Abstract
Plants can sense the direction of gravity and orient their growth to ensure that roots are anchored in soil and that shoots grow upward. Gravitropism has been studied extensively using Arabidopsis genetics, but the exact mechanisms for gravitropism are not fully understood. Here, we demonstrate that five NPY genes play a key role in Arabidopsis root gravitropism. NPY genes were previously identified as regulators of auxin-mediated organogenesis in a genetic pathway with the AGC kinases PID, PID2, WAG1, and WAG2. We show that all five NPY genes are highly expressed in primary root tips. The single npy mutants do not display obvious gravitropism defects, but the npy1 npy2 npy3 npy4 npy5 quintuple mutants show dramatic gravitropic phenotypes. Systematic analysis of all the npy double, triple, and quadruple combinations demonstrates that the five NPY genes all contribute to gravitropism. Our work indicates that gravitropism, phototropism, and organogenesis use analogous mechanisms in which at least one AGC kinase, one NPH3/NPY gene, and one ARF are required.
Keywords: Light signaling, signal transduction, development, root biology, auxin
INTRODUCTION
Plants reorient their growth in response to changes in directional signals such as light and gravity. Both phototropic and gravitropic responses allow plants to orient their roots and shoots in the right directions to ensure their survival. Arabidopsis shoots grow toward a light source whereas roots grow away from the light source. Both shoots and roots can also sense the direction of gravity and adjust their growth directions accordingly. Roots grow toward the direction of gravity while shoots grow against the direction of gravity. Gravitropism allows plants to anchor their roots in soil for nutrient uptake and to grow their stems straight in the air. Phototropism ensures that plants receive sufficient light for photosynthesis and plant growth.
A common factor that affects both gravitropism and phototropism is the plant hormone auxin (Morita, 2010; Zourelidou et al., 2009). Molecular genetic studies in Arabidopsis have demonstrated that auxin biosynthesis, polar transport, and signaling are essential for plants to properly respond to gravity stimuli. The auxin biosynthetic gene TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/SHADE AVOIDANCE3/TRANSPORT INHIBITOR RESPONSE2 (TAA1/SAV3/TIR2) is expressed asymmetrically in response to gravistimuli and TIR2 contributes to a positive regulatory loop required for root gravitropism (Yamada et al., 2009). Mutations in the auxin influx carrier AUX1 completely abolish Arabidopsis root gravitropic curvature (Bennett et al., 1996). Disruption of the auxin efflux carrier PIN-FORMED 2/WAVY GROWTH 6/ETHYLENE INSENSITIVE ROOT 1/AGRAVITROPIC 1 (PIN2/WAV6/EIR1/AGR1) also affects root gravitropism (Chen et al., 1998; Luschnig et al., 1998; Muller et al., 1998). Arabidopsis plants treated with the auxin transport inhibitor N-1-Naphthylphthalamic Acid (NPA) display a complete agravitropic root growth (Marchant et al., 1999). Auxin signaling components INDOLE-3-ACETIC ACID 3/SHORT HYPYCOTYL 3 (IAA3/SHY2) (Tian and Reed, 1999), AUXIN RESPONSE FACTOR 7 (ARF7), and ARF 19 also play a pivotal role in root gravitropism (Li et al., 2006; Okushima et al., 2005; Wilmoth et al., 2005). Auxin is also essential for shoot gravitropic responses because NPA treatments or dominant mutations in IAA19 and IAA7/AUXIN RESISTANT 2 (AXR2) lead to agravitropic shoot growth (Nagpal et al., 2000; Tatematsu et al., 2004). The critical role of auxin in phototropism was demonstrated by many early physiological studies (Holland et al., 2009). Inactivation of the auxin influx carrier AUX1 leads to defects in phototropism in Arabidopsis hypocotyls (Stone et al., 2008). Arabidopsis genetics has also uncovered important auxin signaling components required for proper phototropic responses (Harper et al., 2000; Tatematsu et al., 2004; Yang et al., 2004). Arabidopsis non-phototropic hypocotyl 4 (nph4)/arf7 mutants cannot curve toward directional blue light (Harper et al., 2000). Dominant mutations in IAA19 also abolish phototropism in hypocotyls (Tatematsu et al., 2004). Interestingly, ARF7 is a shared component in gravitropism and phototropism, suggesting that auxin plays a very similar role in both types of tropism-mediated directional growth.
