Abstract
The biochemistry, physiology and behavior of nearly all organisms are influenced by an inherent circadian (24 hr) clock timing mechanism. For mammals, the linchpin of this biological timing process is located in the suprachiasmatic nuclei (SCN) of the hypothalamus. One key feature of the SCN clock is that it is tightly entrained to lighting cues, thus ensuring that the clock is synchronized to the ever-changing seasonal light cycle. Within the field of circadian biology, there has been intense interest in understanding the intracellular signaling events that drive this process. To this end, our recent studies have revealed a role for an evolutionarily conserved translational control kinase, the mammalian target of rapamycin (mTOR), in the SCN clock entrainment process. Here we provide an overview of mechanisms of inducible mTOR activation in the SCN, and describe the effects of mTOR on clock protein synthesis and behavioral rhythmicity. Given that dysregulation of SCN timing has been associated with an array of clinical conditions (e.g., hypertension, obesity, diabetes, depression), new insights into the molecular mechanisms that regulate clock timing may provide new therapeutic treatments for circadian rhythm-associated disorders.
Keywords: Circadian clock, Entrainment, Light, Suprachiasmatic nuclei, mTOR, Rapamycin
General introduction of circadian rhythm
Virtually every aspect of human physiology and behavior is influenced by an inherent circadian (24 hr) clock timing mechanism (1). Within mammals, the master circadian clock is located in the suprachiasmatic nuclei (SCN) of the hypothalamus, a relatively small brain region consisting of ~ 15,000 neurons. Through both synaptic and humoral processes, the SCN communicates internal time to all organ systems of the body, allowing for precise phasing between the SCN clock and peripheral, organ-based, ‘slave’ oscillators. Central to the SCN clock is a cell-autonomous interlocked transcription/translation feedback loop that is centered on the rhythmic expression of the Period (Per) and Cryptochrome (Cry) families of genes. Briefly, a heterodimeric transcription factor formed by CLOCK and BMAL1 binds to an E-Box motif, driving the expression of Pers and Crys. As PERIOD (PER) protein levels increase, they form complexes with CRYPTOCHROMEs (CRYs), translocate to the cell nucleus and repress CLOCK- and BMAL1-mediated transcription. CKIα/CKIδ-mediated phosphorylation targets PER for ubiquitin mediated degradation, thus de-repressing CLOCK and BMAL1, and, in turn, allowing for a new round of Per- and Cry-mediated gene expression to occur (2). Disruption of this 24 hr transcription/translation feedback loop renders animals arrhythmic.
One key feature of the SCN clock is that the phasing of this transcriptional rhythm is tightly regulated by photic input from the retina. Light signaling received by the retina is transduced to the SCN and evokes a series of intracellular signal transduction cascades that ultimately drive resetting of the clock via a process which is thought to be largely dependent on the rapid transcription and translation of the clock proteins PER1 and PER2. Altered PER levels appear to trigger resetting by breaking the dynamic balance of the negative feedback loop (1). Of note, the intracellular signal transduction events that couple light to clock gene transcription and mRNA translation has not been well described.
One key signaling pathway underlying clock entrainment is the p42/44 Mitogen Activated Protein Kinase (MAPK) pathway (3). Along these lines, a brief light entrainment cue triggers robust activation of the MAPK cascade in the SCN (4) and disruption of light-induced MAPK activation by the MEK inhibitors U0126 or SL-327 significantly attenuates light-induced clock entrainment (5). MAPK functions through activation of multiple downstream effectors. For example, photic stimulation has been shown to trigger MAPK-dependent activation of p90 Ribosomal S6 Kinase (RSK) and Mitogen- and Stress-activated protein Kinase 1 (MSK1) (6,7). These light-activated kinases have been shown to stimulate phosphorylation of CREB (cAMP response element-binding protein) at Ser-133, and in turn, activation of cAMP Response Element (CRE)-mediated gene expression (8,9). Importantly, the promoters of many clock entrainment-related genes contain CREs, such as the murine forms of Per1 and Per2. These data provide one route by which MAPK affects clock entrainment. However, our recent work has revealed mammalian Target Of Rapamycin (mTOR) as a second route by which the MAPK pathway influences the clock.
