SUMMARY
The heterogeneity and instability of human tumors hamper straightforward identification of cancer-causing mutations through genomic approaches alone. Herein we describe a mouse model of liver cancer initiated from progenitor cells harboring defined cancer-predisposing lesions. Genome-wide analyses of tumors in this mouse model and in human hepatocellular carcinomas revealed a recurrent amplification at mouse chromosome 9qA1, the syntenic region of human chromosome 11q22. Gene-expression analyses delineated cIAP1, a known inhibitor of apoptosis, and Yap, a transcription factor, as candidate oncogenes in the amplicon. In the genetic context of their amplification, both cIAP1 and Yap accelerated tumorigenesis and were required to sustain rapid growth of amplicon-containing tumors. Furthermore, cIAP1 and Yap cooperated to promote tumorigenesis. Our results establish a tractable model of liver cancer, identify two oncogenes that cooperate by virtue of their coamplification in the same genomic locus, and suggest an efficient strategy for the annotation of human cancer genes.
INTRODUCTION
Tumorigenesis results from a progressive sequence of genetic and epigenetic alterations that promote the malignant transformation of the cell by disrupting key processes involved in normal growth control and tissue homeostasis (Hanahan and Weinberg, 2000). Since complex signaling networks control these processes, mutations in many genes can provide the cell with a specific aberrant capability. Consequently, the combination of genetic alterations that can occur during tumor evolution is enormous, perhaps underpinning the substantial heterogeneity in tumor behavior that occurs even within a particular tumor type. In addition, genomic instability is a common, if not universal, feature of advanced tumors. This instability provides tumor cells with the ability to adapt to new environments but may also increase the rate of bystander mutations that do not contribute to the malignant phenotype.
The completion of the human genome project has enabled new approaches for studying cancer genetics and cancer genomes. For example, gene-expression profiling using microarrays has improved the classification of some tumor types (Segal et al., 2005). Moreover, DNA resequencing has identified unanticipated mutations in onco-genes such as BRAF and EGFR, thus suggesting new drug targets or therapeutic strategies (Davies et al., 2002; Lynch et al., 2004). Finally, genome scanning for gene copy-number alterations has identified many loci harboring candidate cancer genes (Kallioniemi et al., 1993; Lucito et al., 2003). Because of these advances, efforts to catalog all of the mutational events that contribute to human cancer can now be envisioned. Nevertheless, for such information to be efficiently translated into improvements in cancer diagnosis and therapy, cancer-causing mutations must be distinguished from irrelevant alterations linked to complex cancer genotypes. Furthermore, without in vivo validation, there is little stimulus for therapeutic development efforts. Integrative strategies to identify and validate genes with functional relevance for tumor initiation and progression are clearly needed.
Hepatocellular carcinoma (HCC) represents a tumor type where a more complete understanding of the underlying genetics could have a major impact on treatment of the disease. HCC is the fifth most frequent neoplasm worldwide but, owing to the lack of effective treatment options, represents the third leading cause of cancer death (Parkin et al., 2001). The only curative treatments for HCC are surgical resection or liver transplantation, but most patients present with advanced disease and are not candidates for surgery. To date, systemic chemotherapeutic treatment is ineffective against HCC, and no single drug or drug combination prolongs survival (Llovet et al., 2003).
The development of HCC is invariably associated with liver damage caused by chronic hepatitis, extensive alcohol intake, or toxins, sequentially resulting in liver cirrhosis, dysplastic lesions, and finally invasive liver carcinoma. Recent studies suggest that these agents can target liver progenitor cells (“oval cells” in rodents and “hepatic progenitor cells” in humans), leading to their expansion and transformation (for review, see Alison and Lovell, 2005). One key target in liver carcinogenesis is p53, which is functionally attenuated by hepatitis B virus X protein (Wang et al., 1994) and is a mutational target of aflatoxin B1 (Aguilar et al., 1994). Other established lesions in liver cancer include activation of the c-myc, CCND1 (cyclin D1), or c-met oncogenes, as well as mutations in components of the Ras/PI3 kinase pathways (Lee et al., 2005; Feitelson et al., 2002; Suzuki et al., 1994). Still, the molecular genetics of liver cancer, and how specific lesions interact to produce its aggressive characteristics, remain poorly understood.
Mouse models of human cancer provide powerful tools to investigate cancer biology, genetics, and therapy. Here we used a flexible mouse model of hepatocellular carcinoma to search for spontaneous mutations arising in tumors initiated by different oncogenic lesions and then compared these to alterations observed in human cancers. This approach enabled us to pinpoint “driver genes” that might contribute to human liver carcinogenesis and, using our model, to validate these changes in an appropriate in vivo context. Consequently, our study not only identified two oncogenes in the same focal amplification that cooperate during tumorigenesis but, more broadly, highlights the utility of integrating mouse models and cancer genomics for the functional annotation of cancer genes.
