Abstract
Background
The human dystrobrevin-binding protein 1 (DTNBP1) gene has been linked to risk for schizophrenia. Recent studies indicate that several single nucleotide polymorphisms (SNPs) in the DTNBP1 gene may also influence general cognitive ability in both schizophrenic patients and healthy controls. We examined the relationship between DTNBP1 SNPs and general cognitive ability in non-psychiatric healthy samples via meta-analysis.
Methods
Medline search (12/31/2009) yielded 11 articles examining DTNBP1 variation and general cognitive ability, of which 8 studies had data available encompassing 10 independent cohorts (total n=7,592). The phenotype was defined as either the first principal component score from multiple neuropsychological tests (Spearman's g) or full scale IQ. Meta-analyses were conducted for 9 SNPs for which cognitive data were available from at least 3 cohorts. For each SNP in each cohort, effect size (ES) was computed between major allele homozygotes and minor allele carriers; ES was then pooled across studies using a random effect model.
Results
Pooled ES's from 2 of the 9 SNPs (rs1018381 and rs2619522) were -0.123 and -0.083, p's<0.01, respectively, suggesting that the minor allele carriers of these SNPs had lower cognitive ability scores than the major allele homozygotes. Results remained significant after examining heterogeneity among samples and potential publication biases. Other SNPs did not show significant effects on general cognitive ability.
Conclusion
Genetic variation in DTNBP1 modestly influences general cognitive ability. Further studies are needed to elucidate the biological mechanisms that may account for this relationship.
Keywords: DTNBP1, genetics, general cognitive ability, meta-analysis, human, healthy control
Introduction
The human dystrobrevin-binding protein 1 (DTNBP1) gene is located on chromosome 6p22.3, and contains the coding region for the dysbindin protein, part of the dystrophin-associated protein complex(1). Because DTNBP1 is widely expressed in the brain(2), and is present in presynaptic, postsynaptic, and microtubule locations(3), it is believed to play a role in a number of brain functions. Animal studies have demonstrated that dysbindin plays a critical modulatory role in synaptic transmission(4, 5), as well as subserving neurite outgrowth in the developing organism(6, 7). In humans, variation in DTNBP1 was first reported to influence risk for schizophrenia in a large linkage study of Irish families(8), with subsequent association studies also implicating this locus(9). A recent meta-analysis of nine association studies indicated that several single nucleotide polymorphisms (SNPs) in DTNBP1 were associated with schizophrenia(10), and a comprehensive meta-analysis of schizophrenia susceptibility studies identifies DTNBP1 as one of four genes with the strongest evidence for association to illness(11).
In addition to influencing risk for schizophrenia, DTNBP1 variation has also been linked with human cognitive function. Burdick et al(12) reported that DTNBP1 alleles previously associated with schizophrenia were also associated with lower general cognitive ability (Spearman's g, defined as the first principal component of a factor analysis of multiple neurocognitive measures) in 213 schizophrenia patients, with the same relationship noted in 126 healthy controls. DTNBP1 genetic variation also influenced cognitive decline over time in patients(13). Subsequent studies have reported similar effects(14-17), although several negative studies(18-20) have also been reported. These studies vary in sample size, ethnic composition, age, sex, and educational background, adding complexity to the genotype-phenotype association.
In order to systematically synthesize these disparate studies, a potentially useful methodology is to utilize meta-analytic techniques that incorporate results from multiple studies in an unbiased fashion(21). In the present study, we attempted to examine the association between DTNBP1 SNPs and cognitive function in a non-psychiatric, healthy sample via a meta-analysis. We chose to focus our meta-analysis on healthy samples because of the substantial data available in this population. Fewer studies have included patient samples, with little overlap in the SNPs genotyped, making a meta-analysis in a combined schizophrenia cohort less feasible.
A challenge for meta-analysis of this literature is the use of differing neuropsychological measures assessing a wide range of cognitive domains, including working memory, verbal recall, language abilities, and full scale IQ. Because the specific measures used in each study differ, individual tests or domains can not be directly compared across multiple studies. Therefore, in the present meta-analysis, we elected to assess general cognitive ability, or Spearman's g, as the phenotype. The concept of g has been employed for more than a century(22) as a measure of general cognitive ability(23), and is based on the significant covariance among many cognitive processes such as working memory, verbal ability, and spatial processing ability. Genetic factors play a substantial role in the inter-individual variation in g, with heritability estimates ranging from 40 to 80%(24), making it suitable for molecular genetic studies. An example of the utility of general cognitive ability as a phenotype is provided by a recent meta-analysis of the COMT Val158/108Met polymorphism and cognition, which failed to find significant associations with any specific cognitive tests/domains, but did demonstrate a small but significant effect on IQ in a cohort comprised of 21 independent samples(25).
