Abstract
A cross sectional study was conducted on 906 apparently healthy camels slaughtered at Akaki and Metehara abattoirs to investigate the pathology of camel tuberculosis (TB) and characterize its causative agents using postmortem examination, mycobacteriological culturing, and multiplex polymerase chain reaction (PCR), region of difference-4 (RD4)-based PCR and spoligotyping. The prevalence of camel TB was 10.04% (91/906) on the basis of pathology and it was significantly higher in females (χ2 = 4.789; P = 0.029). The tropism of TB lesions was significantly different among the lymph nodes (χ2 = 22.697; P = 0.002) and lung lobes (χ2 = 17.901; P = 0.006). Mycobacterial growth was observed in 34% (31/91) of camels with grossly suspicious TB lesions. Upon further molecular characterization using multiplex PCR, 68% (21/31) of the colonies showed a positive signal for the genus Mycobacterium, of which two were confirmed Mycobacterium bovis (M. bovis) by RD4 deletion typing. Further characterization of the two M. bovis at strains level revealed that one of the strains was SB0133 while the other strain was new and had not been reported to the M. bovis database prior to this study. Hence, it has now been reported to the database, and designated as SB1953. In conclusion, the results of the present study have shown that the majority of camel TB lesions are caused by mycobacteria other than Mycobacterium tuberculosis complex. And hence further identification and characterization of these species would be useful towards the efforts made to control TB in camels.
Introduction
Pastoral production system accounts for the livelihood of 50–100 million people in developing countries and 60% of this population lives in more than 21 African countries confined to the most arid regions of the continent [1], [2]. In eastern Africa, Ethiopia has the largest pastoralist population (7–8 millions) representing around 20 ethnic groups [3]. The major ethnic groups in Ethiopia are Somalis, Afar, Kereyu and Borena pastoral communities occupying the Eastern and southern lowlands of the country. Pastoralist depends on livestock for their livelihood, moving seasonally from place to place in search of water and pasture for their animals [4]. The dromedary camel (Camelus dromedarius), which is a versatile animal capable of living in harshly semi-arid and arid areas of the world, is extremely important for livelihood of pastoral communities through provision of milk, meat and draft power for transportation of goods. In pastoral communities of Afar, Somali and Borena, camels are kept almost entirely for milk production [5]. In these communities, camel milk is consumed raw, and this habit combined with close physical contact with their animals create a potential public health concern for transmission of zoonotic diseases such as tuberculosis (TB) from animals to the pastoralist.
Although, the extent of TB has been well documented in humans and most domestic animals, very little is known about the pathology and cause of camel TB in pastoral areas of the world. Camel TB has been reported in Egypt [6], United Arab Emirates [7], [8], Pakistan [9], and Australia [10]. Mycobacterium tuberculosis (M. tuberculosis), Mycobacterium bovis (M. bovis), and atypical mycobacteria such as Mycobacterium kansasii (M. kansasii), Mycobacterium aquae (M. aquae), Mycobacterium fortuitum (M. fortuitum) and Mycobacterium smegmatis (M. smegmatis) have been isolated in camel as causative agents of camel TB [8], [11]. In Ethiopia, except one report indicating the existence of camel TB [12], there is a large paucity of information on the pathology and the causative agent of TB in camels of pastoral regions of the country. Therefore, investigation of the pathology of camel TB and identification of its causative agents is important to encourage the effort in the control of the disease and reduce its risk of zoonosis to the pastoralist community of Ethiopia. The present study, therefore, was designed to investigate the pathology of camel TB and identify the causative agent using molecular tools.
Materials and Methods
Study Animals
The cross sectional study was carried out on 906 apparently healthy male (n = 535) and female (n = 371) slaughtered camels. The camels slaughtered were brought to Akaki (Addis Ababa) and Metehara Abattoirs from the two main pastoral regions of Ethiopia, namely Awash-Fentale pastoral area (Kereyu and Afar in the Middle Awash region) and Borena pastoral area (southern Ethiopia). The catchment areas possess large number of camels, in Fentale pastoral area (Middle Awash region) there are 68,331 camels and in Borena pastoral area of Oromia Regional State which border with Kenya possesses an estimated population of 97,131 camels [13]. After arriving at the abattoir, the camels were staying for 2–7 days undergoing physical examination. On average 6–8 camels were slaughtered per day depending on the request from customers. The main consumers of camel meat in Addis Ababa are the Somali immigrants residing in the city.
Post mortem inspection and pathology scoring
Postmortem inspection was performed following the procedure as previously described [14]. Mandibular, retropharyngeal, bronchial, mediastinal, mesenteric and hepatic lymph nodes were examined and organs including lungs, liver, small intestine and kidneys were examined in detail during post-mortem in the abattoir under a bright-light source. The lobes of the left and right lungs were inspected and palpated externally. Then, each lobe was sectioned into about 2-cm-thick slices to facilitate the detection of lesions with sterile surgical blades. Similarly, lymph nodes were sliced into thin sections (about 2mm thick) and inspected for the presence of visible lesions. Whenever gross lesions suggestive of TB were detected in any of the tissue, the tissue was classified as having lesions.