RESULTS
Generation and Transplantation of Genetically Altered Liver Progenitor Cells
Most mouse models for liver cancer are based on germline transgenic approaches that direct expression of an onco-gene to the liver using a tissue-specific promoter (Sandgren et al., 1989; Murakami et al., 1993). Although these models continue to provide important insights into the pathogenesis of liver cancer, they express the oncogene throughout the entire liver, a situation that is distinct from spontaneous tumorigenesis. Moreover, incorporation of additional lesions, such as a second oncogene or loss of a tumor suppressor, requires genetic crosses that are time consuming and expensive. Finally, traditional transgenic and knockout strategies do not specifically target liver progenitor cells, one proposed “cell of origin” of the disease (Alison and Lovell, 2005).
Based on our previous work in the hematopoietic system (e.g., Schmitt et al., 2002), we reasoned that the study of genetic interactions in liver tumorigenesis would be facilitated by ex vivo genetic manipulation of liver progenitors followed by their retransplantation into the livers of recipient mice (Figure 1A). Embryonic hepatoblasts express high E-cadherin levels, enabling these cells to be isolated to high purity from E12.5-15 fetal livers using magnetic bead selection (Nitou et al., 2002). These cells also expressed markers characteristic of bipotential oval cells (see Figure S1A in the Supplemental Data available with this article online), one presumed target of transformation in the adult liver (Alison and Lovell, 2005). Although they proliferated poorly in initial experiments, the introduction of defined medium (Block et al., 1996), feeder layers, and gelatin-coated plates to the culture conditions enabled the hepatoblasts to be expanded without loss of their defining characteristics (data not shown). These conditions also allowed efficient gene transfer using MSCV-based retroviral vectors expressing green fluorescent protein (GFP) (Figure S1B) or short-hairpin RNAs (shRNAs) capable of suppressing gene expression through RNA interference (Zender et al., 2006).
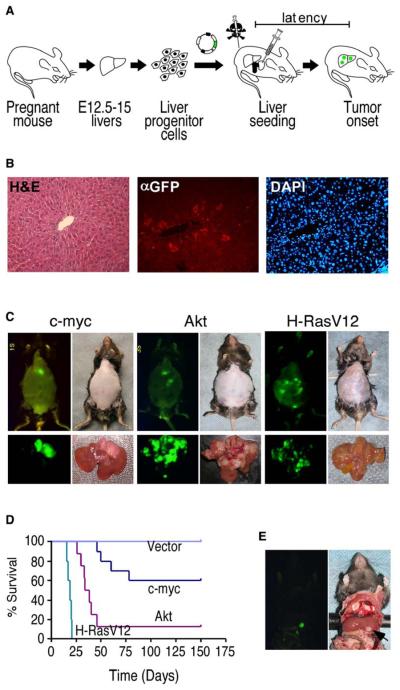
Figure 1. Development and Characterization of a Genetically Tractable, Transplantable Mouse Model of HCC.
(A) Schematic diagram showing the generation of in situ liver carcinomas following retroviral transduction of purified E-cadherin+ hepato-blasts (see Figure S1).
(B) Analysis of liver sections from mice seeded with GFP-expressing hepatoblasts 1 week postreconstitution. Left, H&E; middle, anti-GFP immunofluorescence; right, DAPI.
(C) External GFP tumor imaging (top panels) or direct imaging of the respective explanted tumor-bearing livers (bottom panels) of mice reconstituted with p53−/− hepatoblasts transduced with the indicated oncogene.
(D) Survival curves of mice after intrahepatic seeding of p53−/− liver progenitor cells transduced with the indicated oncogene or control vector.
(E) Explanted murine liver carcinomas (p53−/−; myc) were grown briefly in culture and then directly injected into the left liver lobe. Shown is a GFP-expressing (left) in situ tumor (right) 42 days postinjection.
To determine whether genetically modified hepato-blasts could colonize recipient livers, we used a protocol that optimizes engraftment of transplanted cells (Guo et al., 2002). Animals were pretreated with retrorsine, an alkaloid that exerts a strong and persistent block of native hepatocyte proliferation and increases the competitive advantage of transplanted cells (Laconi et al., 1998). Ten days after the last retrorsine treatment, 2 3 106 GFP-tagged E-cadherin+ liver progenitor cells were delivered to the liver by intrasplenic injection. One week later, immunohistochemical analysis of liver sections revealed that approximately one percent of the host liver consisted of “seeded” GFP-positive cells that were embedded within the normal liver architecture (Figure 1B).
