Abstract
Assembly of virus capsids and surface proteins must be regulated to ensure that the resulting complex is an infectious virion. In this review, we examine assembly of virus capsids, focusing on hepatitis B virus and bacteriophage MS2, and formation of glycoproteins in the alphaviruses. These systems are structurally and biochemically well-characterized and are simplest-case paradigms of self-assembly. Published data suggest that capsid and glycoprotein assembly is subject to allosteric regulation, that is regulation at the level of conformational change. The hypothesis that allostery is a common theme in viruses suggests that deregulation of capsid and glycoprotein assembly by small molecule effectors will be an attractive antiviral strategy, as has been demonstrated with hepatitis B virus.
Is regulation of self-assembly required?
The common structural denominator for a typical small virus is a genome surrounded by a shell composed of dozens of copies of the capsid protein. Many small viruses also have lipid envelopes studded with glycoproteins that can facilitate cell entry. Even a small virus is complex, yet its formation is a canonical example of self-assembly. Given the right conditions, the capsid proteins of many viruses will assemble to capsid-like structures, rapidly, in high yield and with high fidelity (Table 1 ). This observation opens up two distinct fields of study: the process of self-assembly and regulation of assembly. The process of self-assembly is best described by physical chemistry, for example self-assembly can be modeled by a system of differential equations or emulated with small molecules. Regulating assembly, by contrast, is fundamentally biochemical and ultimately biological in nature. Consider our hypothetical virus where unregulated assembly could yield capsids that did not contain the viral genome or toxic accumulation of the fusion protein. These spontaneous assembly reactions must be delayed until the right time and place. Regulation is probably at the level of allostery, which is defined as a conformational change in a molecule, usually a protein, that alters its activity, induced by an effector molecule. In addition to allostery, regulation could require viral or host factors. Regulation of the large event of virus assembly by a small reaction, such as inducing conformational change of a single protein, provides the leverage that controls virus replication in vivo. Disruption of regulation is an ideal target for antiviral therapeutics. Defining the regulation that directly controls the physical chemistry of assembly is a field in its infancy and the focus of this review.
Table 1.
An incomplete list of in vitro icosahedral capsid assembly systems
| Species | Family | Number of proteins | Refs |
|---|---|---|---|
| Plant viruses | |||
| Cowpea chlorotic mottle virus, Brome mosaic virus | Bromo | 180, 120 and 60 | 71, 72 |
| Southern bean virus | Sobemo | 180 and 60 | [73] |
| Turnip crinkle virus, Sesbania mosaic virus | Tombus | 180 | [74] |
| Physalis mottle virus | Tymo | 180 and 60 | [75] |
| Animal viruses | |||
| Herpes simplex virus | Herpes | ∼2000 | [76] |
| Hepatitis B virus | Hepadna | 240 | [6] |
| Sindbis virus, Ross River virus | Toga | 240 | 77, 78 |
| Human papillomavirus 16 | Papilloma | 360 | [79] |
| Polyomavirus SV40 | Polyoma | 360 | 80, 81 |
| Poliovirus, Foot and mouth disease virus | Picorna | 180 | 82, 83 |
| Rous sarcoma virus HIV | Retro | ∼2000 | 22, 23 |
| Hepatitis E virus | Hepe | 180 | [84] |
| Bacteriophages | |||
| HK97, Lambda | Sipho | 420 | 12, 85 |
| P22 | Podo | 420 | [13] |
| MS2, R17 | Levi | 180 | 33, 35 |
| øX174 | Micro | 300 | [16] |
In this review, we will separately examine the assembly of virus capsids and the membrane-bound glycoprotein complexes present on the exterior of enveloped viruses. Both the capsid and glycoprotein are protein oligomers; from a physical–chemical perspective, their assembly is similar; from a biological perspective they demonstrably have similar elements of regulation. Although there are numerous experimental systems (Table 1, Table 2 ), the discussion of capsid assembly will focus on hepatitis B virus (HBV) with significant references to retroviruses and bacteriophage MS2. Furthermore, small molecules that dramatically affect HBV assembly have demonstrable antiviral activity. The discussion on glycoprotein assembly will focus on the formation of the spike complex of alphaviruses as the spike is composed of two proteins which intricately interact and are required for viral entry. Finally, we will discuss potential steps in assembly that could be targeted for designing antiviral therapeutics.
Table 2.
An incomplete list of enveloped virus structuresa
| Example | Family | Refs | Method(s) |
|---|---|---|---|
| Viruses with an icosahedral core and icosahedral glycoprotein layer | |||
| Sindbis virus, Venezuelan equine encephalitis virus | Toga | [51] | Single-particle averaging |
| Viruses with an icosahedral core and local symmetry in the glycoprotein layer | |||
| Hepatitis B virus | Hepadna | [86] | Single-particle averaging |
| Tula virus | Bunya | [87] | Tomography |
| Viruses with an icosahedral core and no symmetry in the glycoprotein layer | |||
| Epstein–Barr virus, herpes virus | Herpes | [88] | Tomography |
| Viruses with local symmetry in the core and no symmetry the glycoprotein layer | |||
| Simian immunodeficiency virus, HIV, murine leukemia virus | Retro | 31, 89 | Tomography |
| Viruses with no symmetry in the core but icosahedral glycoprotein layer | |||
| Dengue virus, West Nile virus, tick-borne encephalitis virus | Flavi | [90] | Single-particle averaging |
| Rift Valley fever virus, Uukuniemi virus | Bunya | [91] | Tomography |
| Viruses with helical symmetry in core and glycoprotein symmetry undetermined | |||
| Vesicular stomatitis virus, measles virus | Rhabdo | [92] | Single-particle averaging |
| Viruses with no symmetry observed in either the core nor the glycoprotein layer | |||
| Mouse hepatitis virus, severe acute respiratory syndrome virus | Corona | [93] | Single-particle averaging |
| Vaccinia virus | Pox | [94] | Tomography |
| Influenza virus | Orthomyxo | [95] | Tomography |
| Ebola virus | Filo | [96] | Tomography |
| Parainfluenza virus, Hendra virus, Sendai virus | Paramyxo | [97] | Tomography |
The structures of most enveloped viruses, either entire particles or portions of the particle, have been determined using cryo-electron microscopy either by single-particle averaging or tomography.