Pathology scoring was conducted on tissues with abscesses and tubercle lesions to determine the severity of the lesions based on semi quantitative procedure developed previously [15], [16]. Briefly, lesions in the lobes of the lungs were scored separately as follows: 0 = no visible lesions; 1 = no gross lesions but lesions apparent on slicing of the lobe; 2 = fewer than five gross lesions; 3 = more than five gross lesions; 4 = gross coalescing lesions. The scores for the individual lobes were summed and generated lung score. Similarly, the severity of gross lesions in individual lymph nodes was scored as follows: 0 = no gross lesions; 1 = small lesion at one focus; 2 = small lesions at more than one focus; 3 = extensive necrosis. Individual lymph node scores were summed and generated the lymph node score. Total pathology score per animal was obtained from the sum of the two total scores.
Mycobacterial isolation from tissue lesions
For mycobacteriological isolation tuberculous lesions from slaughtered camels were aseptically collected into sterile universal bottles with about 5 ml of 0.9% saline solution and also kept in icebox with solid packs to keep the cold chain. Then the samples were transported to Aklilu Lemma Institute of Pathobiology (ALIPB) and stored at +2 to +8°C until mycobacteriological culturing was carried out in TB laboratory.
The samples were further processed for isolation of mycobacteria in accordance with the Office International des Epizooties [17], [18]. The specimens were sectioned using sterile blades, minced with scissors and homogenized with a sterile mortar and pestle under a biological safety cabinet. The homogenates were decontaminated by adding an equal volume of 4% NaOH on the sample in order to remove contaminants. Thereafter, centrifuged at 3,000 rpm for 15 minutes to concentrate the mycobacteria. The supernatant was discarded, and the sediment was neutralized by 1% (0.1 N) HCl acid using phenol red as an indicator. Neutralization was achieved when the color of the solution changed from purple to yellow [17]. Next, 0.1 ml of suspension from each sample was spread onto a slant of Löwenstein Jensen (LJ) medium. Duplicate slants were used, one enriched with sodium pyruvate and the other enriched with glycerol. Cultures were incubated aerobically at 37°C for 8–12 weeks with weekly observation for growth of colonies. Positive cultures were confirmed with Ziehl Nelseen staining and preserved with freezing media while at the same time heat killed in water bath at 80°C for 45 minutes. The frozen and heat killed isolates were stored at (−20°C) for further mycobacteriology and molecular typing analysis.
Mycobacterial genus typing
The multiplex PCR differentiate M. tuberculosis complex from M. avium complex, M. intracellularae and other mycobacterial species. Mycobacterial genus typing was conducted as described previously [19]. Heat killed AFB positive samples were used as source of DNA template.
DNA amplifications was done in thermocycler with 20 µl reaction volumes consisting: 5 µl of genomic DNA as a template, 8 µl HotstarTaqMasterMix (MgCL2, dNTP, Taq polymerase and PCR buffer) (Qiagen, United Kingdom) for each sample, 0.3 µl internal primer per sample, 0.3 µl forward and reverse primer per each sample and 5.2 µl per sample of Qiagen water. The primers used for amplification were MYCGEN-F, 5′AGA GTT TGA TCC TGG CTC AG 3′ (35ng/µl); MYCGEN-R, 5′TGC ACA CAG GCC ACA AGG GA 3′ (35ng/µl); MYCAV-R, 5′ ACC AGA AGA CAT GCG TCT TG 3′(35ng/µl); MYCINT-F, 5′CCT TTA GGC GCA TGA TGT CTT TA 3′(75ng/µl); TB1-F, 5′ GAA CAA TCC GGA GTT GAC AA 3′ (20ng/µl); TB-1-R, 5′ AGC ACG CTG TCA ATC ATG TA 3′ (20ng/µl). M. tuberculosis strains (H37Rv) and M. avium were used as positive control while Qiagen water was used as negative control. The reaction mixture was then heated in Programme Thermal Controller (Applied biosystem; PTC- 100™) cycle using the following amplification program: 95°C for 10 minutes for enzyme activation; 95°C for 1 minute for denaturation; 61°C for 0.5 minute for annealing; 72°C for 2 minutes for extension, involving 35 cycles all in all; and final extension at 72°C for 10 minutes.
The products were electrophoresed in 1% agarose gel in 10× TAE running buffer. Ethidium bromide at ratio of 1∶ 10, 100bp DNA ladder, and orange 6× loading dye were used in gel electrophoresis. All members of the genus Mycobacterium produce a band of 1030bp, M. avium or subspecies such as M. avium subspecies paratuberculosis produces a band of 180bp, M. intracellularae a band of 850bp while members of M. tuberculosis complex produce a band with 372bp.
RD4 deletions typing
PCR analysis on the basis of RD regions has been found to be an important differentiating tool between members of the M. tuberculosis complex. RD4 is 12.7 kb genetic segment that is deleted from M. bovis BCG strain, but present in M. microti, M. africanum, and M. tuberculosis [20].