The first step of root gravitropic response is to sense the direction of gravity—a process that is still poorly understood. Gravity sensing is believed to take place in the specialized cells called statocytes. Statocytes are located at the root cap and contain dense, starch-filled amyloplasts that may provide information of the direction of gravity when changing positions inside the statocytes in response to gravity changes (Morita, 2010). However, it is not clear how gravity signals are converted to chemical signals that eventually lead to the differential growth. In contrast, the molecular mechanism of phototropism is much better defined. Arabidopsis molecular genetic studies have defined several non-phototropic hypocotyl loci (nph1, nph2, nph3, and nph4) important for phototropism in hypocotyls (Liscum and Briggs, 1995). As discussed above, NPH4 encodes the ARF7 (Harper et al., 2000). NPH1 and NPH2 encode the photoreceptor PHOTOTROPIN1 (PHOT1) and PHOTOTROPIN2 (PHOT2), respectively (Sakai et al., 2001). PHOT1 and PHOT2 share 58% amino acid sequence identity and are responsible for the perception of directional blue light. Phototropins contain two light-sensing LOV (light, oxygen, or voltage) domains at the N-terminal region and a Ser/Thr kinase domain at the C-terminal region (Christie et al., 1998; Huala et al., 1997). Upon photo-excitation of the LOV domains in phototropins, the C-terminal kinase domain is activated and consequently the photoreceptor is autophosphorylated (Christie et al., 1998). Therefore, the phototropins define the starting point of a signaling pathway ultimately responsible for the phototropic curvature. PHOT1 is localized on plasma membrane, but upon blue light treatment, a small fraction of PHOT1 is released to the cytoplasm (Sakamoto and Briggs, 2002; Wan et al., 2008). NPH3, which encodes a plant-specific, plasma membrane-bound protein, has been shown to play an essential role in phototropic response (Motchoulski and Liscum, 1999). Inactivation of NPH3 completely abolishes the curvature of hypocotyls to directional blue light (Motchoulski and Liscum, 1999). NPH3 belongs to a large family with 32 members in the Arabidopsis genome. Interestingly, RPT2 (Inada et al., 2004), an NPH3 homolog, is required for root phototropic responses. Both NPH3 and RPT2 physically interact with PHOT1 (Inada et al., 2004; Motchoulski and Liscum, 1999), but the exact mechanisms for NPH3 and RPT2 in phototropism are not understood. However, it is known that NPH3 is phosphorylated in the dark and dephosphorylated in light (Pedmale and Liscum, 2007). It has been suggested that an unidentified phosphatase is activated by phototropins directly or indirectly upon light perception. It is also suggested that NPH3 is phosphorylated by an unknown kinase other than PHOT1 and PHOT2 (Pedmale and Liscum, 2007). Although the entire phototropism signaling pathway has not been solved, it is clear that PHOT1/PHOT2, NPH3/RPT2, and ARF7 are important for the phototropic pathway.
We previously reported the isolation of an Arabidopsis mutant called npy1 (naked pins in yucca (yuc) mutants 1) that formed pin-like inflorescences in the yuc1 yuc4 double-mutant background, but not in wild-type background (Cheng et al., 2007). Mutant npy1 is allelic to enhancer of pinoid (pid) 1 (enp1) (Treml et al., 2005) and macchi-bou 4 (mab4) (Furutani et al., 2007), which were isolated as pid enhancers. The yuc1 yuc4 double mutants are defective in auxin biosynthesis (Cheng et al., 2006; Zhao et al., 2001). PID is involved in regulating auxin transport/signaling (Benjamins et al., 2001; Christensen et al., 2000). Interestingly, NPY1 is homologous to NPH3, which is known to participate in phototropism. Although npy1 alone does not have obvious developmental defects, simultaneous, inactivation of NPY1 and two of its close homologs NPY3 and NPY5 leads to the formation of pin-like inflorescences (Cheng et al., 2008), a phenotype also observed in pin1 (Galweiler et al., 1998), pid (Christensen et al., 2000), yuc1 yuc4 npy1 triple mutants (Cheng et al., 2007), and monopteros (mp) (Przemeck et al., 1996). PID is a Ser/Thr kinase that is homologous to PHOT1 and PHOT2 and belongs to the AGC kinase super-family. AGC kinase is the collective name for cAMP-dependent protein kinase A, cGMP-dependent protein kinase G, and phospholipids-dependent protein kinase C (Galvan-Ampudia and Offringa, 2007). MP encodes ARF5 (Hardtke and Berleth, 1998), a close homolog of NPH4/ARF7. We proposed that auxin-regulated organogenesis and phototropism use analogous mechanisms for which at least one NPH3-like gene, one AGC kinase, and one ARF are required (Cheng et al., 2007, 2008).