Assembly of virus capsids
The capsids of icosahedral viruses have tens to hundreds of copies of the capsid protein(s). The simplest vision of virus capsid assembly is one where rigid assembly units (AUs) collide by Brownian motion, interact with perfect geometry and associate irreversibly [1]. Like any utopia, this vision fails examination: assembly simulations are incredibly sensitive to kinetic traps consisting of partial capsids that have a negligible chance of completion due to depletion of subunits (Figure 1 ).
Figure 1.
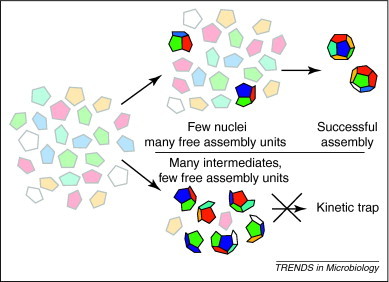
Successful assembly and kinetic traps. Assembly of a dodecahedron from pentagonal assembly units (faded colors) is more robust if regulated by nucleation. In nucleated assembly (top path), the initial association reactions are relatively slow, resulting in rare nucleation centers (brightly colored oligomer) and plenty of assembly units to allow completion of capsids. If assembly is unregulated (lower path), many partial capsids will form and very few assembly units will remain, kinetically trapping these intermediates. Of course, this trap might resolve if stochastic dissociation of some intermediates allows others to go to completion; alternatively, partial capsids can associate incorrectly, resulting in aberrant structures.
Instead, assembly simulations, from master equations that treat assembly reactions as a well-mixed solution 2, 3 to molecular dynamics that describe stochastic formation of single capsids 4, 5, concur on three generalizations (Box 1 ). First, weak interactions are necessary to minimize errors and kinetic traps. Weak interactions also contribute to a defined nucleation step. Second, nucleation minimizes the initiation of assembly, decreasing kinetic trapping of intermediates due to depletion of AUs. Third, the initial kinetic phase where there is little capsid formation is due to the time required to build the steady state of intermediates that supports subsequent assembly. More sophisticated mathematical models that incorporate the biological details of individual viruses (e.g. nucleic acid, scaffolding and allostery) will help to identify opportunities to interfere with assembly.
Box 1. An analytical description of capsid assembly.
Probably the simplest way to describe assembly of a capsid of N assembly units (AUs) is as a progressive series of intermediates [100]. To quantify this description, one can assume that all contacts between AUs have the same energy, energies are additive, microscopic forward rates are identical (subject to statistical considerations) and that backward rates are the product of forward rates, dissociation constants and statistical terms. The result is a series of rate equations each with terms for assembly and disassembly. Two equations in the series are unique: the monomer equation references all intermediates and the final equation is a dead-end with only one build-up and one build-down term.
Incorporating weak interactions between AUs and a slow nucleation step to limit the initiation of assembly results in robust reactions with minimal kinetics traps where errors and (meta)stable intermediates accumulate 4, 39. This model necessarily leads to sigmoidal kinetics and, at equilibrium, a pseudocritical concentration of free AU 5, 100. The resulting model recapitulates most of the features observed in vitro (Figure I ).
Figure I.
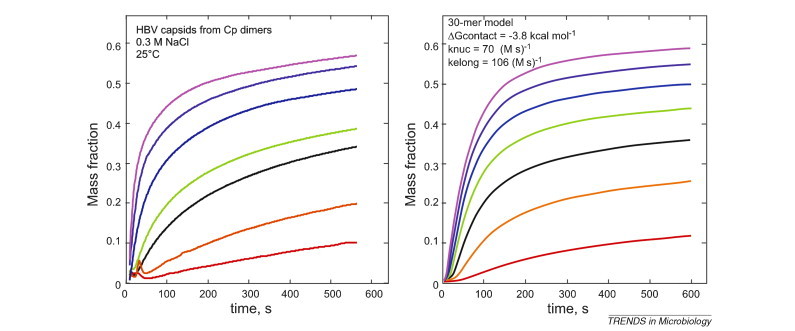
Comparison of HBV capsid assembly with simulations of capsid assembly. Assembly of a concentration series of HBV core protein (left) compared to assembly simulations of a 30-mer model (right), where subunits have a HBV-like geometry and the nucleus is trimeric [1]. The protein concentration increases from 4 μM (red) to 10 μM (purple) in 1 μM increments. The calculation is parameterized to resemble HBV assembly. For the simulation, the bimolecular rates for nucleation (knuc) and elongation (Kelong) were 70 M−1 s−1 and a diffusion-limited 105 M−1 s−1, respectively; the pairwise association energy was –3.8 kcal/mol.
Assembly can be described in much greater detail as stochastic reactions using discrete event simulators or coarse-grained molecular dynamics [4]. These molecular simulations provide detail, showing conditions where association of intermediates could be an important path, where reversibility is crucial [5], and how subunit geometry can affect the size and shape of assembly products [4]. This statistical mechanical view of assembly leads to a view that is fundamentally similar to the thermodynamic–kinetic view described above [4]. However, the detail provides additional insights into the reactions and the behaviors that can be anticipated by biological molecules.
Experimental observation of assembly of empty HBV capsids is in good agreement with the predictions of theoretical models [6] (Box 1). HBV has a T = 4 icosahedral capsid composed of 120 AUs, the homodimeric core protein (Cp) [7]. Cp can be assembled in vitro in response to ionic strength [6] and the kinetics are sigmoidal [6]. The average association energy between two subunits is –3 to –4 kcal/mol, corresponding to a millimolar dissociation constant. Because subunits are tetravalent, the weak association energy corresponds to a micromolar pseudocritical concentration [8]. In vitro HBV assembly is resistant to, but not altogether immune to, kinetic traps 9, 10.
