°/s, Nm | 0.50 | 0.64 | 0.41 | 0.23 | 0.021 | 0.39 | 0.42 |
Significant P-values are marked in boldj;
Four selected tagging SNPs.
Replication analyses
We replicated the associations with putatively functional polymorphisms, detected in the second stage of the fine mapping, in men selected from two independent studies. Given the limited evidence for association for the INHBC gene, we focused the replication effort on rs10783485, rs10783486 and rs2854464 in ACVR1B.
All three polymorphisms were genotyped in 192 men from the BLSA study.19 In the SPAH individuals22 (n=74), only rs10783485 and rs2854464 were genotyped because of the technical difficulties. All genotype distributions were in HWE (P>0.001).
Replication analyses tested whether individuals homozygous for the strength increasing allele – as determined in stage 2 of the fine mapping – differed significantly from other individuals.
Because of testing protocol differences, strength measurements were normalized per sample before the analyses. Age was included as a covariate in the analyses because of the large age range of the individuals. Rs10783485 only showed significant association with moderate velocity torque of the knee extensors (GG: 0.074±0.079 versus GT+TT: −1.15±0.070; P=0.38). Individuals homozygous for the rs2854464 strength increasing A allele had significantly higher dynamic knee extensor strength than GG and AG individuals (all P<0.036; Table 4).
Table 4. Muscle strength characteristics according to rs2854464 genotype in the combined sample.
| AA | AG+GG | ||
|---|---|---|---|
| N | 81 | 64 | P-value |
| Knee flexion | |||
| Isometric torque at 120° | 0.14±0.15 | −0.19±0.18 | 0.17 |
| Slow velocity torque | 0.021±0.094 | 0.062±0.11 | 0.77 |
| Moderate velocity torque | −0.028±0.096 | −0.0070±0.11 | 0.88 |
| Knee extension | |||
| Isometric torque at 140–150° | −0.015±0.099 | 0.087±0.11 | 0.50 |
| Isometric torque at 120° | 0.061±0.096 | 0.0077±0.11 | 0.72 |
| Slow velocity torque | 0.13±0.080 | −0.12±0.086 | 0.036 |
| Moderate velocity torque | 0.16±0.078 | −0.17±0.085 | 0.0053 |
All values are least square mean±SEM from an analysis of covariance with age as covariate; significant P-values are marked in bold.
Quantification of mRNA expression
Effects at the mRNA level were tested only for rs2854464, located in a putative miR-24-binding site 985–1011 bp downstream of the stop codon of the ACVR1B gene, and inducing a higher strength in AA compared with AG+GG individuals (Tables 2, 3 and 4).
We genotyped this SNP in a limited sample (N=16) from an independent study conducted in our research center from which muscle tissue biopsies were available.24 Relative expression levels were 0.93±0.18 and 0.95±0.25 for AA homozygotes and G-allele carriers, respectively, and did not differ significantly (P=0.80).
Discussion
We described the results of a two-staged gene-centered fine mapping approach applied on a previously identified linkage peak in the 12q12-14 region, and identified genetic variants in the activin receptor 1B (ACVR1B) gene to be associated with human muscle strength in a sample form the Leuven Genes for Muscular Strength study. A replication effort in a (size-limited) sample consisting of 266 men from two independent studies partly supports this association.
This is the first report that demonstrates a possible genetic link between knee strength and ACVR1B, which is a member of the TGFβ superfamily of proteins – as is myostatin36 – and is known to be involved in the molecular pathway regulating myostatin and activin signaling. These signal pathways are triggered by the binding of the ligand to a type II receptor, the recruitment of a type I receptor and the formation of a heteromeric, active receptor complex. The type I receptor is the center of the signaling as it is essential for signal specificity as well as for signal propagation beyond the plasma membrane.15, 16
Cross-linking studies demonstrated that activin receptor IIB (ACVR2B) is the primary type II receptor for myostatin.37, 38 Blocking the activity of this receptor in mice models leads to dramatic increases in muscle mass, comparable with those seen in myostatin knock out mice.37, 39 Additional evidence for a role of activin receptor signaling in determination of human muscle mass and strength comes from Walsh et al,40 who showed a genetic association between ACVR2B and follistatin variants and skeletal muscle mass and strength. Moreover, resistance training could downregulate ACVR2B expression, suggesting a role for ACVR2B and the activin pathway in muscle response to strength training.41, 42
Although it has been recognized that activin receptors are important in myostatin signaling, published studies have focused on the initial binding of myostatin to ACVR2B, whereas no studies reported on the subsequent steps of the signaling cascade. Cross-linking studies, however, showed that after binding of myostatin to ACVR2B, a type I receptor (ACVR1B (ALK4) or TGFβ receptor 1 (ALK5)) is recruited.38
Combined linkage and association, and family-based association results show that genetic variation in ACVR1B can influence human muscle strength. Of particular interest is the association between rs2854464 and muscle strength, with ∼2% lower knee strength for G-allele carriers compared to AA homozygotes in the LGfMS sample. Follow-up replication analyses in an independent but size-limited study sample also show an enhanced strength for AA homozygotes with regard to dynamic knee extensor strength.