mTOR signaling
mTOR is a serine/threonine protein kinase that can be bound and inhibited by the antifungal metabolite rapamycin, which is produced by a bacterial strain originally found in soil from Easter Island (locally known as Rapa Nui)(10). The most well recognized role of the mTOR pathway is to coordinate the metabolic activity of a cell with changes in cellular energy and stress levels. It executes its function by forming two distinct multi-protein complexes: the rapamycin-sensitive mTOR Complex 1 (mTORC1), which contains Raptor, and the rapamycin-insensitive mTORC2, which contains Rictor (11). mTOR functioning within mTORC1 regulates translational control while mTOR functioning within mTORC2 controls cytoskeleton organization and, in turn, regulates spatial aspects of cell growth (10,11).
mTORC1-regulation of mRNA processivity occurs through two distinct signal transduction effectors: S6 kinase1 (S6K1, including two isoforms, p70 S6K and p85 S6K) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) (12). Both S6K1 and 4E-BP1 bind to the mTORC1 component raptor and are subject to phosphorylation at multiple sites. Asubset of phosphorylation sites regulate enzymatic activity, and thus, phosphorylation site-specific antibodies can be used to monitor the activation states of the two arms of mTOR pathway. Along these lines, S6K1 regulates the phosphorylation-dependent activation of a number of proteins involved in regulation of mRNA translation and processing, including ribosomal protein S6 (phosphorylated at Ser240/244), eIF4B (phosphorylated at Ser322), eEF2K (phosphorylated at Ser366) and Pdcd4 (phosphorylated at Ser67) (12). In the absence of stimulation, 4E-BP1 binds to eIF4E, thereby blocking its ability to stimulate CAP-dependent mRNA translation initiation (13). Upon stimulation, mTORC1 phosphorylates 4E-BP1 at Thr-37 and Thr-46, which inhibits its association with eIF4E, thereby allowing CAP-dependent mRNA translation to occur (14,15).
Although there has been a large number of studies exploring the biochemical and physiological effects of mTOR as it relates to cell growth and metabolism, relatively little work has examined mTOR’s functionality within the Central Nervous System (CNS). Key findings thus far have revealed roles for mTOR in developmental processes such as neuronal survival and differentiation, axon growth and navigation, dendritic arborization, and synaptogenesis (16,17). In the adult CNS, mTOR has been shown to influence axonal regeneration, as well as hippocampal plasticity (18). Further, within the hypothalamus, mTOR functions as an metabolic sensor to control food intake and regulate energy balance (19). Our work has revealed the presence of Light-actuated MAPK/mTOR signaling cassette in the SCN circadian clock (20).
mTOR signaling and the SCN clock
Robust mTOR expression in the SCN
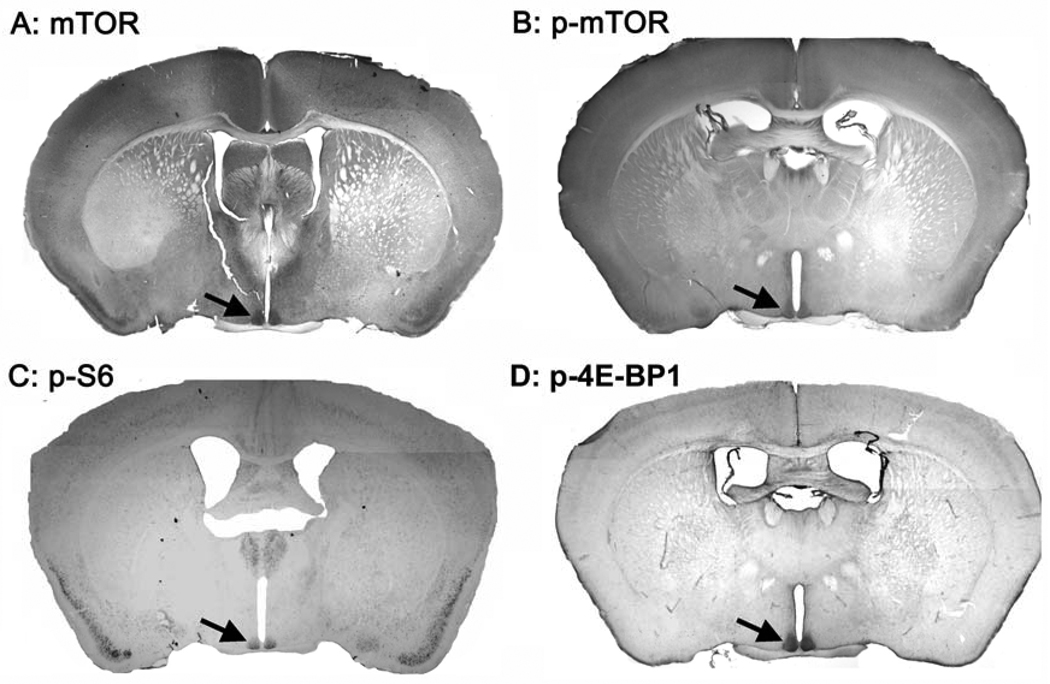
