Abstract
Surfactant protein-A (SP-A) plays an important role in the clearance of surfactant from the lung alveolar space and in the regulation of surfactant secretion and uptake by type II pneumocytes in culture. Two pathways are important for the endocytosis of surfactant by type II cells and the intact lung, a receptor-mediated clathrin-dependent pathway and a non-clathrin, actin-mediated pathway. The critical role of the clathrin/receptor-mediated pathway in normal mice is supported by the finding that SP-A gene-targeted mice use the actin-dependent pathway to maintain normal clearance of surfactant. Addition of SP-A to the surfactant of the SP-A null mice “rescued” the phenotype, further emphasizing the essential role of the SP-A/receptor-mediated process in surfactant turnover. This review presents an overview of the structure of SP-A and its function in surfactant turnover. The evidence that the interaction of SP-A with type II cells is a receptor-mediated process is presented. A newly identified receptor for SP-A, P63/CKAP4, is described in detail, with elucidation of the specific structural features of this 63 kDa, nonglycosylated, highly coiled, transmembrane protein. The compelling evidence that P63 functions as a receptor for SP-A on type II cells is summarized. Regulation of P63 receptor density on the surface of pneumocytes may be a novel approach for the regulation of surfactant homeostasis by the lung.
Key Words: Lung surfactant, Surfactant protein-A, P63/CKAP4, Type II alveolar pneumocytes, Receptors, Receptor-mediated interactions
Introduction
Pulmonary surfactant is a complex mixture of lipids and specific surfactant proteins that forms a thin film over the cells lining the alveolar space. Surfactant functions by reducing surface tension at the air-liquid interface and thereby serves to prevent collapse of the alveoli, allowing even ventilation of the lung. The lipid/protein material contains predominantly phospholipids, especially dipalmitoylphosphatidylcholine (DPPC), with approximately 10% protein. Surfactant proteins consist of surfactant proteins-A (SP-A), -B, -C, and -D with SP-A being the most abundant surfactant-associated protein. The composition and the amount of the heterogeneous lipid-protein mixture that makes up surfactant are tightly regulated. Pathological situations are associated with excesses (alveolar proteinosis) or deficiencies (acute respiratory distress syndrome) in surfactant levels. In situations where there is an absence of surfactant, mammalian life cannot be sustained due to the inability to inflate the lung and defective oxygenation. In view of the absolute requirement of surfactant to maintain viability, it is surprising that our knowledge of the regulation of surfactant turnover is incomplete.
Surfactant concentration within the alveoli is regulated by synthesis, secretion, reuptake, storage, and degradation of the lipid and protein components (for review, [1]). The type II pneumocytes in the lung are responsible for the synthesis and secretion of surfactant into the alveoli as well as contributing to its removal. The major protein component, surfactant protein-A (SP-A), is a 28-36 kDa protein member of the calcium-dependent lectin family of proteins that facilitates the surface tension-lowering properties of surfactant phospholipids in the alveolus, regulates surfactant phospholipid synthesis, secretion, and recycling, and also plays an important role in pulmonary host defense [2]. SP-A is produced by type II cells and released into the alveolar space. In addition, SP-A binds and is internalized by the same pneumocytes. The mechanism of SP-A binding to lung type II cells is not completely understood, yet it appears to be critical for the cell-associated functions of SP-A. Binding of SP-A to type II cells was found to be both saturable and specific, suggesting that this interaction is mediated through a receptor on the cell surface [3, 4]. Several recent reviews deal with the immunological functions of SP-A [5, 6]. This review will focus on the receptor-mediated interactions of SP-A and type II cells that affect surfactant lipid turnover, with an emphasis on the newly described SP-A receptor, P63/CKAP4. P63 is spelled with a capital p in order to clearly differentiate this protein from the p63 transcription factor, a member of the p53 transcription factor gene family that regulates cell proliferation, differentiation and response to stress and has been implicated in tumor formation [7, 8].
SP-A and surfactant metabolism
Role of SP-A in surfactant turnover
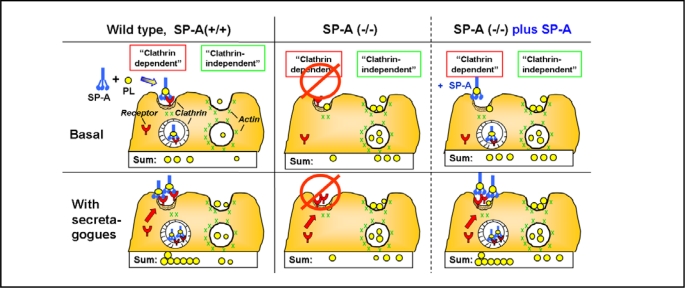
In cell culture systems, SP-A has important effects on two aspects of type II pneumocytes surfactant metabolism, the inhibition of surfactant secretion and the enhancement of surfactant lipid uptake (for review [2]). Thus, it was anticipated that the SP-A knock out mice (SP-A −/−) would suffer from severe respiratory problems; but surfactant levels and turnover in unstressed mice lacking SP-A appeared to be practically unaffected [9, 10, 11]. However, in further studies from our laboratory, differences between the wild type and SP-A gene-targeted mice became apparent with regard to the extent of lipid clearance upon the physiologic challenges of hyperventilation and secretagogue stimulation. Such challenges resulted in changes in the relative use of various clearance pathways [12, 13]. For example, in marked contrast to wild type mice, SP-A knock out mice were unable to increase clearance of liposomes [composed of surfactant lipids and labeled dipalmitoylphosphatidylcholine (DPPC)] upon CO2-induced hyperventilation. Further, perfused lungs isolated from the gene-targeted mice did not respond to secretagogue-stimulated clearance of surfactant-like lipids while uptake by the lungs of the wild type animals was double the unstimulated rate [12]. Upon detailed analysis of the pathways of surfactant lipid removal from the alveolar space by the isolated lungs, we found clear differences between the wild type and SP-A null mice, as shown in Fig. 1. Unlike the wild type mice who utilized an SP-A, receptor-mediated/clathrin coated pit-dependent endocytic pathway for the bulk of surfactant removal under basal conditions (Fig. 1, Basal), the SP-A−/− mice compensated for the loss of SP-A by up-regulation of the alternate actin-dependent, “clathrin-independent” endocytic pathway [13]. Upon challenge by addition of secretagogues (Fig. 1, With secretagogues), the rates of lipid clearance by both pathways were elevated in the wild type mice lungs with the “clathrin-dependent” pathway making a major contribution to lipid clearance. On the other hand, the SP-A −/− mice lungs showed no response to secretagogue exposure. We propose that the non-clathrin clearance mechanism was operating at maximum capacity under basal conditions in these gene-targeted mice, presumably to compensate for the loss of SP-A and, thus, the SP-A receptor-mediated pathway. This “clathrin-independent” pathway could not react to further challenge. Type II cells isolated from either the wild type or SP-A gene-targeted mice and placed in culture demonstrated comparable binding of iodinated SP-A, consistent with the continued presence of SP-A receptors even in the absence of SP-A in the SP-A null mice [13]. Thus, the abnormal phenotype could be “rescued” by the instillation of liposomes containing SP-A into the perfused lungs of the SP-A knock-out mice. Uptake via the “clathrin dependent” pathway and the ability to respond to secretagogue treatment was restored, as shown in Fig. 1, right [SP-A (−/−) plus SP-A] [13]. The results emphasize the critical role of both SP-A and, by inference, the SP-A receptor, for surfactant phospholipid turnover in the lung.
Fig. 1.
Pathways for lipid clearance by wild type (SP-A +/+) and SP-A gene-targeted (SP-A −/−) mice lungs. Data are adapted from reference [13]. 3H-dipalmitoyl-phosphatidylcholine labeled phospholipid (PL) liposomes (circles) were instilled into the trachea of mice and the lungs placed in an isolated perfused lung system under basal (upper panels) or secretagogue-stimulated (lower panels) conditions. Uptake of liposomes sensitive to the clathrin-inhibitor, amantadine, is “Clathrin-dependent.” Uptake of liposomes that is inhibited by cytochalasin D, an actin blocker, but not affected by amantadine is “Clathrin-independent.” (Left) SP-A, present in the surfactant of wild-type mice, associates with the instilled liposomes, while (Middle) SP-A is absent in the SP-A null mice. (Right) SP-A (−/−) plus SP-A: liposomes containing SP-A were instilled into the lungs of SP-A (−/−) mice. The addition of SP-A to the liposomes served to “rescue” the phenotype of the SP-A (−/−) mice by restoring the clathrin-mediated, secretagogue-sensitive process used by the wild type mice which had SP-A present.