Generation of Liver Carcinomas from Transplanted Liver Progenitor Cells that Resemble Human HCC
We next tested whether this approach could produce liver carcinomas in situ. Owing to the importance of p53 inactivation in this disease, we isolated hepatoblasts from p53−/− fetal livers and transduced these cells with retroviruses co-expressing myc (c-myc), activated Akt (Akt1), or oncogenic Ras (H-RasV12) (each of which affects signaling pathways altered in human liver cancer) and GFP. As above, these transduced cell populations were transplanted into retrorsine-treated mice. To further facilitate expansion of the transplanted cells, recipient mice were treated with CCl4 (Guo et al., 2002) and monitored for signs of disease by abdominal palpation and whole-body fluorescence imaging. Although p53−/− hepatoblasts were not tumorigenic during the analysis period, each of the cell populations that also expressed an oncogene eventually produced GFP-positive tumors in the livers of recipient mice (Figure 1C, top).
Gross pathological analysis of explanted livers revealed that Myc-expressing tumors differ significantly from those expressing Akt or Ras (Figure 1C, bottom). First, Myc-expressing tumors grow primarily as unilocular tumors, whereas Akt- and Ras-derived tumors show aggressive, multilocular, and infiltrative intrahepatic growth. Second, the intrinsic tumorigenicity of p53−/− liver progenitor cells expressing Myc was significantly lower than those expressing Akt or Ras (Figure 1D). Of note, p53 loss clearly contributed to tumorigenesis since tumors arising in mice reconstituted with p53+/− hepatoblasts showed further delayed tumor onset and loss of the wild-type p53 allele (Figure S1D). In most instances, GFP-positive cells derived from established tumors could be grown in culture and subsequently formed secondary tumors upon subcutaneous injection into immunocompromised mice (data not shown) or direct intrahepatic injection into syngeneic recipients (Figure 1E).
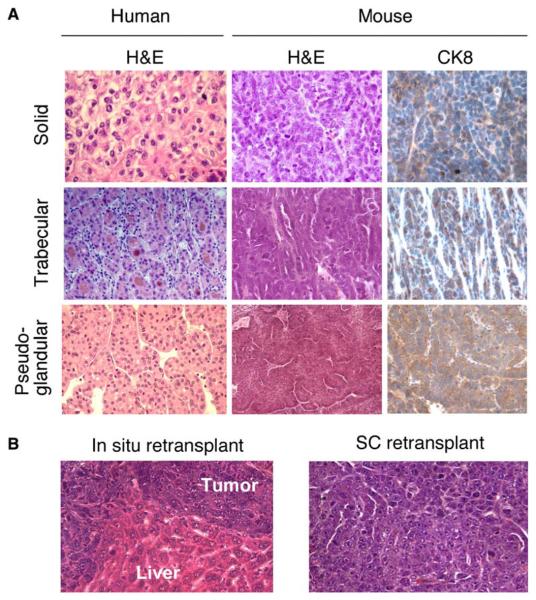
An experienced liver pathologist (P.F.) examined the hematoxylin and eosin (H&E) stained sections derived from Myc-induced murine hepatomas (Figure 2A) and classified most as moderately well to poorly differentiated HCCs with a mostly solid, sometimes mixed solid/trabecular growth pattern. A smaller proportion of tumors displayed growth patterns resembling trabecular or pseudo-glandular HCC. All tumors examined stained positive for cytokeratin 8, confirming their liver origin. However, despite their derivation from cytokeratin 19-positive liver progenitor cells, most HCCs lost this marker during tumor-igenesis (Figure S1C). The tumors also expressed high albumin levels and, similar to the situation in human HCC, about half were positive for α-fetoprotein. Most also expressed moderate levels of vimentin (Figure S1C), a marker linked to aggressive tumor behavior (Hu et al., 2004). Transplanted hepatomas retained their HCC histology when injected orthotopically into the liver or subcutaneously into immunocompromised mice (Figure 2B). Therefore, ex vivo-manipulated liver progenitor cells can produce tumors that recapitulate the histopathology of human HCC.
Figure 2. Murine Liver Carcinomas Derived from E-Cadherin+ Liver Progenitor Cells Histopathologically Resemble Human HCC.
(A) H&E-stained sections of human HCC with the indicated histopathological variegation (left panels) shown adjacent to corresponding histopathologies in murine liver cancers arising from p53−/−;myc hepatoblasts (center panels). Anti-CK8 immunohistochemistry of the murine tumors is shown at right.