An initial examination using immunohistochemical labeling revealed robust mTOR expression in the SCN. The high level of expression is in striking contrast to surrounding hypothalamic regions, where little mTOR or p-mTOR is expressed (Figure 1A and B), and thus, raised the possibility of an important functional role for mTOR in the clock. To this end, we tested the phospho-activation state of the two main downstream effectors of mTOR, 4E-BP1 (phosphorylated at Thr-37/46) and S6 ribosomal protein (phosphorylated at Ser-240/244), which is a direct downstream target of S6K1 and can only be phosphorylated at these two sites by S6K1. Immunolabeling of brain tissue revealed expression of the phospho-activated form of 4E-BP1 throughout the entirety of the SCN (Figure 1D). Likewise, phospho-S6 (p-S6) was also detected in the SCN (Figure 1C). Together, these results support the idea that the SCN exhibits tonically high levels of mTOR activity, and thus raised the possibility that the mTOR pathway plays important roles in the SCN physiology.
Figure 1.
Representative low magnification mouse brain coronal sections immunohistochemically labeled for the expression of mTOR (A), p-mTOR (B), p-S6 ribosomal protein (C) and p-4E-BP1(D). The signal was visualized using nickel-enhanced diaminobenzidine chromogen. Arrow denotes the location of the SCN.
Light-evoked mTOR activation in the SCN
To investigate the role of mTOR signaling in the clock resetting process, mice were transferred from a standard 12 hr light/12 hr dark cycle to constant darkness, and then exposed to brief light pulses (15 min, 400 lux) at different times of the circadian cycle. This protocol is designed to monitored light-evoked changes in SCN physiology. We found that light during either the early or late night triggered robust phosphorylation of S6K1, and its downstream target S6 in the SCN. Light-induced S6K1 expression was highest in the central SCN (corresponding to the SCN region with the strongest axonal input from the retina), with limited expression in the rostral and caudal regions of the SCN. In contrast to the nighttime, S6K1 activity was not stimulated by light treatment during the middle of the subjective day, thus revealing that activation of the mTOR pathway is phase-restricted to the nighttime. This phase-restricted light response phenomenon has been reported for a number of kinase pathways and immediate early genes and is likely to be the molecular mechanism underlying the night time domain-specific effects of light on the clock (21). As with the S6K1 arm of the mTOR pathway, the phosphorylation state of 4E-BP1 (Thr-37 and Thr-46) (14,15) was also significantly increased by light.
To test both the specificity of the light-evoked kinase-activation data described thus far and assess the functional effects of mTOR signaling, we developed an intraventricular infusion approach to deliver rapamycin to the SCN. For these experiments, a cannula was placed in the lateral ventricle and fixed to the skull with dental cement. The infusion of rapamycin (2 µl, 100 µM) led to a potent, yet transient (up to 2 h), suppression of both basal and light-induced S6K1, S6 and 4E-BP1 phosphorylation in the SCN, thus supporting the specificity of our light-evoked phosphokinase data.