The rs2854464 polymorphism is located in a putative miR-24-binding site in the 3′ untranslated region (UTR) of the ACVR1B mRNA. Wang et al43 showed that miR-24 could decrease human ACVR1B expression at mRNA and protein levels. Published evidence shows that genetic variation might influence organismal phenotypes44, 45 by perturbing miRNA-mediated gene regulation. Similarly, we hypothesize that the rs2854464 polymorphism affects muscle strength by interfering with miRNA binding.
Recently, Sun et al46 showed that this miR-24 is involved in the inhibition of skeletal muscle differentiation by TGFβ. Even though miR-24 is not a muscle-specific miRNA, they suggested that miR-24 might function during differentiation and homeostatic maintenance of cardiac and skeletal muscle tissue. We hypothesize that the effect of miR-24 on TGFβ signaling is mediated through a miR-24-binding site in ACVR1B, one of the type I receptors important for TGFβ as well as myostatin signaling.
It should be noted that the significances found for several of the selected ACVR1B SNPs are not completely independent findings, as these SNPs are in (high) LD with each other. The rs2854464 polymorphism is part of a haplotype block consisting of rs746434, rs11612312, rs10783486, rs2172603 and rs2854464 (determined using the four gamete rule in Haploview31). Haplotype analyses using HBAT software34 confirm the single SNP analyses (data not shown). Although rs10783485 is not part of the same haplotype block, it does show considerable LD with SNPs in the haplotype block (r2 range 0.15–0.44) and the observed significant results could therefore be related to rs2854464.
Even though a set of significant association results was observed for INHBC gene variants, these were not consistent over the different strength measurements. Therefore, these associations were not considered to be compelling evidence for a role of INHBC gene variants in determining muscle strength variation.
We find less significant results for associations at higher contraction velocities and different influences on knee flexors and extensors, suggesting a velocity and/or muscle group specific set of genes.11 This is consistent with the observation that the importance of genetic factors decreases with increasing velocities in concentric torques due to genetic variation in contractile and elastic components, which contribute differently to strength production at different velocities.47
It could be argued that the currently applied two-stage strategy infers a bias toward genes with a known function in (the regulation of) muscle strength. Indeed, in both stages literature knowledge was used to prioritize genes for further follow-up. In addition, it should be noted that not all genetic variation in the linkage region was captured using our stage 1 SNP selection method because we only genotyped a limited set of SNPs with a MAF>5%. It can therefore not be ruled out that genes and/or polymorphisms other than the ones we investigated are involved in the linkage signal. Moreover, genes for which no significant associations were found should not be considered negatively associated as not all SNPs within these genes were covered (eg, rare alleles). However, we were aware of these limitations from the beginning of our study and acknowledge that additional research will be necessary to determine which other genes/polymorphisms, in addition to ACVR1B, could be responsible for the linkage signal. These analyses should take into account that since the onset of this study a considerable amount of new information about human genetic variation has become available (eg, the 1000 genomes project; http://www.1000genomes.org).
In summary, we conducted a genetic fine mapping to identify variations in specific genes underlying a previously reported linkage peak for knee muscle strength on chr12q12-14. We are the first to describe, and replicate, a genetic link of ACVR1B, a receptor involved in the myostatin signaling pathway, to the determination of variation in muscle strength phenotypes. A SNP, rs2854644, located within a putative miR-24-binding site in the 3′ UTR of the ACVR1B gene, was found to be significantly associated with human knee muscle strength in three independent non-athletes samples from a wide age range.