The RD4 deletion typing was carried out on isolates that showed band for M. tuberculosis complex by multiplex PCR. For this deletion typing, the procedure described by Cadamus and coauthors was followed [21]. Each sample was tested in a separate PCR tube. Primers directed against the RD4 were used to generate a deletion profile that would allow species identification of the isolate. Primers that were used include RD4intF ACA CGC TGG CGA AGT ATA GC, RD4flankF CTC GTC GAA GGC CAC TAA AG and RD4flankR AAG GCG AAC AGA TTC AGC AT to check for the presence of RD4 locus. The HotStarTaq Master Mix system from Qiagen was used for PCR, with primers described previously. The reaction mixture was 10 µl of HotStarTaq Master Mix, 0.3 µl×3 of each primer (flank R, F and int), 2 µl DNA template and 7 µl distilled water to a final volume of 20 µl. M. tuberculosis H37Rv and M. bovis 2122/97 were used as positive control while Qiagen water was used as negative control. The mixture was heated in Programme Thermal Controller (Applied biosystem; PTC- 100™) using an initial hot start of 95°C for 15 minutes, followed by 35 cycles of 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute; a final extension step of 72°C for 10 minutes to complete the cycle. PCR products were electrophoresed in 1% agarose gel in 1× TAE running buffer, Ethidium bromide at ratio of 1∶ 10, 100bp DNA ladder and orange 6× loading dye were used in electrophoresis. The gel was visualized in Multi–image™ light cabinet using Alpha innotech version 1.2.0.1(Alpha Innotech Corporation). The presence of RD4 (M. tuberculosis, M. africanum) gives a product size of 335bp (RD4int+RD4FlankR) and its absence (M. bovis) gives a product size of 446bp (RD4FlankR+RD4FlankF).
Spoligotyping
Spoligotyping was performed as previously described by Kamerbeek and coauthors [22] and according to the spoligotype kit supplier's instructions (Ocimum Biosolutions Company, Iisselstein, The Netherlands). The direct repeat (DR) region was amplified by PCR using oligonucleotide primers derived from the DR sequence. A total volume of 25 µl the following reaction mixture was used for the PCR: 12.5 µl of HotStarTaq Master Mix (Qiagen: this solution provides a final concentration of 1.5 mM MgCl2 and 200µM of each deoxnucleotides triphosphates), 2 µl of each primer (20 pmol each), 5 µl suspension of heat-killed cells (approximately 10 to 50ng), and 3.5 µl distilled water. The mixture was heated for 15 minutes at 96°C and then subjected to 30 cycles of 1 minute at 96°C, 1 minute at 55°C, and 30 seconds at 72°C. The amplified product was hybridized to a set of 43 immobilized oligonucleotides, each corresponding to one of the unique spacer DNA sequences within the DR locus. After hybridization, the membrane was washed twice for 10 minutes in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA[pH 7.7])-0.5% sodium dodecyl sulfate at 60°C and then incubated in 1∶4000 diluted streptavidin-peroxidase (Boehringer) for 45 to 60 minutes at 42°C. The membrane was washed twice for 10 minutes in 2× SSPE-0.5% sodium dodecyl sulfate at 42°C and rinsed with 2× SSPE for 5 minutes at room temperature. Hybridizing DNA was detected by the enhanced chemiluminescence method (Amersham) and by exposure to X-ray film (Hyperfilm ECL, Amersham) as specified by the manufacturer.
Data management and analysis
Data were classified, filtered and coded using MS Excel 5, and was transferred to STATA version 8 for statistical analysis. Mean and standard error of the mean were used to summarize pathology scores. Similarly, proportions were used to summarize categorical exposure and outcome measures. Friedman test was used to compare pathology score of tropism of TB lesions among lymph nodes as well as among lung lobes. Bivariate and multivariable logistic regression analyses were used to assess the strength of associations of selected factors and prevalence of camel TB. Effects were reported as statistically significant if p-value was less than 5%. Odds ratio and 95% confidence intervals were used to measure the strength of associations.
Results
Prevalence of camel tuberculosis
On the basis of gross pathology, the prevalence of camel TB was 10% (91/906). Culture positivity was confirmed in 34% (31/91) of the camels with suspicious TB lesions. The result of the association of the different risk factors to the pathology showed that having a good body condition has a protective effect against being positive for TB (Table 1).
Table 1. Logistic regression analysis of tuberculous lesions with various host-related risk factors.