Methods
Literature Search
To identify studies eligible for this meta-analysis, we searched Medline for all publications available up to 12/31/2009 that examined the association between the DTNBP1 gene and cognitive measures. The following key words were used in the literature search: cognition, cognitive ability, intelligence, polymorphism, gene, and DTNBP1. We also used the reference lists from identified papers and recent literature review articles to identify additional relevant studies. Furthermore, to find unpublished studies, we searched published meeting abstracts that were likely to contain relevant studies. Each paper included in the meta-analysis met the following criteria: 1) reported the association between DTNBP1 polymorphisms and cognition in human subjects; 2) included healthy control subjects; and 3) used the full scale IQ score, a proxy of IQ, or g. Using this approach, eleven papers were identified; of which two studies produced two papers each(12, 13, 19, 26). One paper reported data on schizophrenia patients only and did not include healthy controls(15). One paper included three independent cohorts(17). Therefore, eight papers with ten independent cohorts were included in the meta-analysis. Figure 1 shows the flow chart of literature search process. Table 1 lists the characteristics of the ten samples.
Figure 1.
Flow chart of literature search.
Table 1.
Characteristics of papers on the association between DTNBP1 and cognition.
| Author, year | Country | N (SCZ) |
N (healthy) |
Sex (% male) |
Race | Age (Mean) |
Cognitive measure | Primary phenotype |
|---|---|---|---|---|---|---|---|---|
| Burdick 2006(12) | USA | 213 | 126 | 38.94% | 100% Caucasian |
51.18 | WRAT-3 Reading test WAIS-R Digit span CPT-I/P CVLT-abridged COWAT Trails making test-A,B |
Spearman's g factor |
| Stefanis 2007(14) | Greece | 0 | 2243 | 100% | 100% Caucasian |
20.69 | Raven Progressive Matrix | IQ |
| Zinkstok 2007(16) | Netherlands | 76 | 31 | 74.2% | 71.0% Caucasian |
21.5 | WAIS-III | IQ |
| Peters, 2008(51) | Australia | 336 | 172 | 59.3% | 75% Caucasian |
Not reported |
The National Adult Reading Test |
IQ |
| Luciano 2009(17) | UK (Scotland) |
0 | 1054 | 50% | 100% Caucasian |
69.6 | WAIS-III subtests WMS-III Digit span |
Spearman's g factor |
| Australia | 0 | 1806 | 49.1% | 98% Caucasian |
16.91 | Multidimensional Aptitude Battery |
IQ | |
| UK (England) |
0 | 745 | 30.1% | 100% Caucasian |
63.1 | Alice Heim 4 Intelligence Test Mill Hill Vocabulary A & B Random Letters Test Cattell Culture Fair Test |
Spearman's g factor |
|
| Kircher 2009(18) | Germany | 0 | 518 | 51.4% | 88.5% European ancestry |
24.75 | MWT-B | IQ |
| Hashimoto 2009(19, 26) |
Japan | 70 | 165 | 32.7% | 100% Asian |
37.28 | WAIS-R | IQ |
| Need 2009(20) | USA | 0 | 701 | 39% | 81% Caucasian |
31.2 | CANTAB | Spearman's g factor |
Note: WRAT-3 = Wide Range Achievement Test-Third Edition; WAIS-R = Wechsler Adult Intelligence Test-Revised; CPT-I/P = Continuous Performance Test-Identical Pairs Version; CVLT = California Verbal Learning Test; COWAT = Controlled Oral Word Association Test; WAIS-III = Wechsler Adult Intelligence Test-Third Edition; WMS-III = Wechsler Memory Scale-Third Edition; MWT-B = Mehrfachwahl-Wortschatz-Intelligenztest-Version B; CANTAB = Cambridge Neuropsychological Test Automated Battery.
Selection of candidate polymorphisms
The human DTNBP1 gene contains 558 non-redundant SNPs in the NCBI dbSNP database (build 129, http://www.ncbi.nlm.nih.gov/SNP/), including 196 HapMap SNPs(1), and multiple SNPs have been tested for association with cognitive ability. To conduct a robust meta-analysis, we therefore selected SNPs reported in at least three cohorts. Ten SNPs fit this criterion. Two of the SNPs, rs2619522 and rs2619528, are perfectly correlated in Caucasian and Asian samples (HapMap Project: http://hapmap.ncbi.nlm.nih.gov), therefore, the data were pooled for these SNPs and only results for rs2619522 are presented here. The remaining nine SNPs are listed in Table 2. The allele frequency in each study was checked against the HapMap data to ensure consistent classification of major/minor alleles. Notably, for rs2619539, the major allele in Japanese samples is C (+ strand) with a frequency of 63.6%, whereas the major allele in Caucasian samples is G (+ strand) with a frequency of 53.4%. Since the Hashimoto et al 2009 paper appear to have genotyped this SNP off the – strand, the data were recoded to ensure consistency of allele assignment with the other reports included in the meta-analysis.