HBV is a simplest-case system, a homopolymer of dimeric AUs. In cowpea chlorotic mottle virus and bacteriophage HK97, different AU oligomers participate in assembly 11, 12. In many viruses, scaffold proteins support assembly. Bacteriophage P22 has a scaffold protein that thermodynamically and kinetically contributes to assembly 13, 14. Excess P22 scaffold can actually block assembly by trapping numerous intermediates [14]. Scaffolds can play roles in switching morphologies, as in bacteriophages P2 and P4 [15], and complex roles in subsequent maturation [16]. Thus, scaffold proteins could direct geometry, stability and a measure of regulation by imposing stepwise assembly.
Viral nucleic acid can also serve as a molecular scaffold. Viral RNA has been considered as an ‘antenna’ to attract free AUs [17] and as a platform that attracts and organizes AUs on its surface [18]. Assembly driven by RNA can be thought of in terms of the McGhee–von Hippel model of nonspecific protein binding to a surface [19], which considers that the association for nucleic acid (NA), K NA, is modified by a cooperativity coefficient, ω, based on the protein–protein association constant. An ω of 1 indicates no cooperativity; an ω >1000 results in steep cooperativity and reactions that appear to be two-state. Cowpea chlorotic mottle virus, which does not assemble under RNA-binding conditions, binds RNA with low cooperativity, displaying gradual assembly and partial capsids [20]. HBV, where the reciprocal of the pseudocritical concentration (equivalent to ω) is ∼105 under physiological conditions, binds RNA tenaciously and with high cooperativity, resulting in quantitative assembly under mild conditions [21]. A NA scaffold can thus concentrate the capsid protein and provide additional association energy.
Assembly and nucleic acids as allosteric effectors
Simple theoretical models fail to describe experimental results where induced conformational changes activate assembly. This behavior fits the definition of allostery. NA-regulated allostery is observed with bacteriophage MS2 and retroviruses, for example human immunodeficiency virus (HIV) and Rous sarcoma virus (RSV).
Retroviral Gag, the ‘capsid protein’ of immature retroviruses, is a multidomain protein. The elements of Gag that are most important for this discussion are the two-domain capsid (CA) segment (the primary mediator of Gag–Gag interactions), a spacer peptide that follows CA, and the RNA-binding nucleocapsid (NC) segment. The earliest suggestion of allostery in retrovirus assembly came from in vitro assembly studies using DNA oligomers and Gag 22, 23. Gag dimerization was crucial for assembly: HIV Gag would not assemble with very short oligos nor if the oligo had high-affinity sequences separated by a low-affinity sequence, suggesting that protein–protein interaction was required to activate assembly [24]. Replacing NC with a leucine zipper promoted dimerization of Gag and was sufficient to drive assembly in cells [25], although other factors were required in vitro [26]. What transitions are driven by protein oligomerization and DNA-binding? Small angle neutron scattering indicated that HIV Gag had a propensity to fold on itself [17], thus straightening might be a crucial assembly transition. Nuclear magnetic resonance (NMR) studies have indicated that the spacer sequence C-terminal to Gag (in HIV and RSV) can refold 27, 28. This refolding transition, driven by binding NA, might expose surfaces on CA to allow interaction [28] and generate a new, potentially helix-rich interaction between the spacer peptides 29, 30. Recent tomographic studies of immature HIV demonstrate CA–CA interactions and spacer peptide–spacer peptide interactions 30, 31. Thus, it appears that Gag undergoes one or more conformational transitions to assemble into an immature lattice. These conformational transitions are involved in Gag assembly and are consistent with a regulatory effect, that is they prevent assembly by obstructing interactions [28] and this obstruction can be removed by binding a cofactor [26].
In the RNA bacteriophage MS2, the role of allostery mediated by NA is unambiguous (Figure 2 ). These phages are organized with T = 3 quasi-symmetry; their capsids comprise 90 chemically identical dimers that fit into AB and CC environments [32]. B half-dimers cluster around icosahedral fivefold vertices, A and C half-dimers alternate around icosahedral threefold vertices (quasi-sixfolds). The A and C half-dimers are very similar, whereas a loop on the B subunits has a different conformation, suggesting a structural switch. Uhlenbeck and co-workers showed that a specific RNA stem–loop from the genome, TR, was necessary and by itself sufficient to induce capsid assembly 33, 34. By NMR, TR-free dimer was shown to be symmetrical in solution, suggestive of a CC dimer, but TR-bound dimer was asymmetric [35]. Pure RNA-free dimers or pure TR-saturated dimers do not assemble rapidly, that is they appeared kinetically trapped [35]. Addition of the other form of coat protein to trapped species resulted in rapid assembly [36]. These results showed that both forms of dimer, symmetric and RNA-bound asymmetric, are needed for efficient assembly and that RNA was an allosteric effector. Non-TR RNA stem–loops also trigger these effects, although they are much weaker binders, implying that multiple RNA–coat protein interactions within the capsids also contribute to conformer switching 36, 37. Normal mode analysis has suggested that the RNA-induced conformational change might be one of dynamics, rather than static dimer conformations [38], and is consistent with the lack of sequence-specificity. Analyzing mixed assembly reactions by mass spectrometry has provided a clear example of assembly by one dimer at a time rather than via coalescence of oligomers [35]. Examination of assembly reactions with TR incorporated into a longer RNA have lead to the surprising conclusion that assembly might be directed by an RNA scaffold [36], funneling the system along only a few of the very many possible assembly paths [36]. Thus, initiation by an allosteric event and limitation by allosteric responses to protein–RNA and protein–protein interaction gives MS2 assembly the appearance of following a deterministic path. An alternative viewpoint suggested by recent theoretical studies is that only a relatively small number of intermediates will ever be used during assembly as a consequence of kinetic availability and thermodynamic stability [39].
Figure 2.