Structure of SP-A
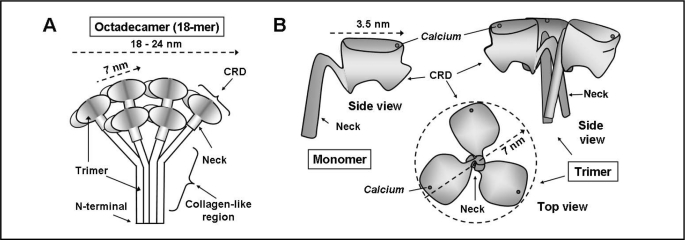
In order to examine the interaction of SP-A and its receptor, a basic knowledge of the structure of SP-A is necessary. The SP-A monomer consists of an N-terminal section, a collagen-like region with a 60° bend, a rigid amphipathic alpha-helical coiled region termed the “neck” region, and a globular carbohydrate recognition domain (CRD) ([14], for review, [15]). The primary structure of SP-A consists of 18 monomers organized into 6 groups of trimers forming an octadecamer, as shown in Fig. 2. The collagen-like regions of the trimers form staggered collagen triple helixes and the neck regions associate into alpha-helical coiled coils. Six trimers associate into the complete hexameric molecule, held together by disulfide bridges in the N-terminal domain [15]. Voss et al [14] used electron microscopy to visualize the macromolecular structure of SP-A and described it as a “flower bouquet like structure” as depicted in the schematic diagram of the structure of the complete 18-mer SP-A protein shown in Fig. 2A. They estimated that the fully assembled molecule was approximately 25-40 nm wide and 25 nm long with each “flower” or trimer carbohydrate recognition domain being approximately 5 nm in diameter [14]. Subsequent studies by Palaniyar et al measured the three-dimensional structure of recombinant rat SP-A trimers with the collagen-like region deleted in combination with phospholipid [16]. They found that the lipid-protein interface was relatively flat, rendering the molecule to appear more like broccoli, with an average dimension that changed from 18 to 24 nm depending on the size of the protein-lipid contact region [17]. The dimension of the lipid-free head was approx. 20 nm [17]. When in contact with lipid, each broccoli floweret, composed of three monomers, was approximately 7 nm in diameter [16].
Fig. 2.
Diagrammatic representation of the structure of SP-A. A. The octadecameric structure of the complete SP-A molecule based on Voss et al [14] and Palanyar et al. [17]. Three monomers form a collagen-like region, a neck region and a globular carbohydrate recognition domain (CRD). Six trimers join to form the “bunch of tulips” or “broccoli” shape. B. Representation of the carbohydrate-recognition domain (CRD) and neck region of SP-A based on the crystal structure from Head, et al [18]. The side view of one monomer and the side and top view of a trimer are shown. The side view of the trimer demonstrates the “T”-like shape and the top view the “boat propeller” configuration. The position of the calcium ion shown in each view is approximated from the crystal structure.
In 2003, Head et al determined the crystal structure of the globular carbohydrate recognition domain (CRD) and the neck domains encompassing residues 81-228 of rat surfactant protein A [18]. Residues 1-80 containing the N-terminal and collagen-like region were deleted and the consensus sequence for glycosylation mutated to prevent asparagine linkage. A diagrammatic outline of three views of the structure of the delta 1-80/N187S fragment of the SP-A monomer and trimer is shown in Fig. 2B. The crystal structure revealed that the angle of the carbohydrate recognition domain relative to the neck region was almost perpendicular giving this region a “T” shape when considering two monomers of the trimer shown in Fig. 2B, trimer, side view. Thus, the region most likely to interact with a cell surface receptor is relatively flat. With the trimeric structure assembled from three SP-A monomers, the top of the trimer molecule would form a structure resembling the shape of a flattened boat propeller, as shown in Fig. 2B, trimer, top view, and Fig. 3A.
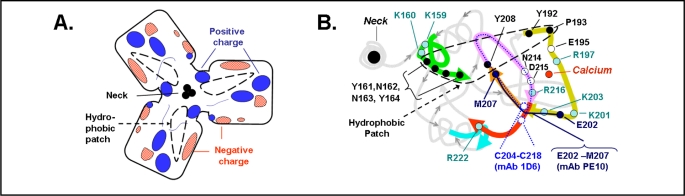
Fig. 3.
Details of the region on the top surface face of the carbohydrate recognition domain of SP-A. A. Diagram of the electrostatic surface charges of an SP-A trimer with calcium bound, based on the data of Head, et al [18]. The area resembles a flattened boat propeller with the location of three neck regions in the center as also depicted in Fig. 2B. The hydrophobic “patch” described by Head, et al [18] is outlined with a dashed black line. B. A string diagram representation of the surface of a monomer of SP-A taken from the RCSR Protein Data Rank Protein Workshop 3.4, protein code 1R13, using the Molecular Biology Toolkit [65] from the data of Head, et al. [18]. The string of amino acids located on the surface is shown in thick lines while the rest of the SP-A structure is in light gray. Hydrophobic amino acids in the area of the hydrophobic patch (dashed black line) are shown in black. Positively charged amino acids postulated to interact with a negatively charged receptor are filled circles. Amino acids recognized by mAb 1D6 are shown in with a dotted line and amino acids recognized by mAb PE10 are shown in with a solid line [19, 20, 21, 22]. Neck region is on the left.
Analysis of the crystal structure revealed two binding sites for calcium [18]. E195, R197, N214, and D215 were determined to be part of the primary calcium binding site of SP-A with E202 participating when calcium is bound at both the primary and secondary sites. As discussed below, binding of SP-A to type II cells has an absolute requirement for calcium. Thus it might be expected that mutating amino acids that participate in calcium binding would adversely affect SP-A-cellular interactions. Previously, an elegant series of studies performed by McCormick and his laboratory, examined the effects of mutating various amino acids in the SP-A molecule to determine the amino acids important for binding of SP-A to type II cells [15]. They found that amino acids E195, E202, N214 and D215, with some contribution by E197, all located in the C-terminal carbohydrate recognition domain (CRD), were the most critical for the maintenance of binding of SP-A to type II cells and regulation of the normal biologic function of SP-A, i.e. inhibition of type II cell surfactant secretion and stimulation of lipid uptake into type II cells. Since these amino acids also are important for calcium binding, they are candidates for participation in the calcium-dependent protein-protein interaction of SP-A binding to the SP-A receptor protein.
The electrostatic state of the surface of the SP-A trimer in the presence and absence of calcium was determined [18]. It was of interest that the binding of calcium to SP-A resulted in a shift to a more positively charged electrostatic profile on this surface domain of the SP-A protein (Fig. 3A). Fig. 3B is a string diagram of an SP-A monomer [Protein Data Bank (code 1R13), Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/)] with the location of the primary calcium binding site shown [18]. The top surface domain that is most likely to be involved in interactions with a cell-surface receptor is emphasized by thick lines. The position of the amino acids which might participate in providing a positive charge to the upper surface of the SP-A monomer, K159, K160, R197, K201, K203, R216 and R222, are indicated. The importance of this region for receptor interaction has been supported by earlier studies using immunological techniques. Monoclonal antibodies to SP-A were produced to identify the protein domains critical for the functional properties of the surfactant protein. Both the monoclonal antibody 1D6, which recognized the region C204 to C218, and the antibody PE10, which recognized the region E202 to M207, were found to suppress the binding and/or the biologic activity of SP-A [19, 20, 21, 22]. These amino acids are located directly in the putative receptor binding domain of SP-A (Fig. 3B). Head et al [18] pointed out this CRD surface also contains a strip of hydrophobic amino acids (Y161-Y164, Y208, Y192 and P193). This hydrophobic “patch” (shown by the black dashed line in Fig. 3A and B) could serve as a site for lipid binding [18] or for interaction with the hydrophobic region of a protein receptor. Thus, the three CRDs together provide a broad 7 nm footprint with a hydrophobic, positively charged planar surface available for interactions with both phospholipids and an SP-A protein receptor.