Table 2.
DTNBP1 SNPs reported in the papers.
| SNP | Gene Location |
Major allele |
Minor allele |
Burdick 2006 |
Stefanis 2007 |
Zinkstok 2007 |
Luciano 2009: Scottish |
Luciano 2009: Australian |
Luciano 2009: English |
Kircher 2009 | Hashimoto 2009 |
Peters 2008 |
Need 2009 |
# samples |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1047631 | UTR | A* | G | X | X | X | 3 | |||||||
| rs909706 | Intron | G | A | X | X | X | X | 4 | ||||||
| rs1018381 | Intron | C* | T | X | X | X | X | X | X | X | Xa | 8 | ||
| rs2619522 | Intron | T* | G | X | X | X | X | X | X | X | 7 | |||
| rs760761 | Intron | C* | T | X | X | X | X | Xb | 5 | |||||
| rs3213207 | Intron | A* | G | X | X | X | X | X | 5 | |||||
| rs1011313 | Intron | G* | A | X | X | X | X | X | X | X | X | 8 | ||
| rs2619539c | Intron | G | C | X | X | X | X | X | 5 | |||||
| rs2619538 | 5′ flanking region |
T | A | X | X | X | 3 | |||||||
| N | 126 | 2243 | 62 | 1054 | 1806 | 745 | 518 | 165 | 172 | 701 | 7592 |
Data was from rs11753919 on Illumina, which is a perfect proxy for rs1018381.
Data was from rs1474605 on Illumina, which is a perfect proxy for rs760761.
According to HapMap, the G allele (+ strand) is the major allele in Caucasians, but not in Japanese subjects. However, for consistency, the G allele (+ strand) homozygotes were compared to C allele (+ strand) carriers in all studies, including that of Hashimoto et al. 2009.
SNPs were genotyped off the – strand.
Definition of phenotype
General cognitive ability was defined as either the full scale IQ score, determined from standardized adult intelligence tests, or Spearman's g defined from the first principal component from a principal component analysis (PCA) of several neuropsychological tests. Although Spearman's g differs from the construct of intelligence (IQ), as it is an empirically derived factor from the specific battery by which it is determined, it is highly correlated with most measures of IQ(27). For studies that did not report either a full scale IQ or g, the authors were contacted to request additional data for computation of g. In order to provide a uniform metric for meta-analysis, independent of the specific tests employed to compute g, a measure of effect size was generated for each SNP within each study, as described in the next paragraph.
Statistical analysis
Data for each SNP were entered into and analyzed separately by the Comprehensive Meta-Analysis software version 2 (Biostat, Eaglewood, NJ). Because the frequency of minor allele homozygotes was low for most SNPs, this group was pooled with heterozygotes as minor allele carriers. Hedge's g was used as the effect size measure, representing standardized differences in cognitive scores between major allele homozygotes and minor allele carriers. Because different studies used different covariates (e.g., sex, age, etc.), raw IQ scores or Spearman's g scores were used to compute effect sizes in each study, whenever possible. Therefore, effect sizes and p-values of the studies may be somewhat different from those in published papers. Pooled effect sizes across studies were computed with a random effects model, which estimates the likely effect size across different populations and takes heterogeneity across studies into account(28). This model is different from a fixed effects model, which estimates the most likely effect size from multiple studies assuming that they are sampled from a single population, but it can be biased by high heterogeneity across studies(21).
Heterogeneity between studies was assessed by the Q and I2 statistics. Q statistic is a chi-square test for heterogeneity. I2 is the proportion of observed variance in effect sizes across studies attributable to true differences among studies. A conventional interpretation of I2 is that it defines bounds for low (<25%), moderate (~50%), and high (>75%) heterogeneity(29).
Publication bias was assessed with the funnel plot, Egger's regression test(30), and the “Trim and Fill” method(31). The “Trim and Fill” method is an iterative procedure to assess whether small extreme included studies and/or potentially unincluded studies can bias the estimate of the true effect size. To examine the influence of potential moderators, meta-regression analysis was conducted with the following predictors: percentage of males in the sample, percentage of Caucasians in the sample, and mean age of the sample. Insufficient data were available to permit a moderator analysis with education levels of the sample.
Sensitivity analysis was conducted to assess potential influences of any one single study on the pooled effect size. Within each meta-analysis, included studies were removed one at a time to check for significant alterations to pooled effect sizes and associated p-values. This method also explicitly tests for a potential “winner's curse”, in which the first study has the largest effect size and tends to bias the meta-analysis(32).