| Characteristics | No. examined | No. of positive (%) | Crude Odds ratio (95% CI) | Adjusted Odds ratio (95% CI) |
| Sex | ||||
| Female | 371 | 46 (12.4) | 1 | 1 |
| Male | 535 | 45(8.4) | 0.62 (0.40–0.95) | 0.64 (0.36–1.2) |
| Age | ||||
| <4 | 99 | 12 (12.1) | 1 | 1 |
| 4–6 | 184 | 14 (7.6) | 0.60 (0.26–1.35) | 0.25 (0.26–1.35) |
| 7–9 | 141 | 10 (7.1) | 0.55 (0.23–1.34) | 0.54 (0.22–1.32) |
| 10–15 | 197 | 19 (9.6) | 0.77 (0.36–1.67) | 0.64 (0.28–1.46) |
| 16+ | 285 | 36 (12.6) | 1.05 (0.52–2.11) | 0.74 (0.32–1.68) |
| BCS | ||||
| Poor | 389 | 44 (11.3) | 1 | 1 |
| Medium | 330 | 36 (10.9) | 0.96 (0.60–1.53) | 0.92 (0.57–1.47) |
| Good | 187 | 11 (5.9) | 0.46 (0.25–.0.97) | 0.42 (0.20–0.86) |
| Origin | ||||
| Kereyu | 609 | 56 (9.2) | 1 | 1 |
| Borena | 297 | 35 (11.8) | 1.32 (0.84–2.06) | 1.24 (0.70–2.2) |
CI = Confidence Interval; BCS = Body Condition Scoring; odds ratio corresponding to different categories of a given variable are adjusted for the remaining three variables.
Pathology scoring
The distribution of lesions and the severity of the disease were established in the 91 camels with suspicious lesions. The tropism of TB lesions to specific lymph nodes and lung lobes was statistically significant among the lymph nodes (χ2 = 22.697; P = 0.002) and lung lobes (χ2 = 17.901; P = 0.006) (Table 2). Lung lesions were detected in 43 camels while 78 camels had at least one lesion in their lymph nodes. The lesions appeared more frequent in the apical and cardiac lobes of both lungs than in the diaphragmatic lobes (Table 2). Similarly, the severity was greater in both right apical and cardiac lobes. Regarding lymph nodes, mesenteric lymph nodes were found the most frequently and severely affected of all the lymph nodes (34%) (Table 2, Figure 1).
Table 2. Distribution and tropism of tuberculous lesions in the lymph nodes and lung lobes in 91 postmortem positive camels with lesions in at least one tissue or organ.
| No (%) of camels with TB lesions | ||||
| Tissue | Total | Positive organs or tissue | χ2 | P-value |
| Lymph nodes | 22.697‡ | *0.002 | ||
| Parotid | 91 | 13 (14.3%) | ||
| Mandibular | 91 | 15 (16.5%) | ||
| Retropharyngeal | 91 | 17 (18.7%) | ||
| Mediastinal | 91 | 30 (33%) | ||
| Left bronchial | 91 | 17 (18.7%) | ||
| Right bronchial | 91 | 21 (23.1%) | ||
| Mesenteric | 91 | 31 (34.1%) | ||
| Hepatic | 91 | 3 (3.3%) | ||
| Lung lobes | 17.901‡ | *0.006 | ||
| Left apical | 91 | 30 (33%) | ||
| Left cardiac | 91 | 27 (29.7%) | ||
| Left diaphragmatic | 91 | 22 (24.2%) | ||
| Right apical | 91 | 25 (27.5%) | ||
| Right cardiac | 91 | 27 (29.7%) | ||
| Right diaphragmatic | 91 | 19 (20.9%) | ||
| Right accessory | 91 | 18 (19.8%) | ||
*Statistically significant.
Chi-square was calculated from the median of pathology score of among tissues examined.
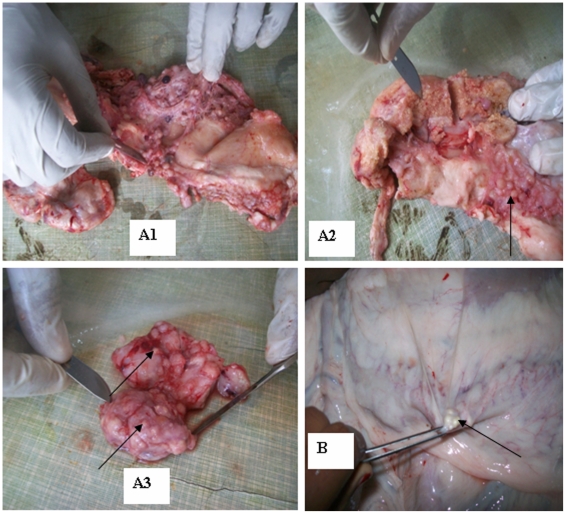
Figure 1. Tuberculous lesions from camels on different organs.
(A1) Disseminated and distinct tuberculous lesions in mediastinal parts of the lung. (A2) Tuberculous lesion in mediastinal lymph node and nodules on other parts as indicated by arrows. (A3) Tuberculous lesions in hepatic lymph node. The arrows show that pea-sized lesions throughout the lymph node. (B) Tuberculous lesion in mesenteric lymph nodes as indicated by arrow.
The mean severity of pathology of camel TB is summarized in Table 3. The mesenteric lymph node constituting the most severely affected lymph node (0.64±0.11; 0.55±0.15) followed by mediastinal lymph node (0.27±0.08).