Root gravitropic responses require both ARF7 and ARF19 because arf7 arf19 double mutants lack gravitropic responses in roots (Li et al., 2006; Okushima et al., 2005). It is also known that PID and its two close AGC kinase homologs WAG1 and WAG2 play important roles in root gravitropic responses (Santner and Watson, 2006; Sukumar et al., 2009). The AGC kinases D6PKs are also important for root gravitropism (Zourelidou et al., 2009). Therefore, it is likely that gravitropism also employs a mechanism analogous to those used in phototropism and auxin-mediated organogenesis. We hypothesized that NPH3-like genes are also required for gravitropic responses because both AGC kinases and ARFs have been demonstrated as regulators of gravitropism. In this paper, we show that five NPY genes (NPY1-5) contribute to Arabidopsis root gravitropic responses. We show that NPY1, NPY2, NPY3, NPY4, and NPY5 are highly expressed in Arabidopsis roots with overlapping patterns. We systematically analyzed root gravitropic responses of all the single, double, triple, quadruple, and quintuple combinations of the npy mutants. It is clear that all of the NPY genes analyzed in this paper play a role in root gravitropism. The single npy mutants did not show obvious defects in gravitropism, but many npy mutant combinations displayed significant defects in root gravitropic responses. The strongest gravitropic defects were observed in the npy1 npy2 npy3 npy4 npy5 quintuple mutants. We conclude that plants use an AGC kinase-NPH3-like protein-ARF module analogous to those in auxin-regulated organogenesis and phototropism to regulate plant root gravitropic responses.
RESULTS
NPY Genes Are Highly Expressed at Arabidopsis Primary Root Tip
The AGC kinases PID, WAG1, and WAG2 were shown to regulate Arabidopsis root gravitropic response (Santner and Watson, 2006; Sukumar et al., 2009). We previously demonstrated that PID, WAG1, and WAG2 also play a critical role in auxin-mediated cotyledon formation (Cheng et al., 2007, 2008). We showed that the PID/WAG kinases work with NPY genes in the same genetic pathway to control organogenesis (Cheng et al., 2007, 2008). Therefore, we hypothesized that NPY genes are also important for gravitropism.
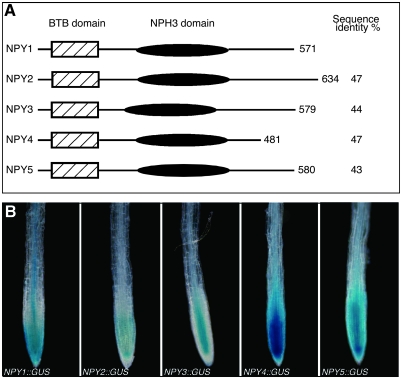
NPY1 and its four closest homologs share similar domain structures with a BTB domain at the N-terminal region and an NPH3 domain in the middle (Figure 1A). NPY1, NPY3, and NPY5 are essential for the formation of flowers whereas the functions of NPY2 and NPY4 are not defined (Cheng et al., 2008). We first investigated whether the NPY genes are expressed in Arabidopsis primary roots. We used transgenic Arabidopsis plants that express the β-D-glucuronidase (GUS) gene under the control of a NPY gene promoter to determine the expression patterns of NPY genes in roots (Figure 1B). All five NPY genes (NPY1 = At4g31820, NPY2 = At2g14820, NPY3 = At5g67440, NPY4 = At2g23050, NPY5 = At4g37590) were expressed in tips of Arabidopsis primary roots, but they displayed unique and overlapping patterns. NPY1::GUS was clearly observed in stele, root cap, and the meristem region (Figure 1B). NPY2::GUS displayed the highest expression in the quiescent center, but less expression in the columella root cap. NPY2::GUS was also expressed in the proximal meristem region (Figure 1B). Among the five NPY::GUS lines, NPY3::GUS expression was the only one whose expression was restricted in the vascular tissue (Figure 1B). We did not observe NPY3::GUS expression in the epidermis and columella root cap. NPY4::GUS had the highest expression levels compared to the other NPY::GUS lines (Figure 1B). NPY4::GUS was expressed in the root cap and in the meristem region, with the highest expression in the central domain (Figure 1B). The expression pattern of NPY5::GUS was very similar to that of NPY2::GUS, with the highest expression in the quiescent center (Figure 1B), but NPY5::GUS was also expressed throughout other tissues at the root tip (Figure 1B). The expression patterns of the NPY genes indicate that they are expressed in root tips and they probably play a role in root development. The expression analysis also suggests that the NPY genes probably have overlapping functions in root development.
Figure 1.
Expression Patterns of NPY::GUS in Arabidopsis Primary Root Tip.
(A) The domain structures of NPY proteins. NPY1 and its closest homologs have a BTB domain at the N-terminal region and an NPH3 domain in the middle.