The MAPK pathway mediates mTOR activation by light
One central question emerging from these studies related to the upstream signaling pathways which couple light to mTOR activation in the SCN. Two candidates merited thorough investigation: the PI3K/AKT pathway, which has been implicated in growth factor-induced mTOR activation, and the MAPK pathway, which has been shown to affect mTOR via multiple mechanisms (11). Our initial analysis revealed that neither PI3K nor AKT was light responsive in the SCN, thus suggesting that PI3K/AKT did not contribute to the light-evoked increase in mTOR activity. Conversely, MAPK exhibits rapid and robust activation by a brief light pulse, thus raising the possibility that this pathway couples light to mTOR. As an initial assessment of the potential role of MAPK signaling in light-induced mTOR activity, SCN sections were double-labeled for activated ERK and S6K1. These assays detected a marked light-evoked temporal and cellular correlation between MAPK pathway activity and S6K1 phosphorylation. Furthermore, ventricular infusion of the MEK1/2 inhibitor U0126 potently repressed mTOR-dependent S6K1 activity and S6 phosphorylation, thus indicating that the MAPK cascade is an essential upstream activator of mTOR in the SCN. Mechanistically, theMAPK pathway could stimulate mTOR activity by several routes: First, ERK can trigger phosphorylation-dependent inactivation of TSC2 GAP activity (22). Second, ERK-evoked RSK1 signaling has been shown to block TSC2 activity (23), and thereby stimulate mTOR1 signaling. Interestingly, we have reported that RSK1 is activated by light in the SCN (6).
Rapamycin modulates the circadian behavioral responses to light
To evaluate the functional significance of mTOR in light-evoked clock entrainment, we infused rapamycin (as described above) and monitored the effects on clock-regulated behavioral (locomotor activity) and physiological (core body temperature) processes. The fact that mTOR signaling is activated by both early night light (a time point when light triggers a phase delay of the clock) and late night light (a time point when light triggers a phase advance of the clock) led us to investigate the potential role of this pathway at both time points. Infusion of rapamycin 30 min before light exposure resulted in striking time-of-night-specific effects on clock entrainment. Thus, disruption of mTOR during the early night led to a significant attenuation (~50 min or 36%) of the early night phase-delaying effects of light, whereas late night disruption of mTOR signaling led to a significant ~2.2-fold lengthening of the light-induced phase advance. Of note, in the absence of light, infusion of rapamycin did not significantly affect the free-running clock rhythm or clock phase, indicating that transient suppression of basal mTOR activity does not significantly alter SCN pacemaker activity. Together, these data raised the possibility that light-evoked mTOR-dependent protein translation regulates clock entrainment.
mTOR facilitates light-induced clock protein expression
To provide a molecular and cellular context for the behavioral effects of rapamycin on clock entrainment, we analyzed the effects of rapamycin on light-induced PER1 and PER2 protein expression. As noted, both of these clock genes are induced by light, and appear to play a central role in the clock entrainment (24,25). The infusion of rapamycin at both early- and late-night time points led to a significant reduction of light-evoked PER1 and PER2 expression. Of note, rapamycin infusion in the absence of light treatment did not alter basal PER1 and PER2 expression, indicating that light-evoked mTOR signaling is required to augment PER protein expression. The precise mechanism by which mTOR regulates PER expressions has yet to be addressed. It is likely that a light-evoked dissociation of 4E-BP1 from eIF4E would facilitate Per mRNA translation. In addition, our work showing that light triggers a rapid, mTOR-dependent, increase in the translation of 5’TOP mRNA eEF1A raises the possibility that mRNA processivity is also upregulated in the SCN via an increase in the expression of elongation factors. Future studies will be aimed at this question.