Results
Eight papers with 10 independent samples (total n = 7,592) met the inclusion criteria and were included in the meta-analysis. As shown in Table 1, the 10 samples consist of two from the United States, five from Europe, two from Australia, and one from Japan. The majority of the subjects were Caucasians (ranging from 71% to 100%, except for the Japanese sample which was 100% Asian). Mean age ranged from 16.9 to 69.6 years. Four samples reported data on Spearman's g, and four samples used IQ scores. One sample(14) reported IQ scores derived from the Raven's Progressive Matrix test (RPM), which measures non-verbal cognitive ability and is highly correlated with WAIS full-scale IQ (r = 0.74)(33). Another sample(18) reported estimated IQ scores converted from a verbal intelligence scale (Mehrfachwahl-Wortschatz-Intelligenztest; MWT-B). This test was developed to assess premorbid intelligence, and was well correlated with global IQ in healthy adults with a median correlation coefficient of 0.72 in 22 samples(34).
Main Analyses
DTNBP1 rs1018381
Eight independent samples with 6,017 healthy control subjects contributed to the meta-analysis. The pooled effect size was −0.123, with 95% confidence interval (CI) of −0.206 to −0.041, p = 0.003, indicating that minor allele carriers of this SNP had significantly lower general cognitive ability than major allele homozygotes (Figure 2). Examination of heterogeneity across studies was not significant, Q-statistic = 9.30, p = 0.23, I2 = 24.76%.
Figure 2.
The association between DTNBP1 rs1018381 and general cognitive ability in healthy samples. Negative effect sizes indicate that minor allele carriers had lower general cognitive ability.
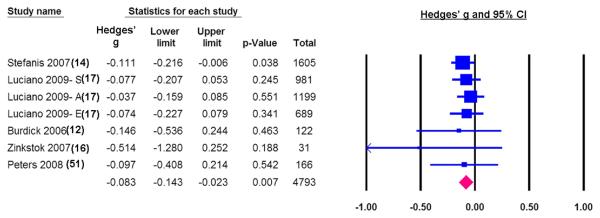
DTNBP1 rs2619522
Six independent samples with a total sample size of 4,793 produced a pooled effect size of −0.083, 95% CI = −0.141 to −0.020, p = .007 (Figure 3). This indicates that subjects carrying at least one copy of the minor allele had significantly lower cognitive ability than those who were homozygotes for the major allele. The forest plot shows a consistent trend among all studies, suggesting that these studies were quite homogeneous, with a Q-statistic = 2.16, p = 0.90, I2 = 0%.
Figure 3.
The association between DTNBP1 rs2619522 and general cognitive ability in healthy samples. Negative effect sizes indicate that minor allele carriers had lower general cognitive ability.
DTNBP1 rs1011313, rs1047631, rs2619538, rs909706, rs3213207, rs2619539, rs760761
Pooled effect sizes for these seven SNPs did not demonstrate any statistical significance in random effects model analysis; there was no significant difference between minor allele carriers and major allele homozygotes in general cognitive ability for any of these SNPs. Tests of heterogeneity among studies were not significant for any SNP. Figure 4 summarizes the pooled effect sizes for each DTNBP1 SNP, along with a linkage disequilibrium (LD) block map.
Figure 4.
Pooled ES of differences in general cognitive ability between major allele homozygotes and minor allele carriers in ten SNPs, with corresponding LD block map (excluding rs909706, which was not sequenced in the HapMap dataset).
Sensitivity Analysis
Sensitivity analysis was conducted for each meta-analysis to assess the influence of any single study. When removing one study at time from each meta-analysis, there was no significant change in the pooled ES's and their associated p values for all SNPs except two: rs2619522 and rs760761. For rs2619522, removing Stefanis et al 2007 (n = 2,243) resulted in a pooled ES of −0.070, p = 0.062; minimally changed from the originally estimated ES of −0.083, p = 0.007 (n = 4,793). For rs760761, removing Need et al 2009 resulted in a significant pooled ES of −0.104, p = 0.004, as compared to the original estimate of −0.066, p = 0.205. There was no evidence of “winner's curse” in any of the analyzed SNPs.
Publication Bias Analysis
There was significant evidence for publication bias for only one SNP: rs3213207. The analysis suggested significant publication bias in favor of non-significance. Duval and Tweedie's “trim and fill” method showed that it was necessary to fill three “missing” studies in the funnel plot, and with the additional studies, the pooled effect size became significant, ES = −0.083, 95% CI = −0.151 to −0.015, p < 0.05. Therefore, results for this SNP are inconclusive given available data.
Moderator Analysis
Meta-regression analysis was conducted for each SNP to assess the effects of the following variables as potential moderators: age, sex, and ethnicity. There was no significant effect of any of the variables on any of the SNPs.