(B) H&E staining of in situ and subcutaneously retransplanted HCCs derived from p53−/− ;myc hepatoblasts.
ROMA Identifies Spontaneous Mutations in a Subset of Murine Liver Carcinomas Including a Recurrent Amplicon at Chromosome 9qA1
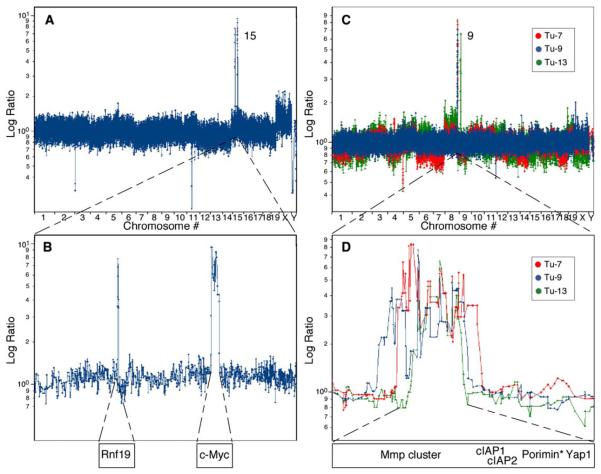
To further molecularly characterize the murine HCCs described above, we searched for spontaneously acquired lesions in these cancers using representational oligonucleotide microarray analysis (ROMA), a genome-wide scanning method capable of identifying copy-number alterations in tumor cells at high resolution (Lucito et al., 2003; B. Lakshmi, I.M. Hall, C. Egan, J. Alexander, J. Healy, L.Z., W.X., M.S.S., S.W.L., M.W., and R.L., unpublished data). Each human or mouse ROMA array consists of 85,000 oligonucleotide probes, allowing genome scanning at a theoretical resolution of ~35 kb. Although we did not detect focal genomic alterations (<5 Mb) in liver cancers induced by Akt, a number were detected in those initiated by Myc or Ras (Table S1). For example, a Ras-expressing tumor harbored two focal amplifications on chromosome 15 (Figure 3A), one containing Rnf19 and the other containing c-myc (Figure 3B). While Rnf19 has not been linked to tumorigenesis, c-myc alterations are common in human liver cancer (Peng et al., 1993), and myc cooperates with oncogenic Ras in transgenic models of HCC (Sandgren et al., 1989). These observations underscored the relevance of our model and suggested that further analyses would reveal other genes involved in human cancer.
Figure 3. ROMA Identifies Focal DNA Amplifications in Murine HCCs.
(A) ROMA profile of a tumor derived from p53−/−;Ras embryonic hepatoblasts. Data plotted are the normalized log ratio for each probe (*EST).
(B) Single-probe resolution of chromosome 15 corresponding to the tumor described in (A).
(C) Genome-wide profiles of three independent HCCs (Tu-7, Tu-9, and Tu-13) derived from p53−/−;myc embryonic hepatoblasts.
(D) Single-probe resolution of chromosome 9qA1 from the tumors described in (C). The minimal overlap region contains the indicated genes.
ROMA analysis of seven independent Myc-expressing HCCs identified a focal amplicon on mouse chromosome 9qA1 in four of these tumors (Figures 3C and 3D; Figure S2 and Table S1). The minimal overlapping region is approximately 1 Mb and contains genes encoding for several matrix metalloproteinases (MMPs), Yap, cIAP1 (Birc2), cIAP2(Birc3), and Porimin (Figure 3D; see Table S1 for breakpoints). Amplification was confirmed by genomic qPCR using a probe within the cIAP1 gene (data not shown). Interestingly, 9qA1 was never found amplified in Ras-or Akt-driven liver carcinomas as assessed by either genomic qPCR analysis or ROMA (n = 21; Table S1 and data not shown). These observations suggest that at least one of the genes in the 9qA1 region cooperates with myc and p53 loss to promote hepatocarcinogenesis.