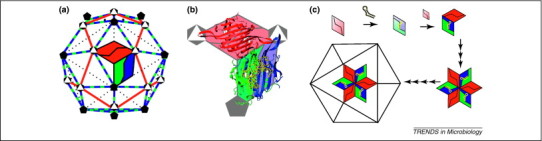
Structure and assembly of bacteriophage MS2. (a) The T = 3 capsid of MS2 has 60 AB dimers (blue and green, respectively) and 30 CC dimers (red), so that A and C subunits alternate around the quasi-sixfold axes. Icosahedral fivefold and quasi-sixfold (icosahedral threefold) vertices are shown in this representation. (b) The CC dimers are necessarily symmetric with an extended loop (highlighted in the open triangles) connecting the F and G β strands at either end. Bound RNA, the TR stem–loop (in yellow), induces asymmetry in the AB dimers so that the FG loop of the B subunit (in the pentagon) adopts a unique conformation that allows it to pack around the fivefold vertices. (c) Pure CC dimers (red) or pure AB dimers (green and blue), with bound RNA, assemble rapidly when mixed, leading to the hypothesis that assembly proceeds in a stepwise reaction. The hexameric threefold cluster is one of the major complexes identified by mass spectrometry, suggesting a highly deterministic assembly path. This hypothesis has been strongly supported by further examination of RNA-driven assembly [98]. This figure was adapted from [35].
Allostery in capsid construction
HBV Cp assembly is also an example of allosterically controlled assembly, even though its assembly superficially resembles association of rigid bodies. For example, HBV Cp binds cooperatively to Zn2+, resulting in increased intrinsic fluorescence (indicating conformational change) and faster assembly kinetics [10]. Conformational differences between free Cp dimer and the same protein in the context of capsid are evident in the crystal structure of an assembly defective mutant (Figure 3b) [40]. Dimers from capsid are symmetrical about the dimer interface [41], whereas free dimer is asymmetric. Comparison of the structures suggests a mechanical linkage of subdomains connected to a central chassis; these subdomains displace one another like so many molecular dominos, leading to movements of up to 9 Å and a conformation that is geometrically incompatible with an icosahedral structure. In comparison to the crystal structure, an NMR study under non-assembly conditions showed a symmetric dimer that was readily asymmetrized by interaction with a Cp-binding peptide [42]. Thus, the dimer is able to adopt multiple conformations, or families of conformations, tentatively categorized as assembly-active and assembly-inactive.
Figure 3.
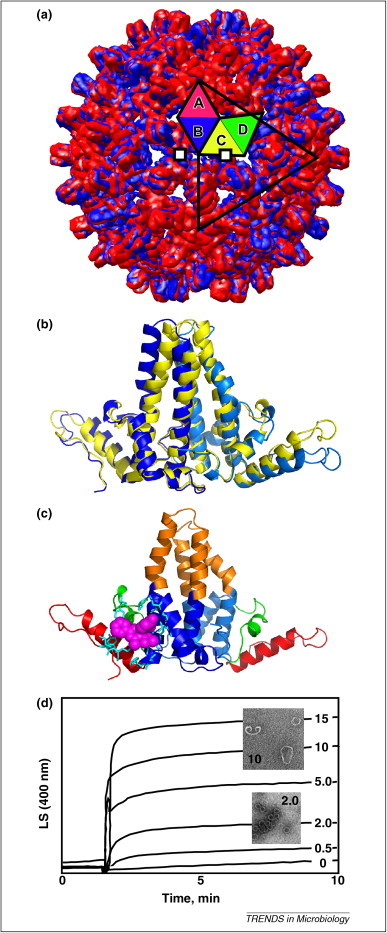
Structure and assembly of hepatitis B virus core. (a) The HBV capsid is constructed from 120 dimers arrayed with T = 4 icosahedral symmetry. One of the 60 icosahedral asymmetric units, an AB and CD dimer, are highlighted on the 20 triangular facets that surface an icosahedron. Shown are two overlaid crystal structures of HBV (blue, PDB 2G33) and HBV with bound HAP1 assembly effector (red, PDB 2G34). More red is visible as HAPs cause the capsid to expand. In total, 60 HAPs are bound with high occupancy to a pocket in the C subunits (two are shown as white squares) [41]. (b) Dimers from the capsid (yellow) are symmetrical and identical. Asymmetric free dimers (blue, PDB 3KXS) are presumably in an assembly-inactive conformation. (c) A free dimer is color coded to show the chassis (blue) that is identical to the dimer from capsid and the three mobile subdomains (red, green and orange). The subdomains are connected to the chassis by flexible joints, either Gly or Pro. Movement of one subdomain will force the others to shift their positions. A HAP molecule (magenta), modeled into its binding site, coordinates mobile subdomains with the chassis, facilitating the transition to an assembly-active state [40]. (d) HAPs accelerate assembly, even without misdirecting the reaction. Assembly of 10 μM Cp initiated by ionic strength in the presence of 0–15 μM HAP1 [9]. The inset micrographs show that 2 μM HAP1 yields spherical virus-like particles very fast, whereas higher HAP concentrations yield aberrant structures that scatter much more light.
Allostery and antiviral strategies based on assembly
For allostery, there must be a conformational change and an allosteric effector. Synthetic HBV assembly effectors have now been identified: the heteroaryldihydropyrimidines (HAPs) and phenylpropenamides. These show promise as antiviral agents. However, the identity of the natural allosteric effector still remains unknown.
HAPs were discovered in a search for non-nucleoside inhibitors of HBV replication and only later shown to affect HBV production by a core protein-dependent mechanism [43]. In vitro experiments showed that HAPs could misdirect assembly and, more crucially, HAPs strengthen dimer–dimer association and accelerate assembly kinetics, sometimes by orders of magnitude 9, 44. A crystal structure of a HAP–capsid complex (Figure 3a) shows that HAPs lead to a change in capsid quaternary structure (but not Cp tertiary structure), bowing out icosahedral fivefold vertices while flattening hexagonal arrangements of dimers [41]. These two effects explain the basis of assembly misdirection by destabilizing fivefold vertices while favoring sheets of Cp dimers [9]. The HAP molecule fills a hydrophobic pocket at the dimer interface, increasing buried surface area and presumably the association constant while slightly distorting the geometry of Cp–Cp interaction [41].
Using a series of HAPs, a strong correlation was observed between antiviral effect and the rate of capsid formation, and a negligible correlation between antiviral effect and HAP stabilization of Cp–Cp interaction [44]. The low concentrations of the most effective HAPs required to suppress HBV replication suggest that misdirection (which requires stoichiometric concentrations of HAP) is not central to antiviral activity [9]. Similarly, phenylpropenamides, which were recently shown to accelerate assembly with little capsid stabilization and no assembly misdirection [45], inhibit virus production in culture to yield empty capsids [46].