Discussion
We conducted meta-analyses on nine SNPs in the DTNBP1 gene and tested for their effects on general cognitive ability in 7,592 healthy subjects. Two SNPs, rs1018381 and rs2619522, were found to have a modest, but significant, effect on general cognitive ability. Individuals carrying the minor alleles of these SNPs demonstrated lower cognitive ability than subjects who were homozygous for the major allele. These results were not influenced by moderator variables, including age, sex, and ethnicity, nor was there any evidence of publication bias. Another SNP, rs3213207, was non-significant in our initial analysis, however, correction for publication bias suggested a possible association with general cognitive ability.
It is noteworthy that the two significant SNPs, rs1018381 and rs2619522, are in moderate LD (D' = 1, R2 = 0.36) when assessing the LD structure of the HapMap CEU sample (Table 2). This raises the possibility that these SNPs may not represent independent signals, as was the case in Burdick et al.(12), where the six-SNP risk haplotype was fully tagged by a single SNP (rs1018381). To date, no marker within DTNBP1 has been shown to be causal to either schizophrenia or to impaired cognition; therefore, the associated SNPs may have an as yet unidentified functional consequence or they may be themselves tagging a functional variant as yet unidentified.
These data implicating variation in the DTNBP1 gene with general cognitive ability converge with other lines of evidence from the recent literature. In animal studies, the DTNBP1 null mutant mouse (sandy) have been shown to have significantly reduced dysbindin-1 protein in many tissues including brain(35). These mice have deficits in spatial learning and memory, novel object recognition, and contextual fear conditioning(36). Similarly, human studies examining domain-specific cognitive performance have found that genetic variation within DTNBP1 affects verbal and visual memory(19, 26), working memory and processing speed(17), executive function(17), selective attention(40), and semantic verbal fluency(41). Our results are also consistent with a recent structural brain imaging study from our group, in which the minor allele of DTNBP1rs1018381 was associated with smaller total brain volumes compared to major allele homozygous subjects(37). Interestingly, Markov et al. reported that minor allele carriers of the same SNP exhibited significantly increased activation of the bilateral middle frontal gyrus compared to noncarriers during a functional MRI working memory task in healthy volunteers(38). The authors suggested that the increased activation in these brain areas may be a consequence of “inefficient” or compensatory prefrontal cognitive control functions. Comparable results were obtained for an episodic memory encoding and retrieval task(39). Finally, Fallgatter et al. used an event-related potential (ERP) paradigm with a PFC-based go-no go task in healthy human subjects and found that the DTNBP1 gene was associated with changes in ERP amplitude during task performance, implicating a role for the DTNBP1 gene in working memory(40).
Despite these converging results, it is not yet clear how DTNBP1 affects cognition through molecular and cellular mechanisms. DTNBP1 is expressed throughout many regions of the brain, and it can affect cognitive functions through different pathways. Dysbindin-1 protein is a key component of biogenesis of lysosome-related organelles complex-1 (BLOC-1), which regulates the trafficking of proteins in the lysosomal pathway(1, 35), and it also provides scaffolding for signaling proteins(42, 43) in the cytoskeleton. It may enrich the associational and commissural pathways in long axon terminals in both neocortex and hippocampal formation, structures that are involved in diverse learning and memory processes(36). Dysbindin also interacts with multiple neurotransmitter systems throughout the brain. Recent animal studies showed that disruption in dysbindin expression led to a marked increase in NMDA receptor subunit 2A and elevated long-term potentiation in hippocampal neurons(44), as well as an increase in dopamine receptor D2(45), and a marked decrease in the excitability of fast-spiking GABA-ergic interneurons in both prefrontal cortex and striatum(46), probably due to a defect in the lysosomal pathway. Glutamatergic synapses in the prefrontal and hippocampal regions have been implicated in working memory and information encoding(47), which is a key component of general cognitive ability. Dysbindin protein is involved in the regulation of neuroplasticity, and interacts with other proteins involved in cell morphology, cellular development, intracellular, and synaptic signaling at its different locations in synaptic vesicles, postsynaptic densities and microtubules(1, 7, 36). Therefore, it is possible that variations in DTNBP1 result in multiple developmental defects at cellular and intracellular levels in multiple brain regions, particularly the prefrontal cortex and hippocampus, leading to compromised overall cognitive capacity.