Comparative Oncogenomics Reveals Lesions in Common between Murine and Human Cancers
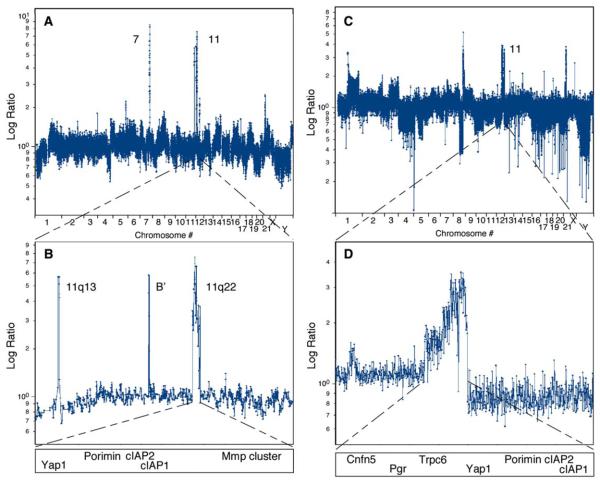
In parallel to our analysis of murine HCCs, we initially conducted ROMA on 25 human HCC samples. Although these tumors contained more alterations than their murine counterparts, using a strict cutoff of <5 Mb, we detected copy-number alterations affecting genes previously linked to HCC (Table S2). For example, three tumors had a chromosome 11 amplification containing CCND1, two had a chromosome 7 amplification containing c-met, two had a focal deletion on chromosome 10 containing the PTEN tumor suppressor, and one had a deletion of chromosome 9 harboring the CDKN2A (INK4a/ARF) locus.
We also detected a focal amplification on chromosome 11q22, a region that is syntenic to the murine 9qA1 locus. This tumor contained a c-met amplification (left peak) on chromosome 7 and three sharply delineated amplifications on chromosome 11 (Figure 4B), including CCND1,B′ (containing no known genes), and 11q22. A second HCC harboring the 11q22 amplicon was identified in a set of 23 additional human HCCs (Figures S2C and S2D), as well as in 4 of 53 human esophageal cancers (data not shown), indicating that it occurs in gastrointestinal malignancies derived from developmentally related organs. ROMA results were verified by genomic qPCR analysis using probes to the cIAP1 and cIAP2 loci (data not shown). Much like the chromosome 9 amplicon in murine HCCs, the boundaries of the 11q22 amplicon in human HCCs and esophageal cancers include genes encoding several matrix metalloproteinases, Porimin, Yap, cIAP1, and cIAP2.
Figure 4. ROMA Identifies Amplification of the Human Syntenic Region 11q22 in HCC and Ovarian Cancer.
(A) Genome-wide profile of a human HCC harboring an amplification on chromosome 7 containing the c-met gene and three regions amplified on chromosome 11.
(B) Single-probe resolution of chromosome 11, with genes contained within each amplicon depicted below.
(C) Genome-wide profile of an ovarian carcinoma containing the 11q22 amplification.
(D) Single-probe resolution of the 11q22 amplicon, with genes contained within the amplified region depicted below.
Cross-Species Expression Analysis of Genes from the Human and Mouse Amplicons Reveals Consistent Overexpression of cIAP1 and Yap
The human 11q22 amplicon is observed in other human cancers (e.g., Figures 4C and 4D), although no driver gene has been decisively identified (Imoto et al., 2001; Dai et al., 2003; Bashyam et al., 2005; Snijders et al., 2005). While it represents only one of many low-frequency events in these tumors and the HCCs evaluated here (Table S2), our cross-species comparison suggests that a gene (or genes) within this recurrent amplified region is crucial for tumorigenesis in some genetic contexts. An essential criterion for establishing whether an amplified gene might contribute to tumorigenesis is that it be overexpressed in the tumors where it is amplified; we further hypothesized this should hold across species. Therefore, we performed a comprehensive expression analysis of overlapping genes from the murine 9qA1 and human 11q22 amplicons.
First, messenger RNA levels for all genes in these regions were measured by real-time quantitative RT-PCR (Figures 5A and 5C; Table S3). All amplicon-positive mouse HCCs displayed elevated mRNA levels for the MMPs except MMP7 and had high variability in maximum expression levels (Table S3). In marked contrast, the mRNA levels for MMP1, MMP3, MMP8, MMP12, MMP13, MMP20, and MMP27 were below detection limit in 25 human HCCs, including a tumor with the 11q22 amplicon (Table S3). However, MMP7 and MMP10 mRNAs were moderately elevated in an 11q22-positive HCC. Therefore, with the possible exception of MMP10, the MMPs are not consistently overexpressed in amplicon-positive murine and human HCCs and probably are not responsible for the selective advantage conferred by this genomic amplification. Furthermore, ROMA analysis on various 11q22-positive human carcinomas identified an ovarian carcinoma harboring an 11q22 amplicon that decisively excluded all of the MMPs (Figures 4C and 4D). Concordantly, low-resolution technologies excluded at least some MMPs from an 11q22 amplification in lung cancer (Dai et al., 2003).