(B) NPY gene expression patterns in root tip as defined by NPY promoter::GUS transgenic plants. The β-glucuronidase gene was under the control of an NPY promoter. The GUS lines of NPY2, NPY3, NPY4, and NPY5 were stained for 15 min. The NPY1::GUS line was stained for 2 h. From left to right: NPY1::GUS, NPY2::GUS, NPY3::GUS, NPY4::GUS, and NPY5::GUS.
NPY Genes Are Required for Root Gravitropic Response
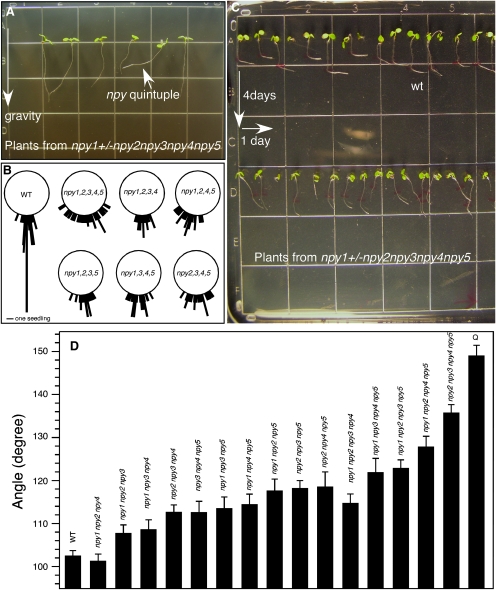
We investigated whether inactivation of NPY genes had any effects on root gravitropic response. Disruption of a single NPY gene did not cause obvious defects in root development (data not shown), probably due to overlapping functions of the NPY genes. We then analyzed whether simultaneous inactivation of the five NPY genes led to any root phenotypes. We germinated seeds from a single plant that was npy1+/– npy2 npy3 npy4 npy5 and grew the seedlings on a vertical plate for 7 d at 23°C with 16 h light/8 h dark cycle. The seedlings were photographed, numbered, and genotyped. The npy1 npy2 npy3 npy4 npy5 quintuple mutants lacked proper gravitropic responses (Figure 2A, arrow). For wild-type, the majority of the plants grew straight down toward the direction of gravity (90°), with a small percentage of seedlings deviating from 85° to 112° (Figure 2B). The roots of the npy quintuple mutants were in random directions within a range from 21° to 150° (Figure 2B).
Figure 2.
NPY Genes Contribute to Root Gravitropism.
(A) Seeds from a plant of npy1+/– npy2 npy3 npy4 npy5 were germinated and grown vertically for 7 d. The npy quintuple mutants (arrow head) showed defects in gravitropism. The direction of gravity is also marked with an arrow.
(B) Directions of root growth of wild-type and npy mutants. Plants were grown vertically for 4 d. The angle between the gravity vector and root were measured and are shown here. The bar refers to one seedling. If two seedlings have the same angle, the line at that angle is twice of the length of the bar. The total number of seedlings analyzed for each genotype were: 49 for WT, 45 for npy1, 2, 3, 4, 5 quintuple, 24 for npy1, 2, 3, 4 quadruple, 27 for npy1, 2, 4, 5 quadruple, 27 for npy1, 2, 3, 5 quadruple, 32 for npy1, 3, 4, 5 quadruple, and 28 for npy2, 3, 4, 5.
(C) Responses to a directional change of gravity. Plants were grown vertically for 4 d and then the plates were turned 90° for 1 d. The top row is wt and bottom row is plants from npy1+/– npy2 npy3 npy4 npy5. Note: wild-type roots bend about 90° while the npy mutants do not bend much.
(D) Quantitative analysis of gravitropism in npy mutant combinations. Plants were grown vertically for 4 d and the plates were turned 90° for 1 d. The angle of root bending was measured. The angle for perfect gravitropism is 90° and the angle for non-gravitropic growth is 180°. Q refers to the npy1 npy2 npy3 npy4 npy5 quintuple mutants. At least 20 seedlings were analyzed for each genotype shown in Figure 2D.
We systematically analyzed all of the double, triple, and quadruple npy mutant combinations by growing them on vertical plates. The double mutants and triple mutants did not display obvious gravitropic defects in this assay, but the quadruple mutants displayed various degrees of gravitropism defects (Figure 2B). Among the quadruple mutants, npy1 npy2 npy4 npy5 and npy2 npy3 npy4 npy5 had the strongest phenotypes (Figure 2B).