Although there were significant associations between two SNPs within the DTNBP1 gene and general cognitive ability, the pooled effect sizes were small, ranging from −0.083 to −0.123, explaining approximately 0.2% to 0.4% of the total variance in general cognitive ability. This is consistent with other genetic studies of human cognitive function(48), including a meta-analysis of the COMT Val158/108Met polymorphism and IQ(25), in which the pooled effect size was 0.06, smaller than the effect sizes in our meta-analysis. These small effect sizes of single SNPs on complex phenotypes are also consistent with data from relatively simple phenotypes such as human height, for which heritability studies have demonstrated that most of the variation in adult height is genetically determined. A recent genome-wide association study reported that 47 SNPs in multiple genes were robustly associated with height(49), however, collectively they explained about 5% of the total variation in adult height(50). Therefore, large samples, as can be achieved by using meta-analytic techniques, may be required to detect such modest effects. It should be noted that this meta-analytic methodology does not allow for direct comparison of relative size between different effect sizes. In addition, the significance level of a pooled effect size depends on the sample size such that a similar effect size may not be significant if the associated n is small. These are some of the limitations of this methodology.
To summarize, we conducted a meta-analysis of the association between DTNBP1 genetic variation and general cognitive ability in over 7,500 healthy individuals. Two SNPs, rs1018381 and rs2619522, significantly influenced cognitive ability, with minor allele carriers demonstrating lower general cognitive ability as compared with major allele homozygotes. Converging evidence from animal models and neuroimaging studies lend support to the notion that variation in the DTNBP1 gene may influence general cognitive ability. Future studies should focus on elucidating the biological pathways underlying this association.
Acknowledgements
The study was partially supported by NIMH grants P50MH080173 and R01MH79800 to Dr. Malhotra, K23MH077807 to Dr. Burdick. We would like to thank the investigators who provided additional data from their studies to make it possible to compute effect sizes for the meta-analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures:
Dr. Malhotra has received consulting fees and/or honoraria from Eli Lilly, BMS, and Merck. Dr. Lencz has received consulting fees and/or honoraria from Eli Lilly, Merck, Clinical Data Inc., GoldenHelix, Inc., Guidepoint Global, and Cowen & Co. Dr. Burdick is a member of Merck's Speaker Bureau. Dr. Zhang reports no biomedical financial interests or potential conflicts of interest.
References
- 1.Guo AY, Sun J, Riley BP, Thiselton DL, Kendler KS, Zhao Z. The dystrobrevin-binding protein 1 gene: features and networks. Molecular psychiatry. 2009 Jan;14(1):18–29. doi: 10.1038/mp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weickert CS, Straub RE, McClintock BW, Matsumoto M, Hashimoto R, Hyde TM, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004 Jun;61(6):544–555. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 3.Talbot K, Cho DS, Ong WY, Benson MA, Han LY, Kazi HA, et al. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Human molecular genetics. 2006 Oct 15;15(20):3041–3054. doi: 10.1093/hmg/ddl246. [DOI] [PubMed] [Google Scholar]
- 4.Dickman DK, Davis GW. The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science. 2009 Nov 20;326(5956):1127–1130. doi: 10.1126/science.1179685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen XW, Feng YQ, Hao CJ, Guo XL, He X, Zhou ZY, et al. DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. J Cell Biol. 2008 Jun 2;181(5):791–801. doi: 10.1083/jcb.200711021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubota K, Kumamoto N, Matsuzaki S, Hashimoto R, Hattori T, Okuda H, et al. Dysbindin engages in c-Jun N-terminal kinase activity and cytoskeletal organization. Biochem Biophys Res Commun. 2009 Feb 6;379(2):191–195. doi: 10.1016/j.bbrc.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Ghiani CA, Starcevic M, Rodriguez-Fernandez IA, Nazarian R, Cheli VT, Chan LN, et al. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Molecular psychiatry. 2009 Jun 23; doi: 10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. American journal of human genetics. 2002 Aug;71(2):337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funke B, Finn CT, Plocik AM, Lake S, DeRosse P, Kane JM, et al. Association of the DTNBP1 locus with schizophrenia in a U.S. population. American journal of human genetics. 2004 Nov;75(5):891–898. doi: 10.1086/425279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, He L. Association study between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia: a meta-analysis. Schizophrenia research. 2007 Nov;96(1-3):112–118. doi: 10.1016/j.schres.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nature genetics. 2008 Jul;40(7):827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 12.Burdick KE, Lencz T, Funke B, Finn CT, Szeszko PR, Kane JM, et al. Genetic variation in DTNBP1 influences general cognitive ability. Human molecular genetics. 2006 May 15;15(10):1563–1568. doi: 10.1093/hmg/ddi481. [DOI] [PubMed] [Google Scholar]