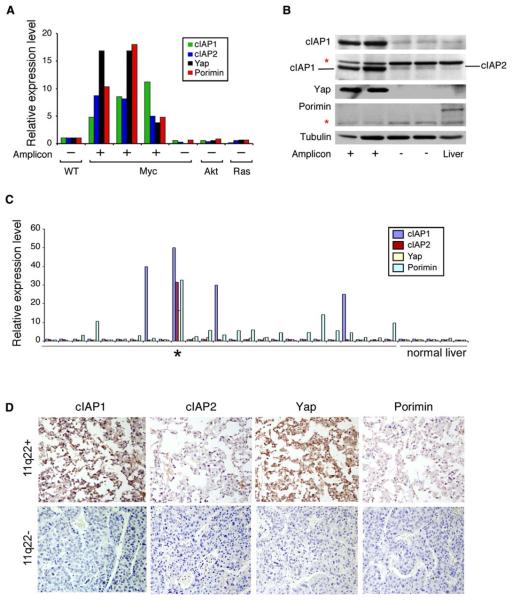
Figure 5. cIAP1 and Yap Are Consistently Overexpressed in Mouse and Human Tumors Containing the 9qA1 or 11q22 Amplicon.
(A) cIAP1, cIAP2, Yap, and Porimin mRNA levels in murine HCCs that contain the 9qA1 amplicon as determined by quantitative real-time RT-PCR analysis.
(B) Protein lysates from 9qA1-positive (+) or -negative (−) liver cancers and adult mouse liver were immunoblotted with antibodies against cIAP1, cIAP1/2, Yap, and Porimin. * denotes nonspecific bands. Tubulin was used as a loading control.
(C) Quantitative real-time RT-PCR analysis of cIAP1, cIAP2, Yap, and Porimin expression in human HCCs. * denotes a tumor with the 11q22 amplification. Cutoff for increased expression was >2-fold of the expression level in nonneoplastic liver (normal liver).
(D) Immunohistochemistry of an 11q22-positive (top) and -negative (bottom) human HCC using antibodies against the indicated protein.
In contrast, cIAP1 and Yap mRNA and protein were elevated in all mouse and human amplicon-containing tumors examined (Figures 5A-5D). Both mRNAs were also found to be overexpressed in some 11q22-positive oral carcinomas (Snijders et al., 2005). Although Porimin and cIAP2 mRNAs were elevated in all amplicon-containing tumors examined, we could not detect overexpression of either protein in 9qA1-positive mouse tumors (Figure 5B) or an 11q22-positive human tumor (Figure 5D). These observations may be explained by reports that many cells express Porimin mRNA without detectable protein (Ma et al., 2001) and that cIAP1 promotes the ubiquitylation and degradation of cIAP2 (Conze et al., 2005; Silke et al., 2005). In fact, we observed that cIAP1 promoted the turnover of cIAP2 in a dose-dependent manner in vitro (Figure S3A) and showed that cIAP2 protein increases in 9qA1-positive murine HCC cells grown in the presence of a proteasome inhibitor (Figure S3B). Based on these aggregate observations, we considered cIAP1 and Yap as the most likely candidate oncogenes in the region.
cIAP1 Has Oncogenic Properties and Is Required for Rapid Tumor Growth
Inhibitor of apoptosis (IAP) proteins were originally identified in baculovirus because of their ability to block cell death of infected cells (Crook et al., 1993). Overexpression of cellular IAPs can inhibit apoptosis induced by different stimuli (Lacasse et al., 1998). Although some IAPs bind and inhibit caspases, their contribution to apoptosis regulation and oncogenesis in mammalian cells is controversial (Wright and Duckett, 2005).
A significant advantage of profiling the genomes of defined murine tumors is that candidate genes can be evaluated in the genetic context where the mutation spontaneously arose. Our studies identified the 9qA1 amplicon in tumors derived from p53−/− hepatoblasts expressing Myc but not in other configurations, suggesting that these cells would be ideal for evaluating the oncogenic properties of cIAP1. Therefore, p53−/−;myc liver progenitor cells expressing cIAP1 or a control vector were produced using retroviral-mediated gene transfer; as predicted, cIAP1 conferred a modest resistance to cell death triggered by serum starvation or confluence (Figure 6A).