We then used a more sensitive and quantitative assay to determine gravitropism defects among the npy mutant combinations. Seedlings were grown vertically for 4 d and then the plates were turned 90° to change the gravity vector. The angle of root bending was then measured (Figure 2C). Perfect gravitropism leads to the formation of a near 90° angle whereas non-gravitropism will have an angle of 180° (Figure 2C). Wild-type roots bent about 90° to follow the direction of gravity (Figure 2C), but npy mutant combinations displayed defects in reorienting their root growth (Figure 2C). In this more sensitive assay, we observed that all of the npy triple mutants except npy1 npy2 npy4 triple mutants displayed significant gravitropism defects (Figure 2D). All of the npy quadruple mutants except npy1 npy2 npy3 npy4 had stronger gravitropism defects than the npy triple mutants (Figure 2D). We conclude that all of the NPY genes contribute to root gravitropic responses.
Effects of npy Mutants on Root Elongation and DR5–GUS Expression
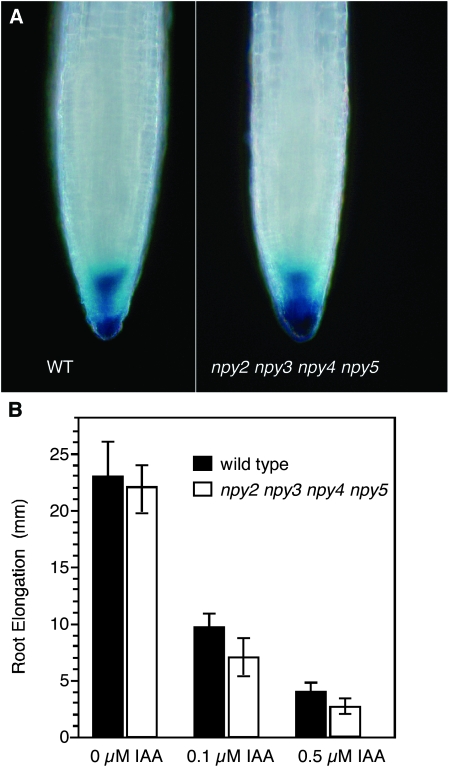
Auxin is known to regulate gravitropism. NPY1, NPY3, and NPY5 are known to participate in auxin-mediated organogenesis (Cheng et al., 2008). We used the auxin reporter DR5–GUS (Sabatini et al., 1999) to analyze auxin responses in the npy mutants. As shown in Figure 3A, DR5–GUS is highly expressed in the root tip in wild-type Arabidopsis. DR5–GUS is expressed in the root cap and quiescent center. The DR5–GUS expression patterns in npy2 npy3 npy4 npy5 are very similar to those in wild-type (Figure 3A).
Figure 3.
The Effects of npy Mutations on Auxin Responses.
(A) The auxin reporter DR5–GUS expression in wild-type (left) and npy2 npy3 npy4 npy5 quadruple mutants (right) are shown. More than 10 seedlings for each genotype were analyzed. The typical staining patterns were shown.
(B) Auxin response in npy2 npy3 npy4 npy5 quadruple mutants. Plants were grown vertically for 4 d and then transferred to media with and without IAA. The root elongation of wild-type and npy2 npy3 npy4 npy5 quadruple mutants were measured after additional 4 d. At least 20 seedlings for each genotype and each auxin concentration were measured.
We also measured root elongation with and without IAA in the media. The npy2 npy3 npy4 npy5 quadruple mutants had slightly shorter root than wild-type (Figure 3B). The npy quadruple mutants were not resistant to exogenous auxin in the root elongation assay (Figure 3B).
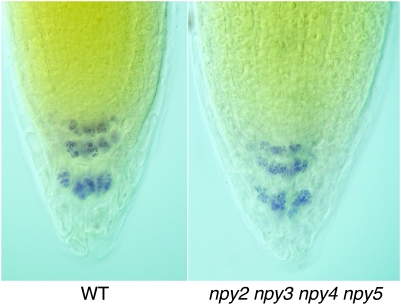
Effects of npy Mutations on the Formation of Starch Granules in Root Tips
Gravity sensing statocytes contain dense, starch-filled amyloplasts that can be visualized by Lugol staining. Lugol solution contains iodine that reacts with starch granules to generate a bright blue color. We studied whether the gravitropic defects of npy mutants were caused by disruption of starch granule formation. In wild-type root tips, differentiated columella cells displayed strong staining (Figure 4). We observed similar staining patterns in npy2 npy3 npy4 npy5 quadruple mutants.
Figure 4.
Formation of Starch Granules in Root Tips.
Starch granules were visualized by Lugol staining. The left root tip is from wild-type and the right is from npy2 npy3 npy4 npy5 quadruple mutants. At least 10 seedlings for each genotype were stained. The typical patterns were shown here.