- 13.Burdick KE, Goldberg TE, Funke B, Bates JA, Lencz T, Kucherlapati R, et al. DTNBP1 genotype influences cognitive decline in schizophrenia. Schizophrenia research. 2007 Jan;89(1-3):169–172. doi: 10.1016/j.schres.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefanis NC, Trikalinos TA, Avramopoulos D, Smyrnis N, Evdokimidis I, Ntzani EE, et al. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biological psychiatry. 2007 Oct 1;62(7):784–792. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Donohoe G, Morris DW, Clarke S, McGhee KA, Schwaiger S, Nangle JM, et al. Variance in neurocognitive performance is associated with dysbindin-1 in schizophrenia: a preliminary study. Neuropsychologia. 2007 Jan 28;45(2):454–458. doi: 10.1016/j.neuropsychologia.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Zinkstok JR, de Wilde O, van Amelsvoort TA, Tanck MW, Baas F, Linszen DH. Association between the DTNBP1 gene and intelligence: a case-control study in young patients with schizophrenia and related disorders and unaffected siblings. Behav Brain Funct. 2007;3:19. doi: 10.1186/1744-9081-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luciano M, Miyajima F, Lind PA, Bates TC, Horan M, Harris SE, et al. Variation in the dysbindin gene and normal cognitive function in three independent population samples. Genes, brain, and behavior. 2009 Mar;8(2):218–227. doi: 10.1111/j.1601-183X.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 18.Kircher T, Markov V, Krug A, Eggermann T, Zerres K, Nothen MM, et al. Association of the DTNBP1 genotype with cognition and personality traits in healthy subjects. Psychological medicine. 2009 Apr 1;:1–9. doi: 10.1017/S0033291709005388. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto R, Noguchi H, Hori H, Ohi K, Yasuda Y, Takeda M, et al. Association between the dysbindin gene (DTNBP1) and cognitive functions in Japanese subjects. Psychiatry and clinical neurosciences. 2009 May 19; doi: 10.1111/j.1440-1819.2009.01985.x. [DOI] [PubMed] [Google Scholar]
- 20.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Human molecular genetics. 2009 Dec 1;18(23):4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004 Sep;20(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Spearman C. General intelligence, objectively determined and measured. American Journal of Psychology. 1904;15:201–292. [Google Scholar]
- 23.Plomin R, Spinath FM. Intelligence: genetics, genes, and genomics. Journal of personality and social psychology. 2004 Jan;86(1):112–129. doi: 10.1037/0022-3514.86.1.112. [DOI] [PubMed] [Google Scholar]
- 24.Plomin R. Genetics and general cognitive ability. Nature. 1999 Dec 2;402(6761 Suppl):C25–29. doi: 10.1038/35011520. [DOI] [PubMed] [Google Scholar]
- 25.Barnett JH, Scoriels L, Munafo MR. Meta-analysis of the cognitive effects of the catechol-O-methyltransferase gene Val158/108Met polymorphism. Biological psychiatry. 2008 Jul 15;64(2):137–144. doi: 10.1016/j.biopsych.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto R, Noguchi H, Hori H, Nakabayashi T, Suzuki T, Iwata N, et al. A genetic variation in the dysbindin gene (DTNBP1) is associated with memory performance in healthy controls. World J Biol Psychiatry. 2009 Apr 7;:1–8. doi: 10.1080/15622970902736503. [DOI] [PubMed] [Google Scholar]
- 27.Deary IJ. Human intelligence differences: towards a combined experimental-differential approach. Trends Cogn Sci. 2001 Apr 1;5(4):164–170. doi: 10.1016/s1364-6613(00)01623-5. [DOI] [PubMed] [Google Scholar]
- 28.Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. Introduction to Meta-Analysis. John Wiley & Sons, Ltd; Chichester, West Sussex, UK: 2009. [Google Scholar]
- 29.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003 Sep 6;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997 Sep 13;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duval SJ, Tweedie RL. A non-parametric “trim and fill” method of assessing publication bias in meta-analysis. Journal of the American Statistical Association. 2000;95:89–98. [Google Scholar]
- 32.Kraft P. Curses--winner's and otherwise--in genetic epidemiology. Epidemiology. 2008 Sep 19;(5):649–651. doi: 10.1097/EDE.0b013e318181b865. discussion 657-648. [DOI] [PubMed] [Google Scholar]