SP-A receptors
SP-A interaction with type II cells is receptor-mediated
It is now well established that SP-A binds to type II cells via a specific cell surface receptor. The binding of SP-A is saturable and calcium-dependent, typical for many receptor-ligand interactions [4, 23, 24, 25]. The binding curve of SP-A to type II cells demonstrated both a saturable component and a non-saturable process [4, 23, 24, 26]. Classically, receptors are clustered within clathrin-coated pits and enter the cell via clathrin-coated vesicles [27]. SP-A has been morphologically localized in clathrin-coated pits in type II cells by microscopic studies suggesting the presence of the receptor at those sites [28]. Further, any perturbation of the formation and/or internalization of clathrin-coated pits/vesicles blocks the uptake of SP-A, including intracellular potassium depletion [29], phenylarsine oxide exposure, which cross-links clathrin and thus prevents the formation of clathrin-coated vesicles [30], or amantadine treatment, which stabilizes clathrin coated pits and thus prevents vesicle budding [30]. Interference with the association of SP-A with the surface of type II cells, for example through the use of a monoclonal antibody specific for the C-terminal domain of SP-A, abrogates the biological activity of SP-A, in this case the inhibitory effect on lipid secretion from type II cells [20, 21]. Mutating amino acids in SP-A critical for SP-A binding to type II cells blocks either or both of the biological activities of SP-A, the inhibition of surfactant secretion or the stimulation of surfactant lipid uptake (for review, [15]). Trypsin treatment of cells at levels sufficient to remove surface protein receptors reduced the binding of SP-A to pneumocytes, consistent with a protein regulated interaction [4, 24]. Recovery of basal SP-A binding capacity occurred within 20 minutes of exposure to protease [24]. Further, secretagogues cAMP and PMA stimulated association of SP-A to the cells by two-fold over the same time frame [24]. The prompt recovery from enzyme treatment together with the rapidity of increase in SP-A binding upon secretagogue exposure indicated that the SP-A receptors were recruited to the cell surface from intracellular stores. These observations are consistent with the accepted paradigm that receptor plasma membrane density is unlikely to be controlled primarily by new protein synthesis. Taken together, the evidence supports the existence of protein on the surface of type II pneumocytes which interacts with SP-A.
History of SP-A receptors and surfactant lipid turnover
As the evidence became convincing that SP-A interacted with type II cells with receptor-mediated characteristics, the search for the elusive SP-A receptor began. Several approaches have been used including the generation of anti-idiotypic antibodies to SP-A [31, 32], SP-A affinity column chromatography [26, 33], and cross-linking of SP-A to proteins on the plasma membrane of type II cells [34]. Work published up to 1998 on putative SP-A receptors that participate in the SP-A-mediated regulation of surfactant metabolism has been summarized in detail in previous reviews [15, 35] and will be outlined only briefly. Table 1 describes the approaches used to isolate the receptors, the molecular weights of the proteins and the effects of the anti-receptor antibodies on SP-A-mediated effects of type II cell surfactant metabolism. Using an anti-idiotypic approach, Strayer and collaborators identified three proteins with apparent Mr of 30, 52, and 60 kDa reduced [32, 36] and 210 kDa non-reduced [37] on the type II cell plasma membrane. The 30-kDa protein was shown to be associated with the regulation of secretagogue-stimulated surfactant secretion [38] and phospholipid uptake [37]. Stevens et al. [31] using similar strategy with SP-A as the immunogen identified a 55-kDa protein, binding protein 55 (BP55). The antibody generated against BP55, Ab-2H5, was able to inhibit the SP-A-induced uptake of liposomes and, thus, this putative receptor protein was felt to be involved in SP-A-induced surfactant endocytosis by type II cells [39]. Using ligand affinity chromatography, proteinswere identified by Kresch et al [33] on the type II cell surface with apparent Mr of 50, 55, and 65 kDa reduced and >200 kDa non-reduced together with an 86 kDa protein present under both conditions. A monoclonal antibody generated against the >200 kDa protein was shown to inhibit SP-A-stimulated phospholipid uptake. Chroneos and colleagues described a SP-A receptor from U937 macrophages with a reduced molecular mass of 210 kDa using SP-A affinity column chromatography [26]. The receptor, surfactant protein receptor 210 (SP-R210), was detected in both alveolar macrophages and type II epithelial cells, and subsequently, on T cells [40, 41]. Anti-SP-R210 antibody was found to block the SP-A-mediated inhibition of phospholipid secretion by type II cells. This receptor was later identified as unconventional myosin 18A [42]. From these studies, it seemed that the SP-A receptor(s) on type II cell surface consists of several polypeptides with a molecular weights of 30, 50-55, 60-65 and 86 kDa, reduced and 86 and > 200 non-reduced, data consistent with a receptor complex. The cDNA and the deduced amino acid sequence of the 30 kDa protein were presented [32, 36]. SP-R210 has been characterized further with regard to its presence on T cells and the role of SP-A and SP-R210 in cell-mediated immunity [40, 41]. However the molecular nature of the other protein components of the putative receptor(s) has not been described.
Table 1.
SP-A Receptors on Type II Cells. Groups presented in chronological order, a, anti-idiotypic antibody, surfactant; b, ligand (SP-A) affinity. column; c, cross-linking; **unconventional myosin 18A; TII, type II pneumocytes; MΦ, macrophages; ND, not done; Ab, antibody; PL, phospholipid; Non-red., non-reduced; Ref, reference; (Yes), SP-A blocked, binding of receptor antibody to type II cells; ^unpublished, Bates.
| Group |
Method of isolation |
Name of antibody or receptor |
Ab to receptor blocks: | ||||||
|---|---|---|---|---|---|---|---|---|---|
| mw (kDa) | On MΦ? |
SP-A binding |
SP-A effect on PL: | Ref. |
|||||
| Non-red. | Reduced | Secretion | Uptake | ||||||
| Strayer | a | ab-A2R/A2C | 210 | 30, 52, 60 | No | Yes | Yes | Yes | [32, 36, 37, 38] |
| Stevens | a | ab-2H5 or Bp55 | 170-200 | 55 | No | Yes | No | Yes | [31, 67] |
| Kresch | b, TII | SPAR | 86 | 86 | |||||
| >200 | 50-65 | No | (Yes) | ND | Yes | [33] | |||
| Chroneos | b, MΦ | SP-R210** | 600 | 210 | Yes | Yes | Yes | ND | [26, 42] |
| Bates/Fisher | c | CKAP4/P63 | 63 | 63 | No | Yes | Yes | Yes^ | [34] |
Our laboratory has conducted a more recent study of SP-A receptors on pneumocytes and used cross-linking of SP-A to the surface of rat type II cells to identify an SP-A binding protein of Mr 63 kDa. Antibodies to this protein blocked the biologic activity of SP-A on surfactant secretion [34] and preliminary evidence indicates inhibition of SP-A-stimulated phospholipid uptake as well (Bates, unpublished). Further details on this SP-A receptor protein, P63/CKAP4, are provided below. However, thus far, it is not understood why SP-A associates with several different proteins on the type II cell membrane or whether the proteins that bind to SP-A are inter-related, as separate entities and/or components of a receptor complex.
P63/CKAP4
P63 as a microtubule-binding protein in the ER
P63 is a 63kDa reversibly palmitoylated, non-glycosylated, type II transmembrane protein. Thus, the amino-terminal region is located in the cytosol and the carboxy-terminal region is in the lumen. The protein is found in many cell types, both transformed and primary cells, from various species and tissue sources including HepG2 liver cells, Caco-2 intestinal epithelial cells, MRC5 fibroblasts, Vero cells, HeLa cells, A549 lung adenocarcinoma cells, vascular smooth muscle cells, bladder epithelial cells and lung type II cells, to name a few, although the protein is not present in rat alveolar macrophages or L2 rat lung cells [34, 43, 44, 45]. P63 is an ER-resident protein that also has been localized to the plasma membrane in vascular smooth muscle cells [44], bladder epithelial cells [45], and type II pneumocytes.