Acknowledgments
AW and WH were funded by the Research Fund of the KU Leuven (OT/04/44 and OT/98/39, respectively). The Research Foundation Flanders (FWO) funded GDM by grant G.0496.05 and MP as post-doctoral researcher. The fine mapping phase of the LGfMS is funded by OT/04/44 and FWO grant G.0496.05. The BLSA research was conducted as a component of the Intramural Research Program of the National Institute on Aging and further supported by AG022791 from the National Institutes of Health. Strength phenotyping of the SPAH cohort was supported by the Flemish Government in the Flemish Policy Research Centre Sport, Physical Activity and Health and genotyping by a FWO Research grant to Martine Thomis. We thank Ivo Salden for his support during the preparation of DNA samples, Ruben van ‘t Slot, Bart Claes, Gilian Peuteman, Ricardo Lima and Andrew Ludlow for the genotyping, Monique Ramaekers and Els Van den Eede for assistance in the biopsy study and Karolina Szlufcik for support during the mRNA expression analyses. We would like to dedicate this paper to Gunther De Mars, who deceased on 28/02/2010 (age 34) and greatly contributed to the LGfMS study and to earlier versions of this manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Rantanen T, Harris T, Leveille SG, et al. Muscle strength and body mass index as long-term predictors of mortality in initially healthy men. J Gerontol A Biol Sci Med Sci. 2000;55:M168–M173. doi: 10.1093/gerona/55.3.m168. [DOI] [PubMed] [Google Scholar]
- Karasik D, Kiel DP. Genetics of the musculoskeletal system: a pleiotropic approach. J Bone Miner Res. 2008;23:788–802. doi: 10.1359/jbmr.080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HM, Alpert JS, Fain M. Frailty, inflammation, and cardiovascular disease: evidence of a connection. Am J Geriatr Cardiol. 2008;17:101–107. [PubMed] [Google Scholar]
- Wijndaele K, Duvigneaud N, Matton L, et al. Muscular strength, aerobic fitness, and metabolic syndrome risk in Flemish adults. Med Sci Sports Exerc. 2007;39:233–240. doi: 10.1249/01.mss.0000247003.32589.a6. [DOI] [PubMed] [Google Scholar]
- Wolfe RR. The underappreciated role of muscle in health and disease. Am J Clin Nutr. 2006;84:475–482. doi: 10.1093/ajcn/84.3.475. [DOI] [PubMed] [Google Scholar]
- Peeters MW, Thomis MA, Beunen GP, Malina RM. Genetics and sports: an overview of the pre-molecular biology era. Med Sport Sci. 2009;54:28–42. doi: 10.1159/000235695. [DOI] [PubMed] [Google Scholar]
- Bray MS, Hagberg JM, Perusse L, et al. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc. 2009;41:35–73. doi: 10.1249/mss.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- Tiainen KM, Perola M, Kovanen VM, et al. Genetics of maximal walking speed and skeletal muscle characteristics in older women. Twin Res Hum Genet. 2008;11:321–334. doi: 10.1375/twin.11.3.321. [DOI] [PubMed] [Google Scholar]
- Huygens W, Thomis MA, Peeters MW, et al. Linkage of myostatin pathway genes with knee strength in humans. Physiol Genomics. 2004;17:264–270. doi: 10.1152/physiolgenomics.00224.2003. [DOI] [PubMed] [Google Scholar]
- Huygens W, Thomis MA, Peeters MW, et al. A quantitative trait locus on 13q14.2 for trunk strength. Twin Res. 2004;7:603–606. doi: 10.1375/1369052042663850. [DOI] [PubMed] [Google Scholar]
- Huygens W, Thomis MAI, Peeters MW, Aerssens J, Vlietinck R, Beunen GP. Quantitative trait loci for human muscle strength: linkage analysis of myostatin pathway genes. Physiol Genomics. 2005;22:390–397. doi: 10.1152/physiolgenomics.00010.2005. [DOI] [PubMed] [Google Scholar]
- De Mars G, Windelinckx A, Huygens W, et al. Genome-wide linkage scan for maximum and length-dependent knee muscle strength in young men: significant evidence for linkage at chromosome 14q24.3. J Med Genet. 2008;45:275–283. doi: 10.1136/jmg.2007.055277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mars G, Windelinckx A, Huygens W, et al. Genome-wide linkage scan for contraction velocity characteristics of knee musculature in the Leuven Genes for Muscular Strength Study. Physiol Genomics. 2008;35:36–44. doi: 10.1152/physiolgenomics.90252.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomis M, De Mars G, Windelinckx A, et al. Genome-wide linkage scan for resistance to muscle fatigue Scand J Med Sci Sports 2010. e-pub ahead of print 10 March 2010, doi: 10.1111/j.1600-0838.2009.01082.x [DOI] [PubMed]