- 33.O'Leary UM, Rusch KM, Guastello SJ. Estimating age-stratified WAIS-R IQS from scores on the Raven's Standard Progressive Matrices. J Clin Psychol. 1991 Mar;47(2):277–284. doi: 10.1002/1097-4679(199103)47:2<277::aid-jclp2270470215>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 34.Lehrl S, Triebig G, Fischer B. Multiple choice vocabulary test MWT as a valid and short test to estimate premorbid intelligence. Acta Neurol Scand. 1995 May;91(5):335–345. doi: 10.1111/j.1600-0404.1995.tb07018.x. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O'Brien EP, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nature genetics. 2003 Sep;35(1):84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talbot K, Ong WY, Blake DJ, Tang J, Louneva N, Carlson GC, et al. Dysbindin-1 and its protein family. In: Javitt D, Kantrowitz J, editors. Schizophrenia. Springer Science+Business Media, LLC; 2009. pp. 110–241. [Google Scholar]
- 37.Narr KL, Szeszko PR, Lencz T, Woods RP, Hamilton LS, Phillips O, et al. DTNBP1 is associated with imaging phenotypes in schizophrenia. Hum Brain Mapp. 2009 Nov;30(11):3783–3794. doi: 10.1002/hbm.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markov V, Krug A, Krach S, Jansen A, Eggermann T, Zerres K, et al. Impact of schizophrenia-risk gene dysbindin 1 on brain activation in bilateral middle frontal gyrus during a working memory task in healthy individuals. Hum Brain Mapp. 2010 Feb;31(2):266–275. doi: 10.1002/hbm.20862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thimm M, Krug A, Markov V, Krach S, Jansen A, Zerres K, et al. The impact of dystrobrevin-binding protein 1 (DTNBP1) on neural correlates of episodic memory encoding and retrieval. Hum Brain Mapp. 2010 Feb;31(2):203–209. doi: 10.1002/hbm.20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fallgatter AJ, Herrmann MJ, Hohoff C, Ehlis AC, Jarczok TA, Freitag CM, et al. DTNBP1 (dysbindin) gene variants modulate prefrontal brain function in healthy individuals. Neuropsychopharmacology. 2006 Sep;31(9):2002–2010. doi: 10.1038/sj.npp.1301003. [DOI] [PubMed] [Google Scholar]
- 41.Markov V, Krug A, Krach S, Whitney C, Eggermann T, Zerres K, et al. Genetic variation in schizophrenia-risk-gene dysbindin 1 modulates brain activation in anterior cingulate cortex and right temporal gyrus during language production in healthy individuals. Neuroimage. 2009 Oct 1;47(4):2016–2022. doi: 10.1016/j.neuroimage.2009.05.067. [DOI] [PubMed] [Google Scholar]
- 42.Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J Biol Chem. 2001 Jun 29;276(26):24232–24241. doi: 10.1074/jbc.M010418200. [DOI] [PubMed] [Google Scholar]
- 43.Ceccarini M, Grasso M, Veroni C, Gambara G, Artegiani B, Macchia G, et al. Association of dystrobrevin and regulatory subunit of protein kinase A: a new role for dystrobrevin as a scaffold for signaling proteins. J Mol Biol. 2007 Aug 31;371(5):1174–1187. doi: 10.1016/j.jmb.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 44.Tang TT, Yang F, Chen BS, Lu Y, Ji Y, Roche KW, et al. Dysbindin regulates hippocampal LTP by controlling NMDA receptor surface expression. Proc Natl Acad Sci U S A. 2009 Dec 15;106(50):21395–21400. doi: 10.1073/pnas.0910499106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iizuka Y, Sei Y, Weinberger DR, Straub RE. Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J Neurosci. 2007 Nov 7;27(45):12390–12395. doi: 10.1523/JNEUROSCI.1689-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji Y, Yang F, Papaleo F, Wang HX, Gao WJ, Weinberger DR, et al. Role of dysbindin in dopamine receptor trafficking and cortical GABA function. Proc Natl Acad Sci U S A. 2009 Nov 17;106(46):19593–19598. doi: 10.1073/pnas.0904289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wall PM, Messier C. The hippocampal formation -- orbitomedial prefrontal cortex circuit in the attentional control of active memory. Behavioural Brain Research. 2001;127(1-2):99–117. doi: 10.1016/s0166-4328(01)00355-2. [DOI] [PubMed] [Google Scholar]
- 48.Plomin R, Owen MJ, McGuffin P. The genetic basis of complex human behaviors. Science. 1994 Jun 17;264(5166):1733–1739. doi: 10.1126/science.8209254. [DOI] [PubMed] [Google Scholar]
- 49.Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, Hammond N, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009 Apr;5(4):e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lettre G. Genetic regulation of adult stature. Curr Opin Pediatr. 2009 Aug;21(4):515–522. doi: 10.1097/MOP.0b013e32832c6dce. [DOI] [PubMed] [Google Scholar]
- 51.Peters K, Wiltshire S, Henders AK, Dragovic M, Badcock JC, Chandler D, et al. Comprehensive analysis of tagging sequence variants in DTNBP1 shows no association with schizophrenia or with its composite neurocognitive endophenotypes. Am J Med Genet B Neuropsychiatr Genet. 2008 Oct 5;147B(7):1159–1166. doi: 10.1002/ajmg.b.30741. [DOI] [PubMed] [Google Scholar]