The discovery of P63 occurred in the early 1990s when Dr. Hauri and his colleagues, interested in the morphogenesis of the endoplasmic reticulum, used monoclonal antibody techniques to identifying proteins specific for the ER-Golgi. They identified a 63 kDa, non-glycosylated, membrane protein [43] that localized to the ER-Golgi intermediate compartment in Vero cells (African green monkey kidney cells). Subsequent studies using polyclonal antibodies established that P63 was located in the rough ER [46]. Pulse-chase experiments in Vero cells demonstrated labeled protein remaining present after 18 hrs of chase, indicative of a fairly stable protein with a prolonged turnover time [43]. Early studies revealed that P63 was reversibly palmitoylated when intracellular transport was inhibited with Brefeldin A [47]. P63 plays an important role in the stability of the ER by connecting the ER membranes to microtubules in the cytoskeletal network and, therefore, has been renamed CLIMP-63 for cytoskeletal-linking membrane protein [48, 49]. Over expression of wild-type P63/CLIMP-63 bundled microtubules due to the P63-dependent increase in association of the ER with the microtubule network [48]. Expression of a mutated P63 that is unable to bind to microtubules results in the collapse of the ER [50]. During mitosis, a process that requires rearrangement of the ER complex, phosphorylation of P63 is enhanced resulting in its inability to bind microtubules in vitro [50]. The luminal segment, produced by recombinant methods, formed large alpha helical oligomers which appeared as curved rods by transmission EM [49]. The calculated length of a two-stranded, alpha-helical coiled coil molecule with 70 heptad repeats would be 69 nm [49]. Similarly sized P63 rods were seen in addition to much larger particles and aggregates giving a final average protein length of 91 nm to the preparation. Formation of such complexes provides a mechanism for the retention of P63 within specific domains of the ER [49, 51]. Thus, P63 plays an important, if not crucial, role in the maintenance of ER structure (for review [52]).
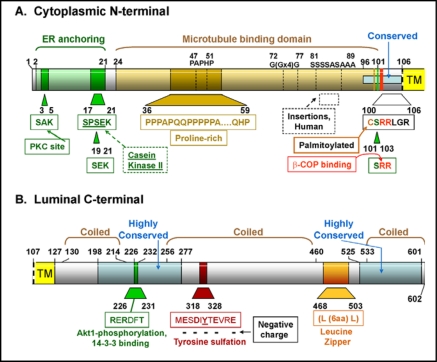
Structure of P63
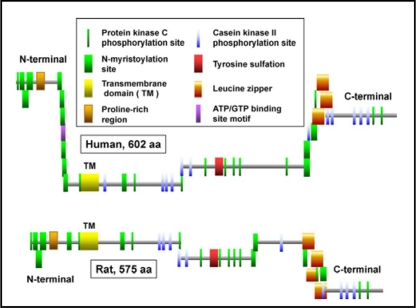
P63 is composed of 602 amino acids in humans and 575 amino acids in mice and rats. The protein was given the gene name of cytoskeletal-linking membrane protein (CKAP4) by the Human Genome Organization in view of the data from the Hauri laboratory [47, 48, 49]. The Rule-based predicted features (ScanProsite, http://expasy.org) of human and rat P63 are shown in Fig. 4 and a schematic diagram of specific features of the cytosolic and luminal domains of human P63 are in Fig. 5A and B. The N-terminal regions (85 amino acids for rat, 106 amino acids for human) differ slightly between species while the C-terminal area is well preserved. Overall, the amino acid sequence homology between rat and mouse P63 is 94% and between rat and human P63 is 84%. Mutational analysis of human P63 demonstrated that amino-teminal amino acids 2-21 were the ER-anchoring domain while amino acids 24-101 represent the microtubule binding domain [51]. Deletion of most of the cytosolic tail (delta 2-101) and changing the charged arginine groups 102 and 103 to alanines results in the translocation of the protein to the plasma membrane [51]. In a series of mutational analysis and chimeric constructs, Schweizer et al [51] demonstrated that all three regions of the protein, the luminal, cytosolic and transmembrane domains contributed to the intracellular localization of P63. In addition, P63 was found to form large aggregates and the extent of self-association between mutant constructs predicted the retention of P63 between the rough ER and the Golgi [51].
Fig. 4.
Rule-based predicted features of P63 comparing the features of the human and mouse protein [66]. The figures are based on the diagrams of P63 from the ScanProsite web page www.expasy.ch/tools/scanprosite whose figures are under copyright and are the property of the Swiss Institute of Bioinformatics. The proteins are aligned by the transmembrane domain. The location of the transmembrane domains and the predicted area of tyrosine sulfation have been added.
Fig. 5.
Structural characteristics of the human P63 protein. A. The N-terminal cytoplasmic portion from amino acids 1-106. B. The C-terminal luminal portion from amino acid 107-602. The enzymes or proteins are listed and the amino acid sequence sites predicted to be involved with them are shown. The amino acid sequences that are highly conserved between human, mouse, and rat are indicated. The data are summarized from the amino acid composition, the ScanProsite predicted features shown in Fig. 4, and the data of Hauri's group [43, 47, 48, 49]. TM, transmembrane domain.
Modification of P63 protein on several epitopes serves to control its function or intracellular localization. There are many possible predicted phosphorylation sites on P63 and phosphorylation of serines 3, 17, and 19 has been shown to be sufficient to prevent binding of P63 to microtubules (Fig. 4 and 5A) [50]. Several amino acid sequences throughout the protein represent consensus sequences for N-myristoylation (ScanProsit) (Fig. 4) but there are no reports of this modification of P63. Human and rodent P63 have a proline-rich region, between amino acids 36 and 59 in humans, whose function is as yet unknown although proline-rich sequences can potentially participate in a host of protein-protein interactions as shown in various cellular processes/systems (for review [53]). Between the proline-rich motif and the transmembrane domain there is an area of polyglycines, polyserines and polyalanines that are present only in the human P63, not in rodents (rat or mouse). The amino-terminal region adjacent to the transmembrane domain is highly conserved between species (Fig. 5A). The cysteine in position 100 (human) is important for palmitoylation of the protein [54]. Site-directed mutagenesis of this region revealed that the critical factor for proper acylation was a six amino acid distance between the cysteine and the transmembrane domain [54]. Recent work identified the enzyme responsible as a member of the palmitoyl acyltransferase family characterized by an Asp-His-His-Cys (DHHC) motif, DHHC2 [55]. Palmitoylation has been shown to have several effects on transmembrane proteins including regulation of protein trafficking and function (for review, [56]). Palmitoylation of P63 may contribute to the localization of P63 on the plasma membrane as knockdown of DHHC2 by siRNA techniques served to reduce levels of surface P63 in HeLa cells as determined by immunolocalization [55]. The di-argenine motif (amino acids 102 and 103 in human P63), a possible β-COP binding site, is also located in this well-conserved area (Fig. 5A). The transmembrane domain is 21 amino acids and is identified as hydropathic by the Kyte Doolittle hydropathy profile of the protein sequence [47]. Mutational analysis revealed that, although this region contributed, it was not sufficient for the correct retention of P63 to an ER-Golgi intermediate compartment [51].
The C-terminal luminal region is predicted to interact with SP-A and determination of the critical epitope/s is the subject of current investigations. This portion of P63 contains three coiled coil regions, two shorter regions on each end and one longer one in the middle of this section of the protein (Fig. 5B). The RERDFT amino acid sequence is located between the first and second coiled region and is in the middle of a highly conserved region (98% conservation between human and rodents). This motif matches the consensus sequence for Akt1 phosphorylation and 14-3-3 binding, RXRXXT/S. Located between coiled regions, it is ideally situated for interactions with other proteins. However, this region is located on the luminal portion of the protein which may preclude any interactions with cytosolic Akt1 or 14-3-3. The putative leucine zipper region is also located between coiled regions, between the second and third coil of P63. Finally, tyrosine 323 (human) located one-third of the way into the large coiled region, is part of a predicted consensus tyrosine sulfation sequence [57] due to the presence of two negatively charged amino acids within two residues of the tyrosine (D321 and E325); four negatively charged amino acids within 5 residues (E319, D321, E325, and E328); only one basic residue (R327); and only three hydrophobic residues (M318, I322, and V326) in this location (Fig. 5B). In addition, this tyrosine is located in a coiled region of the protein without double bonds or N-linked glycans [57]. Tyrosine sulfation is a post-translational modification that involves the addition of a negatively charged sulfate group to an exposed tyrosine residue [58]. The enzymes involved, tyrosylprotein sulfotransferases, are localized in the trans-Golgi and use the universal sulfate donor 3'-phosphoadenosine-5”-phosphosulfate. The resultant tyrosyl O-sulfate can form hydrogen bonding networks and/or salt bridges, especially with positively charged amino acids, thus enhancing protein-protein interactions [57, 58]. This area is an attractive candidate as a possible binding site for the positively charged regions in the carbohydrate-recognition domain of SP-A discussed earlier. Examples of high activity ligand-receptor binding that require tyrosine sulfation of the receptor include the binding of P-selectin glycoprotein ligand-1 [59] to P-selectin, Factor VIII binding of van Willebrand Factor, and the binding of thyroid stimulating hormone to its receptor [61], to name a few (for review [57, 58]).