Table 3. Mean pathology and standard error of the mean scoring of the lungs and lymph nodes of camels.
| Lung lobes | Mean ± SEM | Lymph nodes | Mean ± SEM |
| Left apical lobe | 0.64±0.11 | Parotid | 0.27±0.08 |
| Left cardiac lobe | 0.63±0.12 | Mandibular | 0.34±0.09 |
| Left diaphragmatic lobe | 0.56±0.11 | Retropharyngeal | 0.30±0.08 |
| Right apical lobe | 0.69±0.13 | Mediastinal | 0.55±0.15 |
| Right cardiac lobe | 0.72±0.13 | Left bronchial | 0.31±0.08 |
| Right diaphragmatic lobe | 0.47±0.11 | Right bronchial | 0.44±0.09 |
| Right accessory lobe | 0.41±0.10 | Mesenteric | 0.64±0.11 |
SEM = Standard Error of the Mean.
Mycobacteriology
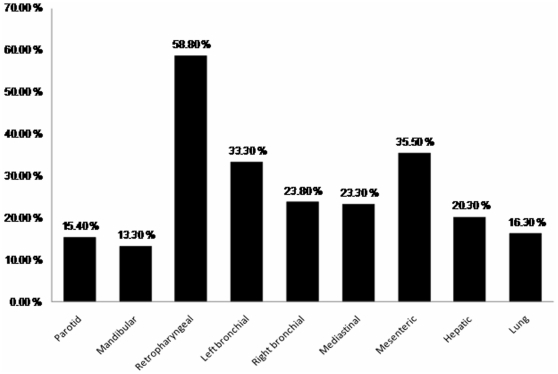
Growth of mycobacteria was observed in 34% (31/91) of camels with suspicious TB lesion (see Figure 2). Culture positivity was highest (58.8%) in the retropharyngeal lymph node followed by the mesenteric lymph node (35.5%). In contrast, isolation from mandibular and parotid lymph nodes were less frequently mycobacterial culture positive with the positivity of 13.3% and 15.4%, respectively.
Figure 2. Proportion of mycobacterial culture positivity of the lymph nodes and lungs of TB suspected camels.
Molecular characterization of the isolates
Multiplex PCR
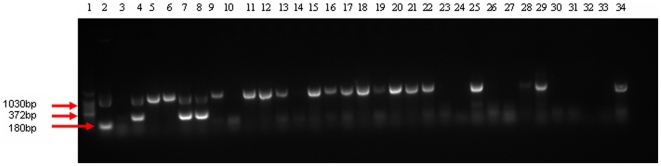
Further Mycobacterium genus typing was conducted on the 31 culture isolates from camels. Based on multiplex PCR using the primers of the M. tuberculosis complex and M. avium complex, 21 isolates gave signal to the genus Mycobacterium. Two of these isolates were confirmed to be members of the M. tuberculosis complex and none of the isolates were M. avium complex (Figure 3).
Figure 3. Gel electrophoresis separation of PCR products by multiplex PCR genus typing of mycobacteria isolated from naturally infected camels.
Lane 1 = 100bp DNA Ladder; Lane 2 = Mycobacterium avium complex (positive control), Lane 3 = Qiagen H2O (negative control), Lane 4 = Mycobacterium tuberculosis complex (positive control), Lanes 5–34 were isolates from individual camels with tuberculous lesions. Lane 7 (sample 63), Lane 8 (sample 62) were positive for Mycobacterium tuberculosis complex and Lane 5, 6, 7, 8, 9, 11, 12–13, 15–22, 25, 28, 29, 34 were positive for genus Mycobacterium, Lane 10, 14, 23, 24, 26, 27, 30–33 were negative for genus Mycobacterium.
RD4 deletion typing
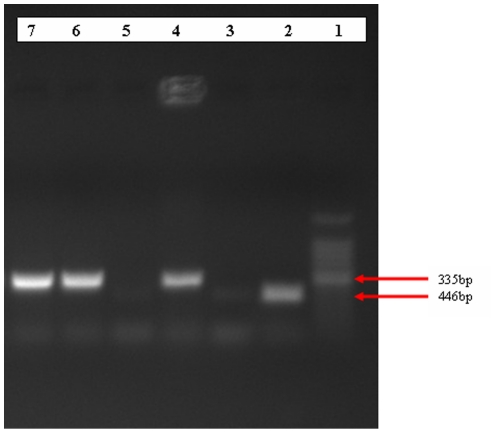
The two isolates that showed signal to M. tuberculosis complex were subjected to RD4 deletion typing for further differentiation of species and they were confirmed to be M. bovis (Figure 4).
Figure 4. Gel electrophoresis separation of PCR products by RD4 deletion typing of mycobacteria isolated from naturally infected camels.
Lane 1 = 100bp DNA ladder, Lane 2 = M. tuberculosis positive control, Lane 3 = Qiagen H2O (negative control), Lane 4 = M. bovis positive control, Lane 5–7 were isolates from camel, Lane 6 and 7 were positive for M. bovis.
Spoligotyping
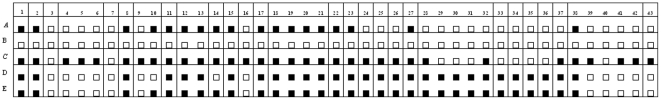
The two isolates that showed signal with RD4 deletion PCR typing were further characterized using spoligotyping. One of these confirmed to be SB0133 and the other one was a new strain which was not reported previously in M. bovis database. The new strain was reported to the global database (http://www.Mbovis.org) and designated as SB1953 (Figure 5). The SB0133 isolate was isolated from camel with generalized and disseminated form of TB.