DISCUSSION
In this report, we provide evidence showing that the five NPY genes contribute to Arabidopsis root gravitropism. This work also demonstrates that gravitropism uses mechanisms analogous to those employed in phototropism and organogenesis. Gravitropism, phototropism, and organogenesis all require the plant hormone auxin, AGC kinases, NPH3-like proteins, and ARFs.
The arf7 arf19 double mutants show agravitropic root growth (Li et al., 2006; Okushima et al., 2005). Both ARF7 and ARF19 are expressed in roots (Li et al., 2006; Okushima et al., 2005). ARF7 is mainly expressed in vascular tissue in roots and ARF19 is highly expressed in the root tip including root cap, root meristem, and elongation Zone (Li et al., 2006; Okushima et al., 2005). Two groups of AGC kinases, PID/WAG kinases and D6PK kinases, have been implicated in gravitropism (Santner and Watson, 2006; Sukumar et al., 2009; Zourelidou et al., 2009). The AGC kinases WAG1 and WAG2 are highly expressed at root tips, but their expression is excluded from the root cap (Santner and Watson, 2006). PID::GUS is mainly expressed in the vascular tissue proximal to the root meristem (Benjamins et al., 2001). PID::GUS is not expressed in the root cap and meristem (Benjamins et al., 2001). D6PK::GUS and D6PKL2::GUS show high expression in the elongation zone and D6PKL1::GUS and D6PKL3::GUS are expressed in columella cells and elongation zone. The five NPY genes discussed in this paper are expressed in root tips (Figure 1B). NPY2 and NPY5 have high expression in the quiescent center while NPY3 is not expressed in the columella root cap (Figure 1B). The root cap, quiescent center, and meristem have at least one expressed NPY gene. The expression patterns of ARF 7, ARF19, AGC kinases, and the NPY genes overlap significantly in the root tip, which is consistent with the idea that these genes function in the same genetic pathway to control gravitropic responses.
The central columella cells in the root cap are believed to be the main sites for gravity sensing. The root curvature takes place at the elongation zone in response to a gravistimulus (Holland et al., 2009). It is proposed that the asymmetric auxin flow from the gravity-sensing cells to the lateral root cap and the elongation zone after gravity stimulation is responsible for the gravitropic curvature (Holland et al., 2009). The auxin efflux carrier PIN3 is expressed in the columella cells and PIN3 protein relocates laterally within columella cells in response to gravistimulation to mediate lateral auxin transport (Friml et al., 2002). Therefore, PIN3 was suggested as a key component to translate directional signal sensed in the statocyte to a directional flow of auxin. However, PIN3-null mutants only have subtle gravitropic defects (Friml et al., 2002). PID has been shown to phosphorylate PIN proteins and to regulate PIN localization (Michniewicz et al., 2007). It is unlikely that PID, WAG1, and WAG2 regulate PIN3 localization in columella cells because the three AGC kinases are not expressed in columella cells. Studies with PID::PID–GFP transgenic lines showed that PID is expressed in the epidermis and endodermis in the elongation zone (Michniewicz et al., 2007). However, PID::GUS showed a completely different expression pattern in roots (Benjamins et al., 2001). PID::GUS is exclusively expressed in vascular tissue, suggesting that either PID proteins move from the site of synthesis or one of the transgenic lines does not reflex the actual PID expression pattern. The D6PK co-localizes with PIN1, PIN2, and PIN4 (Zourelidou et al., 2009). However, it is still an open question whether phosphorylation of PIN proteins by AGC kinases is part of the mechanism for converting gravity signals to auxin flow. The NPY genes are expressed in the right places for regulating gravitropism. It will be very interesting to investigate auxin transport and PIN protein localization in npy mutants.
Gravitropism, phototropism, and organogenesis are involved in differential growth. In phototropism and gravitropism, differential auxin distribution, which is triggered by directional stimuli, causes differential cell elongation and organ curvature. In organogenesis, developmental signals cause the formation of an auxin maximum that initiates the formation of an organ and that is required for organ outgrowth away from the meristem. The three processes all require at least one AGC kinase, one NPH3-like protein, and one ARF. Interestingly, some components are shared whereas others are unique to a particular process. For example, ARF7 is required for both gravitropism and phototropism whereas ARF5 is essential for organogenesis. The PID/WAG kinases are involved in both organogenesis and gravitropic responses, but gravitropism requires additional AGC kinases. The PID/WAG and D6PK kinases are believed to interact with PIN proteins directly to regulate auxin transport whereas the substrates for the AGC kinases PHOT1 and PHOT2 are not known. It is not clear whether AGC kinases PHOT1 and PHOT2 can also interact with and phosphorylate PIN proteins. PHOT1 and PHOT2 are known to physically interact with NPH3 and RPT2 whereas the direct partners of NPY proteins are not known. Interestingly, gravitropism appears to have more genetic redundancy than phototropism and organogenesis. It is known that two AGC kinases (PHOT1 and PHOT2), one NPH3 protein, and one ARF (ARF7) are required for hypocotyl phototropic responses. The formation of flowers needs one AGC kinase (PID), three NPH3-like proteins (NPY1, NPY3, and NPY5), and one ARF (ARF5). Gravitropism uses at least seven AGC kinases (PID, WAG1, WAG2, and four D6PKs), at least five NPH3-like genes, and two ARFs.