Future directions
The findings outlined above provide exciting new insights into the role of inducible translation control via mTOR in SCN clock entrainment and lay the foundation for future studies(20,26). Along these lines, one key unexplored question is whether mTOR signaling contributes to the autonomous SCN clock timing process. As noted above, the clock is driven bytranscriptional and translational feedback loop that appears to be influenced by a range of extracellular cues (27). This, coupled with data showing that the SCN exhibits high level of mTOR activity, raises the possibility that translational regulation via mTOR contributes to clock physiology. Another area of possible inquiry would be to examine the potential contribution of mTOR to peripheral clock entrainment. As noted, peripheral organ clocks maintain tight temporal coordination with the SCN clock, and desynchronization of these clocks from the SCN clock has been implicated in a number of disease states (28). Given the important role that mTOR plays in the SCN clock entrainment, it may be worthwhile to examine the potential contribution of mTOR to peripheral clock entrainment under physiological and pathophysiological conditions.A recent study in Drosophila found that TOR/S6K signaling regulates circadian period length (29).
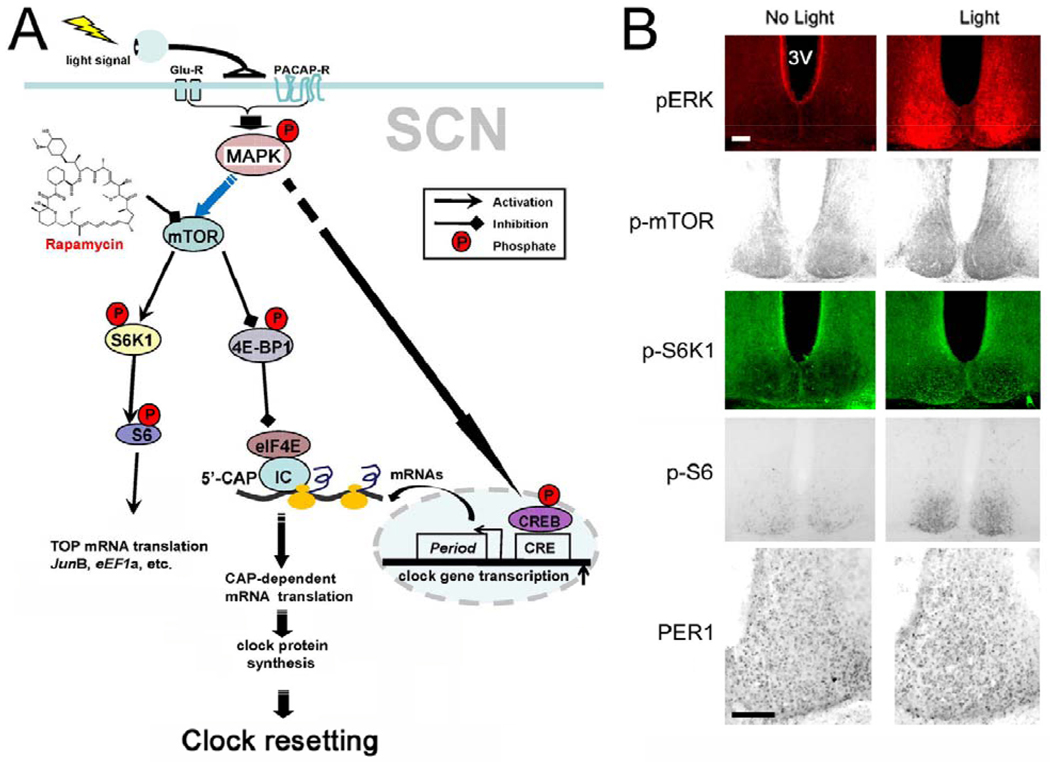
Figure 2.
(A) Schematic overview of the mTOR signaling pathway and SCN clock entrainment. Photic input from the retina drives the release of the excitatory amino acid glutamate and the neuropeptide PACAP. Postsynaptic receptors triggers activation of the MAPK cascade, which, in turn, stimulates mTOR signaling. mTOR, functioning within the mTORC1 complex, causes phosphorylation-dependent activation of the p70 S6K/S6 signaling cassette, which stimulates TOP mRNA translation. mTORC1 also triggers the dissociation of 4E-BP1 from eIF4E, which increases CAP-dependent translation. These two arms of the mTOR pathway work in conjunction to enhance the rate of mRNA processivity. In addition to stimulating mTOR, the MAPK cascade also stimulates the expression of immediate early genes, including Period clock genes. Coordinate upregulation of gene transcription and mRNA translation leads to a robust upregulation of PERIOD protein expression. As a state variable of the clock, the induction of PERIOD leads to a rapid resetting of the molecular oscillator. (B) Representative immunohistochemical and immunofluorescence labeling of SCN tissue for light-induced changes in p-ERK, p-mTOR, p-S6K1, p-S6 and PER1. To better visualize PER1 expression, only one SCN is presented. Scale bar: 100 microns, 3V: third ventricle.