Figure 5. Schematic representation of the spoligotyping patterns of isolates of Mycobacterium bovis from camels with tuberculous lesions.
A = M.bovis SB1176 (positive control); B = Qiagen H2O (negative control); C = M. tuberculosis (positive control); D = sample 63 (SB1953-New strain); E = sample 62 (SB0133). The black rectangles represent positive signals, and the white rectangles indicate negative signals.
Discussion
In general, there is scanty information on TB in camels. Nonetheless, there are few reports published on camel TB in Ethiopia as well as in other countries. The prevalence of camel TB recorded by the present study is similar to the report of previous study in the Afar Region of Ethiopia based on comparative intradermal tuberculin test in camels [23] but it is higher than the report from Dire Dewa Abattoir in camels from eastern Ethiopia [12]. Similarly, it is higher than the prevalence reported in Egypt [6].
The occurrences of TB lesions in camels were relatively higher in the younger and older camels than other age groups. Other researchers have also reported in cattle particularly that older animals are affected by TB [24]–[27] which could be due to the fact that older animals have weaker immune system. The higher frequency of lesion in younger camels could be due to the less developed immunity [28]. Young camels can also be easily infected with higher doses of mycobacteria via colostrums from infected camel in a similar way, as it occurs in cattle [29]. In connection with this, another report mentioned of vertical transmission of M. bovis from an infected dam to her calf through congenital infection in utero [30]. It was observed that lesion was more frequently observed in female camels as compared to male camels. This could be due to the fact that female camels were brought for slaughter at their older age after completion of the reproductive age [26], [31].
The distribution, frequency, and severity of lesions recorded in different tissues of camels were similar with the reports of similar studies in grazing cattle in Ethiopia [18], [32]. In these studies, the frequency and severity of the lesions were higher in the mesenteric lymph nodes than the thoracic lymph nodes, while in other studies under intensive cattle husbandry lesions were predominant in the respiratory tract and thoracic lymph nodes [14], [16].
Tuberculous lesions were subjected to bacteriological culture so as to identify and characterize the causative agents. However, culture positivity of suspicious tissues was 34%, which is lower than what have been reported previously from cattle [18], [32]. The lower culture positivity might be related to the non-optimal condition of the culture for NTM which assumed to be the major isolates causing pathology in camel. Regarding culture positivity of each organ, the highest culture positivity was recorded in the retropharyngeal lymph node followed by mesenteric lymph node, which could suggest that oral route could be the main route of infection. In contrast, other authors have reported that culture positivity was higher in lung tissue and thoracic lymph nodes than in the head and mesenteric lymph nodes [14], [32], [33].
Genus typing of the isolates revealed that out of 21 isolates which showed signals for the genus Mycobacterium, only two isolates were M. bovis as confirmed by RD4 deletion typing and spoligotyping, while the remaining 18 did not show signal to the M. tuberculosis complex, and hence assumed to be members of nontuberculous mycobacteria (NTM). In the present study, the NTM resulted sarcoid-like tuberculous nodules with granulomatous and caseous lesions in lymph nodes, lung and other organs of camel. Previous study reported also the isolation NTM including M. kansasii and M. smegmatis from tuberculous like lesions in camel causing a similar caseous nodules like those caused by M. bovis and M. tuberculosis [11]. In Ethiopia, NTM have been isolated from cattle with tuberculous lesions in different regions of the country [34], which indicates their wider geographic distribution and role as a cause of tuberculous lesions in livestock of the country. Therefore, further identification and characterization of these isolates are necessary.
Spoligotyping of the two M. bovis isolates revealed a distinct spoligopattern. Referring to the global http://www.Mbovis.org database of the spoligopatterns indicated that one of the strains which caused a generalized disseminated TB in camel was SB0133, whereas the other strain was new strain not reported in the database previously. The new strain now has been reported to the database and designated as SB1953. In Ethiopia, a number of studies reported new strains with specific spoligotype pattern in cattle [32], [34]–[36]. The identification of this new M. bovis strain from camel TB case of pastoral area of Ethiopia indicates the need for further research in identifying the circulating strains of M. bovis in various hosts and their distribution in geographical area of the country. On the other hand, the isolation of SB0133 M. bovis strain in present study from camel of pastoral area of Ethiopia inline with the isolation of this strain from cattle of southern Ethiopia [34], [36] and pastoral area of Uganda [37] indicates the predominant localization of the strain to pastoral regions of Eastern Africa and possible interspecies transmission of the strain among livestock of pastoral area. In addition, the development of TB lesions to generalized disseminated form of TB in camel affected by SB0133 strain might imply its high pathogenicity in camels.
In conclusion, the present study has shown that the majority of camel TB lesions were caused by NTM; hence, further identification and characterization of these species would be useful towards the efforts made to control TB in camels. The isolation of M. bovis strain (SB0133) which is similar to cattle strain in pastoral area of East Africa, implies the existence of potential inter-species transmission of the strain among livestock of pastoral area, warrant further investigation to elucidate its epidemiological significance for public health and control of the disease in the region.