The AGC kinase-NPH3-like protein-ARF genetic module may not be limited to phototropism, gravitropism, and organogenesis. Disruption of SETH6/At2g47860, which encodes an NPH3-like protein, greatly affected pollen germination and pollen tube growth (Lalanne et al., 2004). Two Arabidopsis AGC kinases (AGC1.5 and AGC1.7) are critical for polarized growth of pollen tubes (Zhang et al., 2009), suggesting that the two AGC kinases and SETH6 may also participate in the same pathway. The defectively organized tributaries (dot) 3 mutant has defects in vein patterning (Petricka et al., 2008). DOT3/AT5g10250 is also homologous to NPH3, sharing 36% sequence identity and 54% similarity at the amino acid level with NPH3 (Petricka et al., 2008). Interestingly, auxin is also a main regulator for vascular development. Therefore, the AGC kinase–NPH3-like protein–ARF module appears to be widely used in different biological processes to regulate directional growth. This hypothesis is also consistent with the fact that AGC kinases, NPH3-like proteins, and ARFs all belong to large families.
METHODS
Plant Materials
The npy single mutants were all T-DNA insertion mutants (Alonso et al., 2003). The insertion sites of the npy mutants were described previously (Cheng et al., 2007, 2008). The genotyping primers for the npy1, npy2, npy3, npy4, and npy5 mutants were also described previously (Cheng et al., 2007, 2008).
NPY::GUS transgenic lines were constructed as described below. NPY promoters were amplified by PCR and cloned into the vector pBI101.3 to drive the GUS gene expression. The PCR primers for each NPY promoter are:
NPY1P1: 5'-ATTTCTTCGTCTTGTTTACCAAAAGGAAGACAC-3';
NPY1P2: 5'-GGGAGTAAAAGCTGCCGGCGTTGAG-3';
NPY2P1: 5'-GTCTCGAAGATTCAGAAACAGCC-3';
NPY2P2: 5'-CTTCTTCCCCCTAGGATAACTC-3';
NPY3P1: 5'- TTCTTAACGTTGACTGAGTTTGAG-3';
NPY3P2: 5'-TTCTCACAAATAAACAAAGCCAG-3';
NPY4P1: 5'-GGAGTTCTTGAGTCATCGTTTTACG-3';
NPY4P2 : 5'-TTTTTTCTATCTCTTTAATAAGTT-3';
NPY5P1: 5'-GGTTCCATCCTCACAAATGATCAAGC-3';
NPY5P2: 5'-GATGAAGAAGAAACTGATAGAAGAAG-3'.
The NPY::GUS constructs were transformed into wild-type Arabidopsis Columbia background by the floral dipping method (Clough and Bent, 1998). Lines with the most consistent GUS expression patterns were used in this study.
Beta-Glucuronidase (GUS) Staining for NPY::GUS Lines
Five-day-old seedlings grown on 0.5 X MS media in light were used for GUS staining. NPY2::GUS, NPY3::GUS, NPY4::GUS, and NPY5::GUS were highly expressed in root tips and they were stained for only 15 min. The NPY1::GUS lines were relatively weaker and were stained for 2 h. All of the NPY::GUS lines would be over-stained if they were stained for more than 3 h. GUS staining procedure was conducted according to what was described previously (Jefferson et al., 1987). The samples were photographed in dark field using a Leica DM5000 microscope.
Measurements of Gravitropic Responses
Arabidopsis seeds were surface-sterilized with 75% ethanol for 10 min and 100% ethanol for another 10 min. The seeds were allowed to dry on a piece of 3M filter paper in a sterile tissue culture hood. Then the seeds were sown on plates containing 0.5 X MS medium and 0.65% agar. After 2 d at 4°C, the plates were then placed vertically in an incubator at 23°C. After 4 d, the plates were photographed and the angle between gravity and the primary root were determined using NIH Image J, a free program that can be downloaded from http://rsbweb.nih.gov/ij/.