P63/CKAP4 as a receptor
P63 as a receptor for SP-A
The regulation of the turnover of surfactant lipid is an important process. In cell culture, as in the intact lung, SP-A interactions with a receptor play a crucial part in this process. Given the difficulties others have had isolating putative SP-A receptors using affinity chromatography or indirect idiotypic immunological methods; we took an entirely different approach. We directly cross-linked SP-A to isolated plasma membranes as well as intact type II cells. Using two different cross linkers, we identified a 63 kDa protein, under reduced or non-reduced electrophoresis conditions, which subsequent LC-MS/MS analysis and NCBI database search identified as CKAP4/ P63 [34]. Labeling of the cross-linked preparations using Western techniques with antibodies directed against human P63 obtained from the Hauri laboratory confirmed the identity of P63 [43]. Immunoprecipitation experiments with antibodies directed against human P63 or against SP-A [62] demonstrated that the two proteins co-immunoprecipitated from type II cell lysates or isolated plasma membrane preparations. Such data provided firm evidence for specific interaction between SP-A and P63. P63 protein can be identified in the plasma membrane of type II cells and A549 cells, an adenocarcinoma cell line that has been utilized as a type II pneumocyte model system. [34]. Immunocytochemistry of nonpermeabilized type II cells stained with anti-P63 antibody demonstrated punctuate labeling on the cell surface [25, 34]. P63 was identified in type II cells and as well as other cells in mouse lung. Further, SP-A and P63 co-localized in permeabilized type II cells and both proteins were found in the early endosome (EEA-1 positive) compartment of the cells [34]. Using electron microscopic techniques, P63 was localized to the ER, the plasma membrane and the microvilli of type II cells [25]. On microvilli, the P63 is in close approximation to SP-A in the extracellular lining layers of the alveolar space.
SP-A interacts with type II cells through specific, saturable binding that is calcium-dependent and through a non-saturable process that is calcium-independent. Such binding characteristics are typical of many receptor-ligand interactions [23, 24]. Antibody to P63 in competition experiments inhibits the saturable, calcium-dependent binding of SP-A to lung epithelial cells, including both type II pneumocytes and A549 adenocarcinoma cells [25]. In addition, the increase in SP-A binding due to cAMP exposure of type II cells on Transwell membranes is abrogated by the presence of competing P63 antibody. Using siRNA techniques to reduced the P63 content of type II cells results in an inhibition of P63-specific SP-A binding to the cells [25].
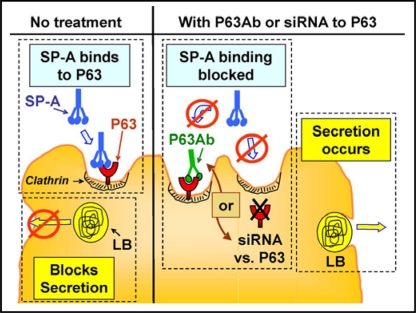
SP-A plays a role in the regulation of surfactant secretion and uptake by type II cells, two processes that involve interaction of SP-A with a cell surface receptor. For P63 to be the important receptor, interference of SP-A interaction with P63 on type II cells should impact type II cell lipid metabolism. That this was the case was shown in experiments measuring phospholipid secretion. Release of radio-labeled surfactant phosphatidylcholine from type II cells is stimulated by secretagogues such as ATP. SP-A markedly reduces both basal and ATP-stimulated secretion. Co-incubation of SP-A and anti-P63 antibody interferes with the biologic activity of SP-A, resulting in a concentration-dependent reversal of the SP-A inhibition of PC secretion [34]. Additional studies using siRNA techniques to reduce the P63 level in the type II cells demonstrated that lowering cellular P63 content prevented SP-A-mediated inhibition of surfactant secretion [25]. The data on the effect of blocking or reducing P63 protein on SP-A/type II cell interactions is diagrammatically summarized in Fig. 6. SP-A binding to type II cells blocks basal and secretagogue-stimulated surfactant secretion (Fig. 6, left). Interfering with the binding of SP-A to P63 on type II cells using either a competitive antibody specific for P63 or siRNA directed against P63 will interfere with the ability of SP-A to obstruct surfactant release (Fig. 6, right). The question as to whether P63 plays a role in the ability of SP-A to enhance surfactant lipid uptake is currently under investigation. Preliminary evidence indicates that P63 also mediates this feature of SP-A's biologic activity, but this issue remains to be explored further (Bates, unpublished).
Fig. 6.
Effect of P63 on SP-A regulation of surfactant secretion from type II cells. (Left) Without treatment, SP-A binds to P63 and blocks surfactant release from lamellar bodies. (Right) Preventing SP-A/P63 interactions using anti P63 antibody (Ab) or reduction of P63 protein through siRNA directed against P63 allows surfactant secretion to proceed. LB, lamellar body.
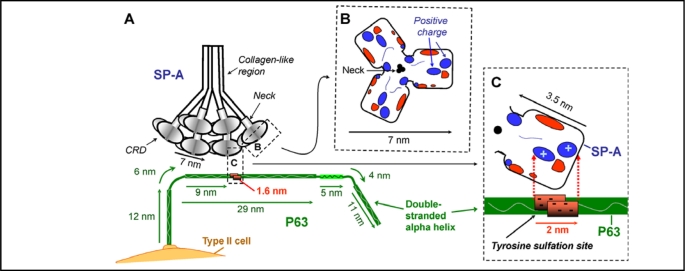
A possible scenario for the interaction of SP-A and P63 is shown in Fig. 7. The size of the two proteins is drawn approximately to scale. The ideal length of a double-stranded alpha-helical coiled coil 602 amino acid protein, 69 nm, was used for the C-terminal luminal portion of P63 since the protein would be anchored in the plasma membrane and less likely to form the large multimers observed when the protein was free in solution [49]. Since the EM micrographs showed curved rods, the protein is drawn in a C shape. In any case, the exact length or shape of P63 is not critical for the hypothesized interaction. It is apparent from Fig. 7A that the interaction of the two proteins is physically possible and that one SP-A molecule could readily bind more that one P63 molecule. Fig. 7B shows the surface of the CRD trimer domain of SP-A that would be exposed to P63 on the cell surface, with the positive charged areas indicated under conditions with calcium present [18]. In the scenario shown in Fig. 7C, the positive portion of the surface of one monomer of SP-A would interact with the negative portion of the P63 protein located in the area of tyrosine sulfation where five negative charges are centralized. This double stranded P63 tyrosine sulfation area would provide a 2 nm area of negative charges available to attract the 3.5 nm CRD area of a monomer of SP-A containing positively charged amino acids. Although this proposed mechanism of interaction is attractive, several important issues remain to be addressed including possible hydrophobic interactions between the proteins and the role of calcium. Significantly more work will be necessary for definitive conclusions.
Fig. 7.
Hypothesized interaction of SP-A and P63. Diagrams are drawn approximately to scale. A. The positively charged areas on the surface of the carbohydrate recognition domain (CRD) of SP-A is hypothesized to interact with the double stranded P63 protein at the negatively charged region of P63 predicted to contain a sulfated tyrosine. The length of the P63 luminal C-terminal was drawn as 69 nm, the predicted size of a double stranded helix of 602 amino acids [49]. Structural features of P63 are based on the amino acid composition, the ScanProsite predicted features, and the data of Hauri's group [43, 47, 48, 49] as shown in Fig. 4 and 5. P63 is drawn as a curved rod as seen in EM photomicrographs [49]. R. View of the top of the 7nm long SP-A trimer CRD region, with the location of the electrostatic surface charges shown after calcium binding [18]. Positive charged regions enlarged in diagram C are indicated. C. View of one SP-A molecule in the trimeric CRD of SP-A, estimated to be approx. 3.5 nm in length. In the P63 molecule, the area from 318 to 328 containing 5 negatively charged amino acids is estimated to be approx. 1.6 nm long forming a patch of 2 nm due to the double strands of the coiled coil of P63. The positively charged area on SP-A could interact with the negatively charged tyrosine sulfation site on P63.