We propose that the kinetic effects of HAPs and phenylpropenamides are the crucial predictor of antiviral activity 44, 45. This hypothesis leads to predictions about the nature of assembly and its allosteric activation. First, as per assembly models, a kinetic effect will be most evident in the nucleation step. Starting capsid assembly at an inappropriate time (e.g. without the viral genetic material) is unfavorable for the virus. Second, faster kinetics indicate that effectors decrease the energy barrier to assembly, which can be best explained by invoking an effector-induced conformational change. The graphic of the HAP site in the context of free and the capsid conformations (Figure 3c) supports this hypothesis. The key point is that only the nucleation step of an assembly reaction needs be affected to gain an overall enhancement of assembly kinetics (Figure 3d). Thus, the few effectors needed for nucleation can be leveraged to consume many copies of Cp to produce failed virus.
As a point of comparison to assembly effectors, there are molecules that inhibit other allosteric transitions in viruses, such as stabilization of picornaviruses to uncoating and inhibition of HIV maturation. The WIN compounds (e.g. pleconaril) bind to a hydrophobic pocket in rhinoviruses and polioviruses and entropically inhibit the structural transition that enables release of the viral genome 47, 48. A peptide inhibitor of HIV, ‘CAI’, inhibits the structural transitions between the N-terminal and C-terminal domains of CA that are associated with maturation, possibly favoring inappropriate geometries [49]. CAI also blocks assembly of immature Gag. However, consideration of assembly models suggests that simply inhibiting assembly could be losing strategy as it is easily overcome by overproduction of the capsid protein(s) [50].
Viral glycoprotein spikes
Although capsid and envelope glycoproteins have different functions, they share common principles that regulate their assembly. Viral glycoproteins, or spikes, are multimers that are assembled sequentially before becoming competent for catalyzing cell entry. The protein–protein interactions within a spike and between spikes are analogous to the interactions between AUs that drive capsid assembly. The processing events of the spikes parallel the nucleation events that initiate capsid assembly. Furthermore, factors such as environmental pH and peptides digested from the spikes as they are folded and refolded function as allosteric regulators and sensors during assembly.
The assembly of viral glycoprotein spikes requires a coordinated, highly regulated, processive mechanism to be successful. The alphavirus system, which is discussed below, demonstrates that even with less than a half-dozen proteins, the virus has developed an assembly mechanism that relies on cellular signals and the environment to initiate conformational or allosteric changes in the assembly process. Stabilizing any of the discrete conformations along the spike assembly path by binding of a small molecule would have tremendous potential for disrupting the timing and compartmentalization of this complex series of reactions.
Assembly of alphavirus spike complexes
Alphaviruses have 80 trimeric spikes, each ∼70 Å in height. Each spike is a trimer of an E2–E1 heterodimer, with the E2 protein at the top of the spike and the E1 protein lying underneath, close to the lipid membrane (Figure 4 ) [51]. In addition to interactions within each heterodimer, there are interactions between heterodimers. An E1 molecule from one heterodimer contacts the stalk of an adjacent E2 molecule, which is part of another heterodimer [51]. Each spike interacts with the nucleocapsid core via the cytoplasmic region of E2, and the E2 and E1 proteins interact uniquely with each other via their transmembrane regions [51].
Figure 4.
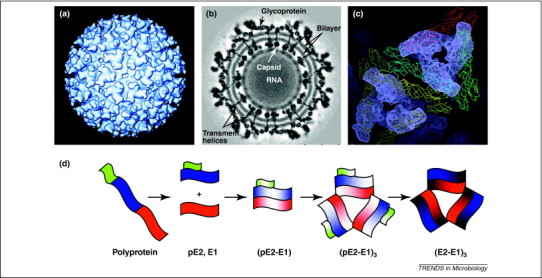
Structure of Sindbis virus. (a) Surface view, looking down the icosahedral twofold axis, of Sindbis virus at 20 Å resolution. The triangular spikes are trimers of the E2–E1 heterodimer. (b) Cross-section through an 11-Å resolution structure of Sindbis virus. The glycoprotein spikes are on the surface of the virus and their transmembrane region is seen traversing the lipid bilayer (transmem. helices). The nucleocapsid core is internal to the lipid bilayer with the RNA appearing disordered. The capsid and glycoproteins of alphaviruses both have icosahedral symmetry and their symmetry axes are aligned. This is in contrast to most enveloped viruses. (c) A surface view of the glycoprotein spikes showing the arrangement of the E2 and E1 proteins. Four monomers of E1 have been fit into the cryo-electron microscopy density of the 9-Å resolution Sindbis map and are shown in yellow, blue, red and green. The yellow monomers surround the icosahedral threefold axis, whereas the blue, red and green monomers surround the quasi-threefold axis. The density corresponding to the E1 proteins has been subtracted from the density of the entire spike. The remaining density, which corresponds to the E2 protein, is shown in gray. The E2 protein covers the E1 protein and makes up the top portion of the spike. Interactions between E2–E1 heterodimers, along with those within the heterodimer, are thought to stabilize the spike complex. (d) A model of the allosteric changes that occur during assembly of the viral glycoproteins. The polyprotein is cleaved by cellular proteases to the individual proteins (green, E3; blue, E2; red, E1; 6K is not shown for clarity). Dimerization of pE2 and E1 to (pE2–E1) is followed by trimerization of the heterodimer (pE2–E1)3. Then, cleavage of E3 from pE2 leaves a fusion competent trimer (E2–E1)3. Changes in color indicate conformational changes in addition to changes in aggregation state. Panels (a), (b) and (c) were adapted from 51, 99.
The E2 protein binds to host cell receptor and the E1 protein mediates fusion between the viral membrane and the host cell membrane. The receptor binding domain in E2 is positioned above the fusion peptide of E1 [51]. One could speculate that upon receptor-binding, unzippering of the E2–E1 dimer must be initiated to ultimately expose the fusion protein to the host membrane. What might control this association and subsequent disassociation? Viral fusion occurs in the endosome and is pH-dependent [52]. pH sensors or amino acid residues that destabilize the E2–E1 dimer or that stabilize the post-fusion E1 trimer conformation have been identified [53]. Interfering with the assembly of the E2–E1 dimer, either through stabilization or destabilization, would severely affect the allosteric regulation by the sensor residues during fusion and, as a result, cell entry would be blocked.