Gravitropic responses were also measured by growing seedlings on 0.5 X MS for 4 d on a vertical plate and then turning the plates 90° so that the seedlings were perpendicular to the gravity vector. The plates were photographed 24 h later. Angles of root curvature were analyzed using the NIH Image J program.
Root Elongation Assay and DR5–GUS Expression
The auxin reporter DR5–GUS was used to study the auxin responses of npy mutants. The DR5–GUS marker was crossed into npy mutants. Both wild-type and npy mutants were grown on 0.5 X MS media vertically for 5 d. Then, the seedlings were stained for GUS activities for 3 h.
Four-day-old light-grown seedlings were transfer from 0.5 X MS agar plates to 0.5 X MS agar plates containing various concentrations of indole-3-acetic acid (IAA). The positions of root tips were marked with a sharpie marker. The seedlings were allowed to grow for four more days vertically. The plates were then photographed. The root elongation was analyzed with the NIH Image J program.
Starch Granule Staining
Four-day-old seedlings were first fixed in FAA (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) at 4°C overnight. The samples were washed once in 50% ethanol. Then, the samples were placed in lugol solution (0.37% iodine, 0.71% potassium iodide) for 1 min, followed with 2 min in chloral hydrate solution (80 g chloral hydrate, 20 ml glycerol, 20 ml water). Pictures were taken immediately.
FUNDING
This work was supported by National Institutes of Health Grant R01GM68631 (to Y.Z.). Yuanting Li was partially supported by a scholarship from the China Scholarship Council.
No conflict of interest declared.
References
- Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development. 2001;128:4057–4067. doi: 10.1242/dev.128.20.4057. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, et al. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl Acad. Sci. U S A. 1998;95:15112–15117. doi: 10.1073/pnas.95.25.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2007;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Christie JM, et al. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Furutani M, et al. The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development. 2007;134:3849–3859. doi: 10.1242/dev.009654. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Offringa R. Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 2007;12:541–547. doi: 10.1016/j.tplants.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Galweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper RM, et al. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JJ, Roberts D, Liscum E. Understanding phototropism: from Darwin to today. J. Exp. Bot. 2009;60:1969–1978. doi: 10.1093/jxb/erp113. [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T. RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell. 2004;16:887–896. doi: 10.1105/tpc.019901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne E, et al. Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics. 2004;167:1975–1986. doi: 10.1534/genetics.104.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dai X, Zhao Y. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 2006;140:899–908. doi: 10.1104/pp.105.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, et al. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Morita MT. Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 2010;61:705–720. doi: 10.1146/annurev.arplant.043008.092042. [DOI] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E. Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science. 1999;286:961–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- Muller A, et al. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998;17:6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, et al. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E. Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J. Biol. Chem. 2007;282:19992–20001. doi: 10.1074/jbc.M702551200. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Clay NK, Nelson TM. Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 2008;56:251–263. doi: 10.1111/j.1365-313X.2008.03595.x. [DOI] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- Sabatini S, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sakai T, et al. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl Acad. Sci. U S A. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner AA, Watson JC. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 2006;45:752–764. doi: 10.1111/j.1365-313X.2005.02641.x. [DOI] [PubMed] [Google Scholar]
- Stone BB, et al. Disruptions in AUX1-dependent auxin influx alter hypocotyl phototropism in Arabidopsis. Mol. Plant. 2008;1:129–144. doi: 10.1093/mp/ssm013. [DOI] [PubMed] [Google Scholar]
- Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 2009;150:722–735. doi: 10.1104/pp.108.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, et al. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell. 2004;16:379–393. doi: 10.1105/tpc.018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- Treml BS, et al. The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development. 2005;132:4063–4074. doi: 10.1242/dev.01969. [DOI] [PubMed] [Google Scholar]
- Wan YL, Eisinger W, Ehrhardt D, Kubitscheck U, Baluska F, Briggs W. The subcellular localization and blue-light-induced movement of phototropin 1-GFP in etiolated seedlings of Arabidopsis thaliana. Mol. Plant. 2008;1:103–117. doi: 10.1093/mp/ssm011. [DOI] [PubMed] [Google Scholar]
- Wilmoth JC, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009;151:168–179. doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, et al. The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1) Plant J. 2004;40:772–782. doi: 10.1111/j.1365-313X.2004.02254.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, He J, McCormick S. Two Arabidopsis AGC kinases are critical for the polarized growth of pollen tubes. Plant J. 2009;58:474–484. doi: 10.1111/j.1365-313X.2009.03792.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zourelidou M, et al. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development. 2009;136:627–636. doi: 10.1242/dev.028365. [DOI] [PubMed] [Google Scholar]