P63 as a receptor for other proteins
Two other groups have found evidence that P63 is a biologically relevant receptor for two different proteins in entirely unique systems. During the course of our studies on P63, the possibility that P63, an ER resident protein, might also function as a plasma membrane receptor on the cell surface was addressed by Razzaq, et al. who were interested in identifying the receptor protein on rat vascular smooth muscle cells that bound tissue plasminogen activator (tPA) [44]. After labeling surface proteins with 125I, affinity chromatographic procedures isolated a 63 kDa protein. Mass spectrometric analysis of the radiolabeled protein band identified the tPA binding protein as P63. Subsequent western blotting procedures using antibodies raised against human P63 provided by Hauri's laboratory [43] confirmed the identity of the protein as P63. Immunocytochemistry of nonpermeabilized cells revealed punctuate labeling on the surface of vascular smooth muscle cells confirming plasma membrane localization. Antibodies directed against P63 were able to inhibit tPA binding and decreased the biologic activity of tPA, plasminogen activation. Finally, transfecting COS cells with a P63 construct known to traffic to the plasma membrane, the N-terminal deletion mutant (delta 2-101AA), lead to a dramatic increase in plasminogen activation which could be blocked by antibody to P63 [44].
A second group was searching for the receptor for the frizzled-8 protein-related antiproliferative factor (APF) on bladder cells [45]. APF is a sialoglycopeptide secreted by the bladder cells of patients with interstitial cystitis that inhibits the growth of the epithelial bladder cells. The peptide sequence of APF is identical to a domain in Frizzled 8, a Wnt ligand receptor [63]. They found that APF bound with high affinity to CKAP4/P63 [45]. P63 was identified in the membrane fraction of bladder epithelial cells from both normal patients and those with interstitial cystitis using anti human CKAP4/P63 antibody. APF and anti-CKAP4/P63 colocalized on bladder cells as determined by confocal microscopy. Roth reducing P63 with siRNA techniques or use of anti-CKAP4/P63 antibodies in competition experiments served to reverse the reduction of cell proliferation which occurs upon exposure to APF, confirming that P63 was a functioning cell membrane receptor for APF [45].
The finding that P63 acts as a receptor in various cell types, such as in smooth muscle cells, bladder cells and type II pneumocytes, for such a diverse group of proteins as tPA, APF and SP-A raises many questions. Do these protein ligands share a common structural domain that renders them able to bind to P63? What structural characteristics of P63 are important for these interactions? Does P63 act as a chaperone for other receptor proteins that are more specific for the ligands? We have shown that incubation of type II cells with secretagogues serves to up-regulate the levels of P63 in the plasma membrane [25]. What factors determine the amount of P63 on the surface of smooth muscle cells or bladder epithelial cells?
Conclusions and future directions/unanswered questions
Evidence continues to accumulate that SP-A provides a critical role in the normal clearance of surfactant lipids from the lung alveoli. SP-A-mediated uptake of phospholipid occurs through a clathrin-mediated mechanism. When SP-A is deficient, as in the gene-targeted mice, an alternate clathrin-independent pathway operates, but this pathway has limited capacity and is unable to respond to physiologic challenges such as hyperventilation or exposure to increased levels of secretagogues. Restoring SP-A to the surfactant rescues the phenotype and restores normal, secretagogue-sensitive surfactant removal from the alveolar space [13]. Since SP-A utilizes a receptor-mediated pathway in the uptake of phospholipids into type II cells, identification of the protein functioning as an SP-A receptor is critical. Despite the many years since the recognition that the interaction of SP-A with type II cells had receptor-ligand characteristics, identification of the “SP-A receptor” has been fraught with difficulties. The “stickiness” of SP-A and resultant non-specific interactions with cell proteins undoubtedly has contributed to the situation. Considerable evidence has accumulated indicating that P63 satisfies the criteria for an SP-A receptor. P63 has been localized to the plasma membrane and the microvilli of type II cells [25]. The close proximity or association of P63 and SP-A has been shown by cross-linking and co-immunoprecipitation studies [34]. Antibodies to P63 block the specific, calcium-dependent binding of SP-A and the subsequent biological effects of SP-A on surfactant secretion [25, 34]. Secretagogue-enhanced binding of SP-A is abrogated by the presence of competing P63 antibodies. Further, reducing P63 levels in type II cells through siRNA techniques adversely affected SP-A binding to the cells and resultant effects on surfactant secretion.
Despite the progress made to date, many key questions remain. Are the previously identified putative receptor proteins that bound SP-A possibly chaperone proteins for P63 or do they affect the properties of P63 in some fashion? Perhaps the 60 kDa protein identified by Strayer [32] and the 65 kDa protein identified by Kresch [33] were, in fact, P63. What mechanisms regulate the levels of P63 or the trafficking of P63 from the ER to the cell surface? P63 is present in many cell types; does it also function as a receptor for proteins important in the other biologic systems? Do the various protein ligands interact with the same domains on P63 or is each interaction unique? What is the domain of SP-A that participates in the binding to the P63 molecule? Although it appears that amino acids E195, E202, N214, D215, and E197 in SP-A participate in binding calcium; do they also participate in binding to P63 via a calcium bridge? Do the hydrophobic regions of SP-A participate in the interaction of the two proteins? SP-A and P63 seem to enter the cell as a unit since both are found in early endosomes. At what point does SP-A and P63 separate or does P63 chaperone SP-A to the lamellar body? How does SP-A bind to both lipid and P63 at the same time? Given the role of P63 in ER structure and mitosis, is it going to be possible to make a P63 deficient mouse? Given the similarity of the structure of the trimeric carbohydrate recognition and neck domains of SP-A and SP-D [18] and the fact that SP-D binds to type II cells [64], does SP-D bind to P63?
The maintenance of lung surfactant homeostasis is vitally important for proper gas exchange. With SP-A playing a central role in this process, control of the regulation of the cell surface levels of P63 provides an opportunity to enhance or inhibit the turnover of surfactant. This would be an important novel approach toward treatment directed at the recovery from the pathological situations of excess or insufficient quantity of surfactant. Further, it has become apparent that understanding the details of the functionally diverse biology of P63 may aid not only in the understanding of the regulation of surfactant clearance but also may have implications in other biological systems.
Acknowledgements
This research has been supported by the National Heart, Lung and Blood Institute Grant HL-19737. I would like to thank Dr. Aron B. Fisher for many helpful discussions and insightful advice and my other colleagues who have contributed to this work over the years.