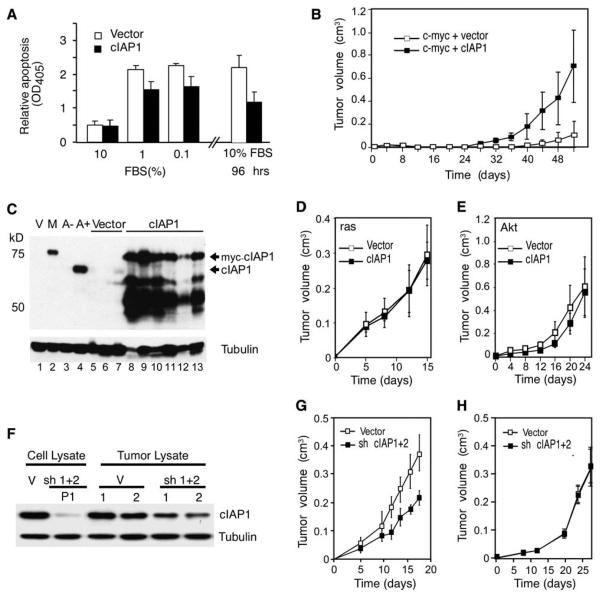
Figure 6. cIAP1 Enhances the Tumorigenicity of myc-Overexpressing p53−/− Hepatoblasts.
(A) Apoptosis measurements (Cell Death Detection ELISAPLUS kit; Roche) from p53−/− hepatoblasts double infected with myc + cIAP1 or myc + vector following culture under the indicated serum conditions for 48 hr (left panel). Cells grown to confluence were cultured for another 48 hr, and cell death was measured (right panel). Error bars represent mean ± SD (n = 3) per data point.
(B) Tumor volume measurements at various times following subcutaneous injection of p53−/− hepatoblasts double infected with myc + cIAP1 or myc + vector into the rear flanks of nude mice (n = 6 for each group). Shown is a representative of three independent experiments, each showing a statistical difference between cIAP1 and control (p < 0.05). Error bars represent ±SD.
(C) Immunoblotting of tumors overexpressing myc-tagged cIAP1 (lanes 8–13) or control vector tumors (lanes 5–7) using antibodies against cIAP1. Samples from cultured myc-tagged cIAP1-expressing hepatoblasts (M, lane 2) or vector alone (V, lane 1) and from 9qA1 amplicon-containing cells (A+, lane 4) were analyzed for comparison. Note that myc-tagged cIAP1 migrates at 75 kDa and endogenous cIAP1 at 65 kDa. A– is lysate from amplicon-negative cells of the same genotype (p53−/−;myc). Tubulin was used as a loading control.
(D and E) p53−/− hepatoblasts coexpressing Ras (D) or Akt (E) with cIAP1 or a control vector were monitored for tumorigenicity following subcutaneous injection into nude mice (n = 6 per group). Error bars represent ±SD.
(F) Immunoblotting of lysates derived from hepatoma cells outgrown from a 9qA1-positive p53−/−;myc tumor transduced with shRNAs targeting cIAP1 and cIAP2 (sh 1+2) or control vectors (V) using antibodies against cIAP1.
(G and H) 9qA1-positive (G) and -negative (H) hepatoma cells expressing cIAP1/2 shRNAs or a control vector were monitored for tumor growth following subcutaneous injection into nude mice. Error bars represent ±SD.
To determine whether cIAP1 could function as an oncogene in vivo, we injected the cells described above subcutaneously into nude mice to facilitate precise measurement of tumor growth. cIAP1 significantly accelerated the growth of p53−/−;myc hepatoblasts (Figure 6B), reducing tumor onset times by half (24 ± 2.3 days for myc + cIAP1 versus 45 ± 12.2 days for myc + vector [p < 0.05]) and greatly increasing tumor burden (myc + cIAP1 versus myc + vector [p < 0.005] at 52 days). The resulting tumors stably overexpressed full-length cIAP1 protein (Figure 6C, compare lane 2 to lanes 8–13) and several degradation products (Silke et al., 2005) and displayed a histopathology that resembled moderately well to poorly differentiated HCC (data not shown). Interestingly, one control tumor that was harvested at a very small size already showed elevated levels of cIAP1 (Figure 6C, lane 7), consistent with a subset of cells acquiring a spontaneous alteration that upregulated the gene. In contrast, cIAP1 did not affect the onset or progression of tumors expressing Akt or Ras (Figures 6D and 6E), even though cIAP1 was efficiently expressed (Figures S4A and S4B). Thus, cIAP1 is selectively oncogenic in the genetic context where its amplification occurs.
To determine whether the cIAP proteins help sustain tumorigenesis, we next examined the impact of reducing cIAP levels on tumor growth. We chose to suppress the expression of cIAP1 and cIAP2 since cIAP2 can be upregulated in response to downregulation of cIAP1 (Conze et al., 2005). shRNAs capable of suppressing cIAP1 and cIAP2 expression by RNA interference (Figure 6F, compare lanes 1 and 2) were cointroduced into outgrown murine hepatoma cells containing or lacking the 9qA1 amplicon. These cells were then injected subcutaneously into immunocompromised mice.