- Joulia-Ekaza D, Cabello G. The myostatin gene: physiology and pharmacological relevance. Curr Opin Pharmacol. 2007;7:310–315. doi: 10.1016/j.coph.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kollias HD, McDermott JC. Transforming growth factor-{beta} and myostatin signaling i. J Appl Physiol. 2008;104:579–587. doi: 10.1152/japplphysiol.01091.2007. [DOI] [PubMed] [Google Scholar]
- Windelinckx A, Vlietinck R, Aerssens J, Beunen G, Thomis MA. Selection of genes and single nucleotide polymorphisms for fine mapping starting from a broad linkage region. Twin Res Hum Genet. 2007;10:871–885. doi: 10.1375/twin.10.6.871. [DOI] [PubMed] [Google Scholar]
- Xu H, Gregory SG, Hauser ER, et al. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics. 2005;21:4181–4186. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock N, Gruelich R, Andres R, et al. Normal Human Aging. The Baltimore Longitudinal Study of Aging. US Government Printing Office, Washington, DC; 1984. [Google Scholar]
- Lindle RS, Metter EJ, Lynch NA, et al. Age and gender comparisons of muscle strength in 654 women and men aged 20–93 yr. J Appl Physiol. 1997;83:1581–1587. doi: 10.1152/jappl.1997.83.5.1581. [DOI] [PubMed] [Google Scholar]
- Lynch NA, Metter EJ, Lindle RS, et al. Muscle quality. I. Age-associated differences between arm and leg muscle groups. J Appl Physiol. 1999;86:188–194. doi: 10.1152/jappl.1999.86.1.188. [DOI] [PubMed] [Google Scholar]
- Matton L, Beunen G, Duvigneaud N, et al. Methodological issues associated with longitudinal research: findings from the Leuven Longitudinal Study on Lifestyle, Fitness and Health (1969–2004) J Sports Sci. 2007;25:1011–1024. doi: 10.1080/02640410600951563. [DOI] [PubMed] [Google Scholar]
- Windelinckx A, De Mars G, Beunen G, et al. Polymorphisms in the vitamin D receptor gene are associated with muscle strength in men and women. Osteoporos Int. 2007;18:1235–1242. doi: 10.1007/s00198-007-0374-4. [DOI] [PubMed] [Google Scholar]
- Vincent B, De Bock K, Ramaekers M, et al. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol Genomics. 2007;32:58–63. doi: 10.1152/physiolgenomics.00173.2007. [DOI] [PubMed] [Google Scholar]
- Vincent B, Windelinckx A, Nielens H, et al. Protective role of {alpha}-actinin-3 in the response to an acute eccentric exercise bout. J Appl Physiol. 2010;109:564–573. doi: 10.1152/japplphysiol.01007.2009. [DOI] [PubMed] [Google Scholar]
- Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64:259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19 (Suppl 1:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth fac. Mol Cell Biol. 2003;23:7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ. Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS One. 2007;2:e789. doi: 10.1371/journal.pone.0000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S, Metter EJ, Ferrucci L, Roth SM. Activin-type II receptor B (ACVR2B) and follistatin haplotype associations with muscle mass and strength in humans. J Appl Physiol. 2007;102:2142–2148. doi: 10.1152/japplphysiol.01322.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulmi JJ, Ahtiainen JP, Kaasalainen T, et al. Postexercise myostatin and activin IIb mRNA levels: effects of strength training. Med Sci Sports Exerc. 2007;39:289–297. doi: 10.1249/01.mss.0000241650.15006.6e. [DOI] [PubMed] [Google Scholar]
- Kim JS, Petrella JK, Cross JM, Bamman MM. Load-mediated downregulation of myostatin mRNA is not sufficient to promote myofiber hypertrophy in humans: a cluster analysis. J Appl Physiol. 2007;103:1488–1495. doi: 10.1152/japplphysiol.01194.2006. [DOI] [PubMed] [Google Scholar]
- Wang Q, Huang Z, Xue H, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- Georges M, Coppieters W, Charlier C. Polymorphic miRNA-mediated gene regulation: contribution to phenotypic variation and disease. Curr Opin Genet Dev. 2007;17:166–176. doi: 10.1016/j.gde.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Callis TE, Chen JF, Wang DZ. MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol. 2007;26:219–225. doi: 10.1089/dna.2006.0556. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhang Y, Yang G, et al. Transforming growth factor-{beta}-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomis MA, Van Leemputte M, Maes HH, et al. Multivariate genetic analysis of maximal isometric muscle force at different elbow angles. J Appl Physiol. 1997;82:959–967. doi: 10.1152/jappl.1997.82.3.959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.