The association of the alphavirus E2–E1 dimer is a regulated process that occurs early in assembly. The alphavirus structural proteins are translated from a subgenomic RNA as a single polyprotein of capsid–E3–E2–6K–E1. The capsid protein autoproteolytically cleaves itself from the rest of the polyprotein in the cytoplasm [54]. The remaining structural polyprotein is targeted to the endoplasmic reticulum and cleaved to pE2 (or p62, corresponding to E3 + E2), 6K and E1 proteins by cellular proteases [55]. In the ER, E1 undergoes several conformations (α, β, γ), each differing in the number of disulfide bond rearrangements. pE2 associates only with the E1β intermediate. As a pE2–E1 dimer and during trimerization, E1 continues to undergo conformational changes until a stable E1ɛ conformation is reached. Late in the secretory pathway E1ɛ becomes metastable, possibly in response to cleavage of pE2 during virus maturation (see below) [56].
One could speculate that pE2 undergoes similar disulfide bond rearrangement as E1 during assembly. Both the E1 and E2 proteins have 12 cysteine residues in their ectodomain, and both interact with resident disulfide isomerases [57]. pE2 has several CXXC motifs, characteristic of disulfide isomerase substrates [58]. It has been suggested that E3 might function as a disulfide isomerase during alphavirus assembly [59]. In the absence of pE2, E1 is not transported to the plasma membrane; in the absence of E1, low amounts of pE2 are transported to the plasma membrane [56]. These results emphasize the necessity of forming a specific pE2–E1 dimer early in assembly.
Recently, a translational frameshift was identified in the 6K protein coding region, producing a TF (for transframe) protein [60]. The synthesis of TF presents a conundrum for maintaining a balance of pE2 and E1 protein during assembly. When the 6K protein is translated, equal amounts of pE2 and E1 are translated. However, when TF is translated, E1 translation is abrogated, presumably drastically impacting the assembly of the pE2–E1 heterodimer.
Activation of viral fusion proteins
Viral glycoproteins fill many roles; the most common is mediating fusion between the viral and host membrane via the fusion protein. The atomic structures of viral fusion proteins in the pre- and post-fusion conformations have been determined and led to their classification based on structural similarities: Class I (orthomyxo-, paramyxo-, retro-, filo-, arena- and coronaviruses), Class II (alpha- and flaviviruses) and Class III (rhabdo-, baculo- and herpesvirus) 61, 62. The three classes are trimers in their fusion-active conformation. For most viruses, the final step of glycoprotein spike assembly is the activation of the fusion protein. This step brings the fusion protein into a metastable state, priming it for rapid fusion with the host cell upon proper activation.
The requirement of a chaperone protein and its cleavage during assembly is a property of the Class II fusion proteins. The cleavage of E3 from pE2 occurs in the trans-Golgi [63] and marks the transition from an immature to mature or fusogenic alphavirus particle. Immature and mature alphaviruses are structurally similar, but immature particles have reduced infectivity levels compared with mature particles because of the presence of E3 and/or because E1 is not in a metastable state able to facilitate fusion [63]. Retaining the E3 protein until the trans-Golgi might serve to prevent premature fusion of the assembled spikes with membranes in the host cell secretory pathway. Thus, E3 is an effector of spike activity. At the plasma membrane the spikes are arranged in large hexagonal arrays [64], creating a platform for virus budding. Host proteins have not been identified within the alphavirus glycoprotein array, suggesting that lateral interactions between the spikes are important, in contrast to vesicular stomatitis virus [65], HIV [66] and influenza virus [67] which also bud from the plasma membrane. Although not usually described as such, pE2 acts as an effector of E1 activation at two different times, as a chaperone and by releasing E3 for activation.
Similar to pE2, the flavivirus prM protein chaperones folding of the flavivirus E protein and cleavage of the pr peptide is required to transition from the immature to the mature flavivirus. In contrast to alphaviruses, immature and mature flaviviruses have radically different structures [68].
Class I fusion proteins do not have a distinct chaperone. Activation of the fusion proteins occurs by cleaving the fusion protein itself. Multiple extracellular and cellular proteases have been proposed to cleave influenza HA0 both intra- and extracellularly, and we suggest that additional proteases for other viruses will also be identified [69]. The Class III fusion proteins do not have a chaperone protein nor do they require a cleavage step to activate the protein into a fusion-active form. The vesicular stomatitis virus G protein is found in three reversible conformations in a pH-dependent equilibrium. The reversible conformations are a unique property among Class III fusion proteins, suggestive of allosteric conformational changes, which also prevents premature fusion during assembly [70]. Thus, the classification of fusion proteins is dependent on the structure of the fusion protein as well as the regulatory mechanism for its activation. Both processes are probably controlled through allosteric mechanisms.
We suggest that most viral families rely on a series of ‘sensing’ events that control protein disulfide bond linkages, glycosylation, acylation, phosphorylation, oligomerization and transport to the appropriate cellular membrane for viral budding. These sensors respond to pH, redox and lipid environment as the proteins are assembled and transported through the host secretory pathway [53] to allosterically enhance assembly and ensure productive spike formation.
Concluding remarks and future directions: interfering with conformational change and control during viral assembly
Although targeting specific conformations is an attractive antiviral strategy, we further propose that one should consider the dynamic nature of virus assembly and that interfering with the allosteric responses to effectors and environmental sensors could be equally beneficial. Both capsid and glycoprotein assembly are carefully orchestrated events requiring activation by known and unidentified effectors. For example, we have learned from the HAP molecules that interfering with assembly is not equivalent to inhibiting assembly. In addition, weak interactions between protein subunits or glycoprotein complexes minimize production of misassembled spikes. Thus, altering the kinetics, misdirecting the intermediates, stabilizing an interaction to prevent disassembly are all viable options that disrupt a highly evolved set of reactions to achieve the same end goal: reduced virus propagation.