References
- 1.Fisher AB. Lung Surfactant Clearance and Cellular Processing. In: Rooney SA, Landes RG, editors. Lung surfactant: cellular and molecular processing Medical Intelligence Unit 5. Austin, TX: Landes Bioscience; 1998. pp. 165–189. [Google Scholar]
- 2.Khubchandani KR, Snyder JM. Surfactant protein A (SP-A): the alveolus and beyond. Faseb J. 2001;15:59–69. doi: 10.1096/fj.00-0318rev. [DOI] [PubMed] [Google Scholar]
- 3.Kuroki Y, Mason RJ, Voelker DR. Chemical modification of surfactant protein A alters high affinity binding to rat alveolar type II cells and regulation of phospholipid secretion. J Biol Chem. 1988;263:17596–17602. [PubMed] [Google Scholar]
- 4.Wright, Borchelt JD, Hawgood S. Lung surfactant apoprotein SP-A (26-36 kDa) binds with high affinity to isolated alveolar type II cells. Proc Natl Acad Sci U S A. 1989;86:5410–5414. doi: 10.1073/pnas.86.14.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastva AM, Wright, Williams KL. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc Am Thorac Soc. 2007;4:252–257. doi: 10.1513/pats.200701-018AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haagsman HP, Hogenkamp A, van Eijk M, Veldhuizen EJ. Surfactant collectins and innate immunity. Neonatology. 2008;93:288–294. doi: 10.1159/000121454. [DOI] [PubMed] [Google Scholar]
- 7.Bourdon JC. p53 Family isoforms. Curr Pharm Biotechnol. 2007;8:332–336. doi: 10.2174/138920107783018444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G, Knight R. p63 in epithelial development. Cell Mol Life Sci. 2008;65:3126–3133. doi: 10.1007/s00018-008-8119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikegami M, Korfhagen TR, Bruno MD, Whitsett JA, Jobe AH. Surfactant metabolism in surfactant protein A-deficient mice. Am J Physiol. 1997;272:L479–485. doi: 10.1152/ajplung.1997.272.3.L479. [DOI] [PubMed] [Google Scholar]
- 10.Ikegami M, Korfhagen TR, Whitsett JA, Bruno MD, Wert SE, Wada K, Jobe AH. Characteristics of surfactant from SP-A-deficient mice. Am J Physiol. 1998;275:L247–254. doi: 10.1152/ajplung.1998.275.2.L247. [DOI] [PubMed] [Google Scholar]
- 11.Korfhagen TR, Bruno MD, Ross GF, Huelsman KM, Ikegami M, Jobe AH, Wert SE, Stripp BR, Morris RE, Glasser SW, Bachurski CJ, Iwamoto HS, Whitsett JA. Altered surfactant function and structure in SP-A gene targeted mice. Proc Natl Acad Sci U S A. 1996;93:9594–9599. doi: 10.1073/pnas.93.18.9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain D, Dodia C, Bates SR, Hawgood S, Poulain FR, Fisher AB. SP-A is necessary for increased clearance of alveolar DPPC with hyperventilation or secretagogues. Am J Physiol Lung Cell Mol Physiol. 2003;284:L759–765. doi: 10.1152/ajplung.00200.2002. [DOI] [PubMed] [Google Scholar]
- 13.Bates SR, Dodia C, Tao JQ, Fisher AB. Surfactant protein-A plays an important role in lung surfactant clearance: evidence using the surfactant protein-A gene-targeted mouse. Am J Physiol Lung Cell Mol Physiol. 2008;294:L325–333. doi: 10.1152/ajplung.00341.2007. [DOI] [PubMed] [Google Scholar]
- 14.Voss T, Eistetter H, Schafer KP, Engel Macromolecular organization of natural and recombinant lung surfactant protein SP 28-36. Structural homology with the complement factor C1q. J Mol Biol. 1988;201:219–227. doi: 10.1016/0022-2836(88)90448-2. [DOI] [PubMed] [Google Scholar]
- 15.McCormack FX. Structure, processing and properties of surfactant protein A. Biochim Riophys Acta. 1998;1408:109–131. doi: 10.1016/s0925-4439(98)00062-3. [DOI] [PubMed] [Google Scholar]
- 16.Palaniyar N, McCormack FX, Possmayer F, Harauz G. Three-dimensional structure of rat surfactant protein A trimers in association with phospholipid monolayers. Riochemistry. 2000;39:6310–6316. doi: 10.1021/bi992793b. [DOI] [PubMed] [Google Scholar]
- 17.Palaniyar N, Ridsdale RA, Holterman CE, Inchley K, Possmayer F, Harauz G. Structural changes of surfactant protein A induced by cations reorient the protein on lipid bilayers. J Struct Biol. 1998;122:297–310. doi: 10.1006/jsbi.1998.4004. [DOI] [PubMed] [Google Scholar]
- 18.Head JF, Mealy TR, McCormack FX, Seaton RA. Crystal structure of trimeric carbohydrate recognition and neck domains of surfactant protein A. J Biol Chem. 2003;278:43254–43260. doi: 10.1074/jbc.M305628200. [DOI] [PubMed] [Google Scholar]
- 19.Kuroki Y, Mason RJ, Voelker DR. Pulmonary surfactant apoprotein A structure and modulation of surfactant secretion by rat alveolar type II cells. J Biol Chem. 1988;263:3388–3394. [PubMed] [Google Scholar]
- 20.Murata Y, Kuroki Y, Akino T. Role of the C-terminal domain of pulmonary surfactant protein A in binding to alveolar type II cells and regulation of phospholipid secretion. Riochem J. 1993;291:71–76. doi: 10.1042/bj2910071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroki Y, McCormack FX, Ogasawara Y, Mason RJ, Voelker DR. Epitope mapping for monoclonal antibodies identifies functional domains of pulmonary surfactant protein A that interact with lipids. J Biol Chem. 1994;269:29793–29800. [PubMed] [Google Scholar]
- 22.Hiraike N, Sohma H, Kuroki Y, Akino T. Epitope mapping for monoclonal antibody against human surfactant protein A (SP-A) that alters receptor binding of SP-A and the SP-A-dependent regulation of phospholipid secretion by alveolar type II cells. Biochim Biophys Acta. 1995;1257:214–222. doi: 10.1016/0005-2760(95)00068-n. [DOI] [PubMed] [Google Scholar]
- 23.Kuroki Y, Mason RJ, Voelker DR. Alveolar type II cells express a high-affinity receptor for pulmonary surfactant protein A. Proc Natl Acad Sci U S A. 1988;85:5566–5570. doi: 10.1073/pnas.85.15.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, Rates SR, Fisher AR. Secretagogues increase the expression of surfactant protein A receptors on lung type II cells. J Biol Chem. 1996;271:25277–25283. doi: 10.1074/jbc.271.41.25277. [DOI] [PubMed] [Google Scholar]
- 25.Rates SR, Kazi AS, Tao JQ, Yu KJ, Gonder DS, Feinstein SI, Fisher AR. Role of P63 (CKAP4) in binding of surfactant protein-A to type II pneumocytes. Am J Physiol Lung Cell Mol Physiol. 2008;295:L658–669. doi: 10.1152/ajplung.90233.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chroneos ZC, Abdolrasulnia R, Whitsett JA, Rice WR, Shepherd VL. Purification of a cell-surface receptor for surfactant protein A. J Biol Chem. 1996;271:16375–16383. doi: 10.1074/jbc.271.27.16375. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein JL, Brown MS, Anderson RG, Russell DW, Schneider WJ. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu Rev Cell Biol. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 28.Ryan RM, Morris RE, Rice WR, Ciraolo G, Whitsett JA. Binding and uptake of pulmonary surfactant protein (SP-A) by pulmonary type II epithelial cells. J Histochem Cytochem. 1989;37:429–440. doi: 10.1177/37.4.2926121. [DOI] [PubMed] [Google Scholar]
- 29.Stevens PA, Wissel H, Zastrow S, Sieger D, Zimmer KP. Surfactant protein A and lipid are internalized via the coated-pit pathway by type II pneumocytes. Am J Physiol Lung Cell Mol Physiol. 2001;280:L141–151. doi: 10.1152/ajplung.2001.280.1.L141. [DOI] [PubMed] [Google Scholar]
- 30.Jain D, Dodia C, Fisher AR, Rates SR. Pathways for clearance of surfactant protein A from the lung. Am J Physiol Lung Cell Mol Physiol. 2005;289:L1011–1018. doi: 10.1152/ajplung.00250.2005. [DOI] [PubMed] [Google Scholar]
- 31.Stevens PA, Wissel H, Sieger D, Meienreis-Sudau V, Rustow R. Identification of a new surfactant protein A binding protein at the cell membrane of rat type II pneumocytes. Biochem J. 1995;308:77–81. doi: 10.1042/bj3080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strayer DS, Yang S, Jerng HH. Surfactant protein A-binding proteins. Characterization and structures. J Biol Chem. 1993;268:18679–18684. [PubMed] [Google Scholar]