Acknowledgements
This work was supported by an National Science Foundation Grant IBN-0090974, National Institutes of Health Grants MH62335 and NS067409, and Ohio State Neuroscience Center Core Grant 5P30NS045758.
Footnotes
Conflicts of Interest
No potential conflicts of interest to disclose.
References
- 1.Reppert S, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Ko C, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 3.Coogan A, Piggins HD. MAP kinases in the mammalian circadian system—key regulators of clock function. J Neurochem. 2004;90:769–775. doi: 10.1111/j.1471-4159.2004.02554.x. [DOI] [PubMed] [Google Scholar]
- 4.Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 5.Butcher G, Dziema H, Collamore M, Burgoon PW, Obrietan K. The p42/44 mitogenactivated protein kinase pathway couples photic input to circadian clock entrainment. J BiolChem. 2002;277:29519–29525. doi: 10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- 6.Butcher G, Lee B, Hsieh F, Obrietan K. Light- and clock-dependent regulation of ribosomal S6 kinase activity in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19:907–915. doi: 10.1111/j.0953-816x.2004.03155.x. [DOI] [PubMed] [Google Scholar]
- 7.Butcher G, Lee B, Cheng HY, Obrietan K. Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism. J Neurosci. 2005;25:5305–5313. doi: 10.1523/JNEUROSCI.4361-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 9.Vermeulen LBW, Beck IM, De Bosscher K, Haegeman G. The versatile role of MSKs in transcriptional regulation. Trends BiochemSci. 2009;34:311–318. doi: 10.1016/j.tibs.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 12.Proud C. mTORC1 signalling and mRNA translation. Biochem Soc Trans. 2009;37:227–231. doi: 10.1042/BST0370227. [DOI] [PubMed] [Google Scholar]
- 13.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pause A, Belsham GJ, Gingras AC, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 15.Gingras A, Gygi SP, Raught B, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao R, Li A, Cho HY. mTOR signaling in epileptogenesis: too much of a good thing? J Neurosci. 2009;29:12372–12373. doi: 10.1523/JNEUROSCI.3486-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swiech L, Perycz M, Malik A, Jaworski J. Role of mTOR in physiology and pathology of the nervous system. BiochimBiophysActa. 2008;1784:116–132. doi: 10.1016/j.bbapap.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of longlasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cota D, Proulx K, Smith KA, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Mol Cell Neurosci. 2008;38:312–324. doi: 10.1016/j.mcn.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meijer J, Schwartz WJ. In search of the pathways for light-induced pacemaker resetting in the suprachiasmatic nucleus. J Biol Rhythms. 2003;18:235–249. doi: 10.1177/0748730403018003006. [DOI] [PubMed] [Google Scholar]
- 22.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 23.Roux P, Shahbazian D, Vu H, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J BiolChem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiyama M, Kouzu Y, Takahashi S, et al. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor andsuprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. MPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 26.Cao R, Li A, Cho HY, Lee B, Obrietan K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J Neurosci. 2010;30:6302–6314. doi: 10.1523/JNEUROSCI.5482-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianchi MM. Collective behavior in gene regulation: metabolic clocks and cross-talking. FEBS J. 2008;275:2356–2363. doi: 10.1111/j.1742-4658.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- 28.Maywood ES, O'Neill J, Wong GK, Reddy AB, Hastings MH. Circadian timing in health and disease. Prog Brain Res. 2006;153:253–269. doi: 10.1016/S0079-6123(06)53015-8. [DOI] [PubMed] [Google Scholar]
- 29.Zheng X, Sehgal A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol. 2010;20:1203–1208. doi: 10.1016/j.cub.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]