Acknowledgments
The authors are grateful to Addis Ababa Abattoir Enterprise (Akaki) and Metahara Abattoir for their support during sample collection. We also thankful for Dr. Stefan Berg for provision of primers and other reagents for PCR, TB laboratory staffs at Aklilu Lemma Institute of Pathobiology and Faculty of Veterinary Medicine of Addis Ababa University for their support during the laboratory work.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was financially supported by the Norwegian Programme for Development, Research and Education (NUFU), PRO-2007/10198. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sheik-Mohamed A, Velema JP. Where Health Care Has No Access; the Nomadic Populations of Sub- Sahara Africa. Trop Med Int Med. 1999;4:695–707. doi: 10.1046/j.1365-3156.1999.00473.x. [DOI] [PubMed] [Google Scholar]
- 2.UNDP. Between a Rock and Hard Place: Armed Violence in Africa Pastoral Communities. 2007. United Nations Development Programme report 2007.
- 3.Markakis J. Pastoralism on the margin. 2004. 15p Report Minority Rights Group International (MRG), 2004.
- 4.Nori M. Sustainable camel milk production and commercialization in Somalia. 2005. Future Livestock Systems APS course, Wageningen-4/2005.
- 5.Getahun T, Belay K. Camel husbandry practices in Eastern Ethiopia: The case of Jijiga and Shinile Zones. Nomadic Peoples. 2002;6:155–176. [Google Scholar]
- 6.Refai M. Bacterial and Mycotic Diseases of Camels in Egypt. 1992. In: ‘Proceedings of the First International Camel Conference’. Newmarket Press. UK.
- 7.Wernery U, Kaaden OR. Infectious Diseases of Camelids. Berlin: Blackwell Science Publisher; 2002. pp. 23–373. [Google Scholar]
- 8.Kinne J, Johnson B, Jahans KL, Smith NH, Ul-Haq A, et al. Camel Tuberculosis-a Case Report. Trop Anim Hlth Prod. 2006;38:38. doi: 10.1007/s11250-006-4366-8. [DOI] [PubMed] [Google Scholar]
- 9.Zubair R, Khan AMZ, Sabri MA. Pathology in camel lungs. J camel science. 2004;1:103–106. [Google Scholar]
- 10.Manefield GW, Tinson AH. Camels. 1997. A Compendium Sydney Post Graduate Foundation Vade Mecum Series C No. 22.
- 11.Elmossalami E, Siam MA, El Sergany M. Studies on tuberculosis-like lesions in slaughtered camels. Zentralblatt für Veterinarmedizia B. 1971;18(4):253–261. doi: 10.1111/j.1439-0450.1971.tb00355.x. [DOI] [PubMed] [Google Scholar]
- 12.Mamo G, Kassaye A, Sanni M, Ameni G. A cross sectional study of camel tuberculosis in Ethiopia. Bull Anim Hlth Prod Afr. 2009;57:1. [Google Scholar]
- 13.CSA. Federal Democratic Republic of Ethiopia Central Statistical Agency. Agricultural Sample Survey Report on Livestock and Livestock Characteristics, Addis Ababa. Statistical bull. 2008;2:16. [Google Scholar]
- 14.Corner LA. Post-mortem Diagnosis of Mycobacterium bovis Infection in Cattle. Vet Microbiology. 1994;40:53–63. doi: 10.1016/0378-1135(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 15.Vordermeier HM, Chambers MA, Cockle PJ, Whelan AO, Simmons J, et al. Correlation of ESAT-6-Specific Gamma Interferon with Pathology in Cattle Following Mycobacterium bovis BCG Vaccination Against Experimental Bovine Tuberculosis. Infect Immunol. 2002;70:3026–3032. doi: 10.1128/IAI.70.6.3026-3032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ameni G, Aseffa A, Engers H, Young D, Hewinson G, et al. Cattle Husbandry in Ethiopia is a Predominant Factor Affecting the Pathology of Bovine Tuberculosis and Gamma Interferon Responses to Mycobacterial Antigens. Clin Vaccine Immunol. 2006;13:1030–1036. doi: 10.1128/CVI.00134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.OIE. Bovine Tuberculosis. 2004. pp. 451–463. OIE Manual of Standards for Diagnostic Tests and Vaccines. 5th ed.
- 18.Ameni G, Aseffa A, Engers H, Young D, Gordon S, et al. High Prevalence and Increased Severity of Pathology of Bovine Tuberculosis in Holsteins Compared to Zebu Breeds under Field Cattle Husbandry in Central Ethiopia. Clin Vaccine immunol. 2007;14:1356–1361. doi: 10.1128/CVI.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilton S, Cousins D. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube PCR. Meth App. 1992;1:269–273. doi: 10.1101/gr.1.4.269. [DOI] [PubMed] [Google Scholar]
- 20.Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, et al. Identification of Variable Regions in the Genomes of Tubercle Bacilli Using Bacterial Artificial Chromosome Arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 21.Cadmus S, Palmer S, Okker M, Dale J, Gover K, et al. Molecular Analysis of Human and Bovine Tubercle Bacilli from a Local Setting in Nigeria. J Clin Microbiol. 2006;44:29–34. doi: 10.1128/JCM.44.1.29-34.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. Simultaneous Detection and Strain Differentiation of Mycobacterium tuberculosis for Diagnosis and Epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussen N. Study on Mycobacterium tuberculosis Complex Infection in Livestock and Humans in Amibara District of Afar Region. 2009. MSc thesis, Addis Ababa University, Faculty of Veterinary Medicine, Debre Zeit, Ethiopia.