- 33.Kresch MJ, Christian C, Lu H. Isolation and partial characterization of a receptor to surfactant protein A expressed by rat type II pneumocytes. Am J Respir Cell Mol Biol. 1998;19:216–225. doi: 10.1165/ajrcmb.19.2.3061. [DOI] [PubMed] [Google Scholar]
- 34.Gupta N, Manevich Y, Kazi AS, Tao JQ, Fisher AR, Bates SR. Identification and characterization of p63 (CKAP4/ ERGIC-63/CLIMP-63), a surfactant protein A binding protein, on type II pneumocytes. Am J Physiol Lung Cell Mol Physiol. 2006;291:L436–446. doi: 10.1152/ajplung.00415.2005. [DOI] [PubMed] [Google Scholar]
- 35.Tino MJ, Wright Interactions of surfactant protein A with epithelial cells and phagocytes. Biochim Riophys Acta. 1998;1408:241–263. doi: 10.1016/s0925-4439(98)00071-4. [DOI] [PubMed] [Google Scholar]
- 36.Strayer DS. Identification of a cell membrane protein that binds alveolar surfactant. Am J Pathol. 1991;138:1085–1095. [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Q, Fisher AB, Strayer DS, Bates SR. Mechanism for secretagogue-induced surfactant protein A binding to lung epithelial cells. Am J Physiol. 1998;275:L38–46. doi: 10.1152/ajplung.1998.275.1.L38. [DOI] [PubMed] [Google Scholar]
- 38.Strayer DS, Pinder R, Chander A. Receptor-mediated regulation of pulmonary surfactant secretion. Exp Cell Res. 1996;226:90–97. doi: 10.1006/excr.1996.0206. [DOI] [PubMed] [Google Scholar]
- 39.Wissel H, Looman AC, Fritzsche I, Rustow B, Stevens PA. SP-A-binding protein BP55 is involved in surfactant endocytosis by type II pneumocytes. Am J Physiol. 1996;271:L432–440. doi: 10.1152/ajplung.1996.271.3.L432. [DOI] [PubMed] [Google Scholar]
- 40.Samten B, Townsend JC, Sever-Chroneos Z, Pasquinelli V, Barnes PF, Chroneos ZC. An antibody against the surfactant protein A (SP-A)-binding domain of the SP-A receptor inhibits T cell-mediated immune responses to Mycobacterium tuberculosis. J Leukoc Biol. 2008;84:115–123. doi: 10.1189/jlb.1207835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borron P, McCormack FX, Elhalwagi BM, Chroneos ZC, Lewis JF, Zhu S, Wright, Shepherd VL, Possmayer F, Inchley K, Fraher LJ. Surfactant protein A inhibits T cell proliferation via its collagen-like tail and a 210-kDa receptor. Am J Physiol. 1998;275:L679–686. doi: 10.1152/ajplung.1998.275.4.L679. [DOI] [PubMed] [Google Scholar]
- 42.Yang CH, Szeliga J, Jordan J, Faske S, Sever-Chroneos Z, Dorsett B, Christian RE, Settlage RE, Shabanowitz J, Hunt DF, Whitsett JA, Chroneos ZC. Identification of the surfactant protein A receptor 210 as the unconventional myosin 18A. J Biol Chem. 2005;280:34447–34457. doi: 10.1074/jbc.M505229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer A, Ericsson M, Bachi T, Griffiths G, Hauri HP. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J Cell Sci. 1993;104:671–683. doi: 10.1242/jcs.104.3.671. [DOI] [PubMed] [Google Scholar]
- 44.Razzaq TM, Bass R, Vines DJ, Werner F, Whawell SA, Ellis V. Functional regulation of tissue plasminogen activator on the surface of vascular smooth muscle cells by the type-II transmembrane protein p63 (CKAP4) J Biol Chem. 2003;278:42679–42685. doi: 10.1074/jbc.M305695200. [DOI] [PubMed] [Google Scholar]
- 45.Conrads TP, Tocci GM, Hood BL, Zhang CO, Guo L, Koch KR, Michejda CJ, Veenstra TD, Keay SK. CKAP4/p63 is a receptor for the frizzled-8 protein-related antiproliferative factor from interstitial cystitis patients. J Biol Chem. 2006;281:37836–37843. doi: 10.1074/jbc.M604581200. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer A, Rohrer J, Slot JW, Geuze HJ, Kornfeld S. Reassessment of the subcellular localization of p63. J Cell Sci. 1995;108:2477–2485. doi: 10.1242/jcs.108.6.2477. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer A, Rohrer J, Jeno P, DeMaio A, Buchman TG, Hauri HP. A reversibly palmitoylated resident protein (p63) of 59 an ER-Golgi intermediate compartment is related to a circulatory shock resuscitation protein. J Cell Sci. 1993;104:685–694. doi: 10.1242/jcs.104.3.685. [DOI] [PubMed] [Google Scholar]
- 48.Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. Embo J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, Hauri HP. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J Cell Biol. 2001;153:1287–1300. doi: 10.1083/jcb.153.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vedrenne C, Klopfenstein DR, Hauri HP. Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell. 2005;16:1928–1937. doi: 10.1091/mbc.E04-07-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweizer A, Rohrer J, Hauri HP, Kornfeld S. Retention of p63 in an ER-Golgi intermediate compartment depends on the presence of all three of its domains and on its ability to form oligomers. J Cell Biol. 1994;126:25–39. doi: 10.1083/jcb.126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vedrenne C, Hauri HP. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic. 2006;7:639–646. doi: 10.1111/j.1600-0854.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 53.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweizer A, Rohrer J, Kornfeld S. Determination of the structural requirements for palmitoylation of p63. J Biol Chem. 1995;270:9638–9644. doi: 10.1074/jbc.270.16.9638. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Planey SL, Ceballos C, Stevens SM, Jr., Keay SK, Zacharias DA. Identification of CKAP4/p63 as a major substrate of the palmitoyl acyltransferase DHHC2, a putative tumor suppressor, using a novel proteomics method. Mol Cell Proteomics. 2008;7:1378–1388. doi: 10.1074/mcp.M800069-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006;2006 doi: 10.1126/stke.3592006re14. re14. [DOI] [PubMed] [Google Scholar]
- 57.Monigatti F, Hekking B, Steen H. Protein sulfation analysis-A primer. Biochim Biophys Acta. 2006;1764:1904–1913. doi: 10.1016/j.bbapap.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 58.Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem. 2003;278:24243–24246. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- 59.Wilkins PP, Moore KL, McEver RP, Cummings RD. Tyrosine sulfation of P-selectin glycoprotein ligand-1 is required for high affinity binding to P-selectin. J Biol Chem. 1995;270:22677–22680. doi: 10.1074/jbc.270.39.22677. [DOI] [PubMed] [Google Scholar]
- 60.Leyte A, van Schijndel HB, Niehrs C, Huttner WB, Verbeet MP, Mertens K, van Mourik JA. Sulfation of Tyr1680 of human blood coagulation factor VIII is essential for the interaction of factor VIII with von Willebrand factor. J Biol Chem. 1991;266:740–746. [PubMed] [Google Scholar]
- 61.Costagliola S, Panneels V, Bonomi M, Koch J, Many MC, Smits G, Vassart G. Tyrosine sulfation is required for agonist recognition by glycoprotein hormone receptors. Embo J. 2002;21:504–513. doi: 10.1093/emboj/21.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher AB, Arad I, Dodia C, Chander A, Feinstein SI. cAMP increases synthesis of surfactant-associated protein A by perfused rat lung. Am J Physiol. 1991;260:L226–233. doi: 10.1152/ajplung.1991.260.4.L226. [DOI] [PubMed] [Google Scholar]
- 63.Keay SK, Szekely Z, Conrads TP, Veenstra TD, Barchi JJ, Jr., Zhang CO, Koch KR, Michejda CJ. An antiproliferative factor from interstitial cystitis patients is a frizzled 8 protein-related sialoglycopeptide. Proc Natl Acad Sci U S A. 2004;101:11803–11808. doi: 10.1073/pnas.0404509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herbein JF, Savov J, Wright Binding and uptake of surfactant protein D by freshly isolated rat alveolar type II cells. Am J Physiol Lung Cell Mol Physiol. 2000;278:L830–839. doi: 10.1152/ajplung.2000.278.4.L830. [DOI] [PubMed] [Google Scholar]
- 65.Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wissel H, Looman AC, Fritzsche I, Rustow B, Stevens PA. SP-A-binding protein BP55 is involved in surfactant endocytosis by type II pneumocytes. Am J Physiol. Lung Cell Mol Physiol. 1996;271:L432–440. doi: 10.1152/ajplung.1996.271.3.L432. [DOI] [PubMed] [Google Scholar]