Note added in proof
The X-ray crystal structures of the alphavirus glycoproteins have recently been solved 101, 102.
Acknowledgments
The lists of assembly systems in Table 1 and structures in Table 2 are necessarily incomplete; we apologize for examples of excellent work that we have failed to include. We thank Drs. Alan Rein, Thomas Gallagher, Peter Stockley and Stephen Stray for their comments. This effort was supported by National Institutes of Health grants R01-AI067417 and R01-AI077688 to A.Z.
Contributor Information
Adam Zlotnick, Email: azlotnic@indiana.edu.
Suchetana Mukhopadhyay, Email: sumukhop@indiana.edu.
References
- 1.Porterfield J.Z., Zlotnick A. An overview of capsid assembly kinetics. In: Stockley P.G., Twarock R., editors. Emerging Topics in Physical Virology. Imperial College Press; 2010. pp. 131–158. [Google Scholar]
- 2.Endres D., Zlotnick A. Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys. J. 2002;83:1217–1230. doi: 10.1016/S0006-3495(02)75245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keef T. Master equation approach to the assembly of viral capsids. J. Theor. Biol. 2006;242:713–721. doi: 10.1016/j.jtbi.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Elrad O.M., Hagan M.F. Mechanisms of size control and polymorphism in viral capsid assembly. Nano Lett. 2008;8:3850–3857. doi: 10.1021/nl802269a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapaport D.C. Role of reversibility in viral capsid growth: a paradigm for self-assembly. Phys. Rev. Lett. 2008;101:186101. doi: 10.1103/PhysRevLett.101.186101. [DOI] [PubMed] [Google Scholar]
- 6.Zlotnick A. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry. 1999;38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 7.Seeger C. Hepadnaviruses. In: Knipe D.M., editor. Fields Virology. Lippincott Williams and Wilkins; 2007. pp. 2977–3029. [Google Scholar]
- 8.Ceres P., Zlotnick A. Weak protein–protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 9.Stray S.J. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stray S.J. Zinc ions trigger conformational change and oligomerization of hepatitis B virus capsid protein. Biochemistry. 2004;43:9989–9998. doi: 10.1021/bi049571k. [DOI] [PubMed] [Google Scholar]
- 11.Zlotnick A. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 12.Hendrix R.W., Duda R.L. Bacteriophage HK97 head assembly: a protein ballet. Adv. Virus Res. 1998;50:235–288. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- 13.Prevelige P.E. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parent K.N. Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology. 2005;340:33–45. doi: 10.1016/j.virol.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Wang S. Assembly of bacteriophage P2 and P4 procapsids with internal scaffolding protein. Virology. 2006;348:133–140. doi: 10.1016/j.virol.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Fane B.A., Prevelige P.E., Jr. Mechanism of scaffolding-assisted viral assembly. In: Chiu W., Johnson J.E., editors. Virus Structure. Academic Press; 2003. pp. 259–299. [DOI] [PubMed] [Google Scholar]
- 17.Datta S.A. Conformation of the HIV-1 Gag protein in solution. J. Mol. Biol. 2007;365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagan M.F. A theory for viral capsid assembly around electrostatic cores. J. Chem. Phys. 2009;130:114902. doi: 10.1063/1.3086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGhee J.D., von Hippel P.H. Theoretical aspects of DNA–protein interactions: cooperative and non-cooperative binding of large ligands to a one dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 20.Johnson J.M. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of cowpea chlorotic mottle virus. J. Mol. Biol. 2004;335:455–464. doi: 10.1016/j.jmb.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 21.Porterfield J.Z. Full-length HBV core protein packages viral and heterologous RNA with similar high cooperativity. J. Virol. 2010;84:7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell S., Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell S., Vogt V.M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y.X. Reversible binding of recombinant human immunodeficiency virus type 1 Gag protein to nucleic acids in virus-like particle assembly in vitro. J. Virol. 2002;76:11757–11762. doi: 10.1128/JVI.76.22.11757-11762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson M.C. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 2002;76:11177–11185. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crist R.M. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 2009;83:2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman J.L. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein Sci. 2004;13:2101–2107. doi: 10.1110/ps.04614804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor G.M. NMR relaxation studies of an RNA-binding segment of the rous sarcoma virus gag polyprotein in free and bound states: a model for autoinhibition of assembly. Biochemistry. 2010;49:4006–4017. doi: 10.1021/bi902196e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 2002;76:11729–11737. doi: 10.1128/JVI.76.22.11729-11737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright E.R. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briggs J.A. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valegard K. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J. Mol. Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 33.Beckett D. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J. Mol. Biol. 1988;204:939–947. doi: 10.1016/0022-2836(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 34.Carey J. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983;22:2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- 35.Stockley P.G. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T = 3 viral capsid. J. Mol. Biol. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basnak G. Viral genomic single-stranded RNA directs the pathway toward a T = 3 capsid. J. Mol. Biol. 2010;395:924–936. doi: 10.1016/j.jmb.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toropova K. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 38.Dykeman E.C. Dynamic allostery controls coat protein conformer switching during MS2 phage assembly. J. Mol. Biol. 2010;395:916–923. doi: 10.1016/j.jmb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 39.Moisant P. Exploring the paths of (virus) assembly. Biophys. J. 2010;99:1350–1357. doi: 10.1016/j.bpj.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Packianathan C. Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J. Virol. 2010;84:1607–1615. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourne C. Global structural changes in hepatitis B capsids induced by the assembly effector HAP1. J. Virol. 2006;80:11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freund S.M.V. Moving towards high-resolution descriptions of the molecular interactions and structural rearrangements of the human hepatitis B core protein. J. Mol. Biol. 2008;384:1301–1313. doi: 10.1016/j.jmb.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 43.Deres K. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 44.Bourne C. Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. J. Virol. 2008;82:10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katen S.P. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem. Biol. 2010 doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feld J.J. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res. 2007;76:168–177. doi: 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 47.Tsang S.K. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J. Mol. Biol. 2000;296:335–340. doi: 10.1006/jmbi.1999.3483. [DOI] [PubMed] [Google Scholar]