- 24.Kazwala RR, Kambarage DM, Daborn CJ, Nyange J, Jiwa SFH, et al. Risk Factors Associated with the Occurrence of Bovine Tuberculosis in Cattle in the Southern Highlands of Tanzania. Vet Res Commun. 2001;25:609–614. doi: 10.1023/a:1012757011524. [DOI] [PubMed] [Google Scholar]
- 25.Cleaveland S, Shaw DJ, Mfinanga SG, Shirima G, Kazwala RR, et al. Mycobacterium bovis in Rural Tanzania: Risk Factors for Infection in Human and Cattle Populations. Tuberculosis. 2007;87:30–43. doi: 10.1016/j.tube.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Inangolet FO, Biffa D, Oloya J, Opuda-Asibo J, Skjerve E. A Cross-Sectional Study of Bovine Tuberculosis in the Transhumant and Agro-Pastoral Cattle Herds in the Border Areas of Katakwi and Moroto Districts, Uganda. Trop Anim Hlth Prod. 2008;40:501–508. doi: 10.1007/s11250-007-9126-x. [DOI] [PubMed] [Google Scholar]
- 27.Munyeme M, Muma JB, Skjerve E, Nambota AM, Phiri IGK, et al. Risk Factors Associated with Bovine Tuberculosis in Traditional Cattle of the Livestock/Wildlife Interface Areas in the Kafue Basin of Zambia. Prev Vet Med. 2008;85:317–328. doi: 10.1016/j.prevetmed.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Menzies FD, Neill SD. Cattle-to-Cattle Transmission of Bovine Tuberculosis. Vet J. 2000;160:92–106. doi: 10.1053/tvjl.2000.0482. [DOI] [PubMed] [Google Scholar]
- 29.Phillips CJC, Foster CRW, Morris PA, Teverson R. The Transmission of Mycobacterium bovis Infection in Cattle. Res Vet Sci. 2003;74:1–15. doi: 10.1016/s0034-5288(02)00145-5. [DOI] [PubMed] [Google Scholar]
- 30.Ozyigit MO, Senturk S, Akkoc A. Suspected Congenital Generalised Tuberculosis in a Newborn Calf. Vet Rec. 2007;160:307–308. doi: 10.1136/vr.160.9.307. [DOI] [PubMed] [Google Scholar]
- 31.Munyeme M, Muma JB, Samui KL, Skjerve E, Nambota AM, et al. Prevalence of Bovine Tuberculosis and Animal Level Risk Factors for Indigenous Cattle under Different Grazing Strategies in the Livestock/Wildlife Interface Areas of Zambia. Trop Anim Hlth Prod. 2008;41:345–352. doi: 10.1007/s11250-008-9195-5. [DOI] [PubMed] [Google Scholar]
- 32.Ameni G, Desta F, Firdessa R. Molecular typing of Mycobacterium bovis isolated from tuberculous lesions of cattle in north eastern Ethiopia. Vet Rec. 2010;167:138–141. doi: 10.1136/vr.b4881. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly LM, Dabron CJ. Epidemiology of Mycobacterium bovis Infections in Animals and Man: A Review. Tuber Lung Dis. 1995;76(Suppl. 1):1–46. doi: 10.1016/0962-8479(95)90591-x. [DOI] [PubMed] [Google Scholar]
- 34.Berg S, Firdessa R, Habtamu M, Gadisa E, Mengistu A, et al. The Burden of Mycobacterial Disease in Ethiopian Cattle: Implications for Public Health. PLoS ONE. 2009;4(4):e5068. doi: 10.1371/journal.pone.0005068. doi: 10.1371/journal.pone.0005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ameni G, Aseffa A, Sirak A, Engers H, Young D, et al. Effect of skin testing and segregation on the incidence of bovine tuberculosis, and molecular typing of Mycobacterium bovis in Ethiopia. Vet Rec. 2007;161(23):782–786. [PMC free article] [PubMed] [Google Scholar]
- 36.Biffa D, Skjerve E, Oloya J, Bogale A, Abebe F, et al. Molecular characterization of Mycobacterium bovis isolates from Ethiopian cattle. BMC Vet Res. 2010;6:28. doi: 10.1186/1746-6148-6-28. doi: 10.1186/1746-6148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oloya J, Kazwala R, Lund A, Opuda-Asibo J, Biffa D, et al. Characterisation of mycobacteria isolated from slaughter cattle in pastoral regions of Uganda. BMC Microbiol. 2007;7:95. doi: 10.1186/1471-2180-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]