- 48.Speelman B. Molecular dynamics simulations of human rhinovirus and an antiviral compound. Biophys. J. 2001;80:121–129. doi: 10.1016/S0006-3495(01)75999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartonova V. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J. Biol. Chem. 2008;283:32024–32033. doi: 10.1074/jbc.M804230200. [DOI] [PubMed] [Google Scholar]
- 50.Zlotnick A. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J. Virol. 2002;76:4848–4854. doi: 10.1128/JVI.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukhopadhyay S. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kielian M., Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez-San Martin C. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 2009;17:514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aliperti G., Schlesinger M.J. Evidence for an autoprotease activity of sindbis virus capsid protein. Virology. 1978;90:366–369. doi: 10.1016/0042-6822(78)90321-5. [DOI] [PubMed] [Google Scholar]
- 55.Garoff H. Isolation and characterization of the membrane proteins of Semliki Forest virus. Virology. 1974;61:493–504. doi: 10.1016/0042-6822(74)90285-2. [DOI] [PubMed] [Google Scholar]
- 56.Carleton M. Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J. Virol. 1997;71:1558–1566. doi: 10.1128/jvi.71.2.1558-1566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molinari M., Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 58.Woycechowsky K.J., Raines R.T. The CXC motif: a functional mimic of protein disulfide isomerase. Biochemistry. 2003;42:5387–5394. doi: 10.1021/bi026993q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parrott M.M. Role of conserved cysteines in the alphavirus E3 protein. J. Virol. 2009;83:2584–2591. doi: 10.1128/JVI.02158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Firth A.E. Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol. J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White J.M. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X. Furin processing and proteolytic activation of Semliki Forest virus. J. Virol. 2003;77:2981–2989. doi: 10.1128/JVI.77.5.2981-2989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Bonsdorff C.H., Harrison S.C. Hexagonal glycoprotein arrays from Sindbis virus membranes. J. Virol. 1978;28:578–583. doi: 10.1128/jvi.28.2.578-583.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown E.L., Lyles D.S. Pseudotypes of vesicular stomatitis virus with CD4 formed by clustering of membrane microdomains during budding. J. Virol. 2005;79:7077–7086. doi: 10.1128/JVI.79.11.7077-7086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hammarstedt M. Minimal exclusion of plasma membrane proteins during retroviral envelope formation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7527–7532. doi: 10.1073/pnas.120051597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nayak D.P. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mukhopadhyay S. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 69.Steinhauer D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 70.Roche S., Gaudin Y. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 2002;297:128–135. doi: 10.1006/viro.2002.1429. [DOI] [PubMed] [Google Scholar]
- 71.Bancroft J.B. The self-assembly of spherical plant viruses. Adv. Virus Res. 1970;16:99–134. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- 72.Bancroft J.B., Hiebert E. Formation of an infectious nucleoprotein from protein and nucleic acid isolated from a small spherical virus. Virology. 1967;32:354–356. doi: 10.1016/0042-6822(67)90284-x. [DOI] [PubMed] [Google Scholar]
- 73.Savithri H.S., Erickson J.W. The self-assembly of the cowpea strain of southern bean mosaic virus: formation of T = 1 and T = 3 nucleoprotein particles. Virology. 1983;126:328–335. doi: 10.1016/0042-6822(83)90482-8. [DOI] [PubMed] [Google Scholar]
- 74.Lokesh G.L. A molecular switch in the capsid protein controls the particle polymorphism in an icosahedral virus. Virology. 2002;292:211–223. doi: 10.1006/viro.2001.1242. [DOI] [PubMed] [Google Scholar]
- 75.Sastri M. Identification of a discrete intermediate in the assembly/disassembly of physalis mottle tymovirus through mutational analysis. J. Mol. Biol. 1999;289:905–918. doi: 10.1006/jmbi.1999.2786. [DOI] [PubMed] [Google Scholar]
- 76.Newcomb W.W. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tellinghuisen T.L. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mukhopadhyay S. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 2002;76:11128–11132. doi: 10.1128/JVI.76.21.11128-11132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kirnbauer R. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salunke D.M. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 81.Colomar M.C. Opening and refolding of simian virus 40 and in vitro packaging of foreign DNA. J. Virol. 1993;67:2779–2786. doi: 10.1128/jvi.67.5.2779-2786.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rombaut B. In vitro assembly of poliovirus empty capsids: antigenic consequences and immunological assay of the morphopoietic factor. Virology. 1984;135:546–550. doi: 10.1016/0042-6822(84)90209-5. [DOI] [PubMed] [Google Scholar]
- 83.Goodwin S. Foot-and-mouth disease virus assembly: processing of recombinant capsid precursor by exogenous protease induces self-assembly of pentamers in vitro in a myristoylation-dependent manner. J. Virol. 2009;83:11275–11282. doi: 10.1128/JVI.01263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li T.C. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 2005;79:12999–13006. doi: 10.1128/JVI.79.20.12999-13006.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaussier H. Building a virus from scratch: assembly of an infectious virus using purified components in a rigorously defined biochemical assay system. J. Mol. Biol. 2006;357:1154–1166. doi: 10.1016/j.jmb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 86.Dryden K.A. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol. Cell. 2006;22:843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 87.Huiskonen J.T. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 2010;84:4889–4897. doi: 10.1128/JVI.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grunewald K. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 89.Forster F. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang W. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Freiberg A.N. Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J. Virol. 2008;82:10341–10348. doi: 10.1128/JVI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ge, P. et al. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327, 689–693 [DOI] [PMC free article] [PubMed]
- 93.Beniac D.R. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cyrklaff M. Cryo-electron tomography of vaccinia virus. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harris A. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Welsch, S. et al. Electron tomography reveals the steps in filovirus budding. PLoS Pathog. 6, e1000875 [DOI] [PMC free article] [PubMed]
- 97.Loney C. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of sendai virus. J. Virol. 2009;83:8191–8197. doi: 10.1128/JVI.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rolfsson O. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J. Mol. Biol. 2010;401:309–322. doi: 10.1016/j.jmb.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang W. Placement of the structural proteins in Sindbis virus. J. Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 101.Li L. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Voss J.E. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]