Tumors arising from 9qA1-positive cells expressing cIAP1 and cIAP2 shRNAs showed a reduced growth rate compared to controls (Figure 6G; p < 0.005 for tumor burden “vector; vector” versus sh cIAP1;sh cIAP2 at day 18). Although tumor inhibition was incomplete, the efficiency of cIAP knockdown was greatly reduced in the outgrown tumors compared to the injected cells (Figure 6F, compare lane 2 and lanes 5 and 6), implying that cells retaining high cIAP levels were selected during tumor expansion. These same shRNAs had no impact on the growth of amplicon-negative tumors expressing either Myc or oncogenic Ras (Figure 6H; Figure S4D), suggesting that only cells selected for cIAP overexpression are sensitive to cIAP inhibition and ruling out off-target effects of these shRNAs on tumor growth (see also Figure S4C). Therefore, the cIAP genes are required for the efficient growth of tumors harboring the 9qA1 amplicon.
Yap Has Oncogenic Properties and Contributes to Rapid Tumor Growth
In addition to cIAP1, Yap was also overexpressed at the RNA and protein level in every tumor harboring the mouse 9qA1 or human 11q22 amplicon. Yap (synonyms Yap65 or Yap1) was originally identified due to its interaction with the Src-family kinase Yes (Sudol, 1994) and acts as a transcriptional coactivator that can bind and activate Runx and TEAD/TEF transcription factors (Yagi et al., 1999). Inconsistent with an oncogenic role, mammalian Yap also interacts with the p53 family member p73 (Strano et al., 2001) and potentiates apoptosis in a manner that is suppressed by Akt (Basu et al., 2003). However, recent studies suggest that yorkie, the Drosophila homolog of Yap, promotes tissue expansion as an effector of the Lats/ Warts pathway by activating cyclin E and the Drosophila inhibitor of the apoptosis gene dIAP (Huang et al., 2005). Interestingly, we also noted that murine tumors harboring the 9qA1 amplicon overexpressed cyclin E (Figure 7B).
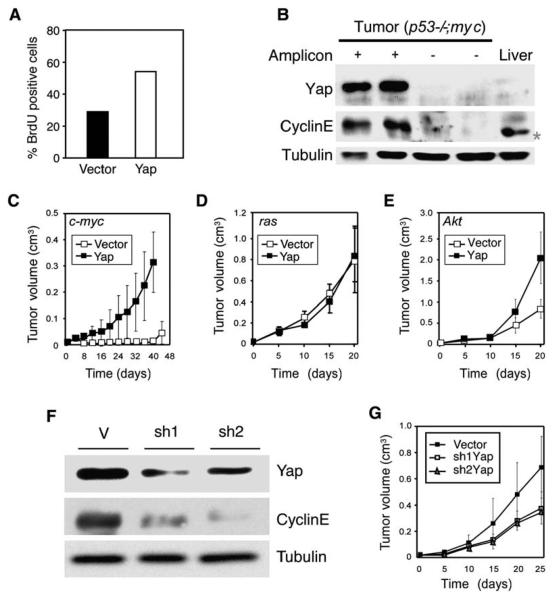
Figure 7. Yap Confers a Proliferative Advantage, Has Oncogenic Properties, and Is Required for Liver Tumor Progression.
(A) The proliferation rates of p53−/−;myc hepatoblasts expressing Yap or a control vector were assessed by the fraction of nuclei incorporating BrdU after a 1 hr pulse.
(B) Protein lysates from two 9qA1 amplicon-positive tumors (+), two amplicon-negative tumors (−), and adult normal mouse liver were immunoblotted with antibodies against Yap and cyclin E. Tubulin was used as a loading control. * denotes a nonspecific band in the liver lysate.
(C–E) The tumorigenicity of p53−/− liver progenitor cells coexpressing the indicated oncogene (upper left) with a control vector or Yap was assessed by caliper measurement following subcutaneous injection into the rear flanks of nude mice (n = 4 per group). Error bars represent ±SD.
(F) Protein lysates from hepatoma cells (p53−/−;myc) stably overexpressing Yap were infected with control vector (V) or two different short-hairpin RNAs targeting Yap (sh1 and sh2) and were analyzed for Yap and cyclin E levels. Tubulin was used as a loading control.
(G) Tumorigenicity of 9qA1-positive cells infected with retroviral vectors expressing shRNAs targeting Yap (sh1Yap or sh2Yap) or control vector (n = 6 per group). Error bars represent ±SD.
To determine whether Yap could also contribute to the transformation of liver progenitor cells, we conducted functional studies that paralleled our analysis of cIAP1. p53−/−;myc hepatoblasts expressing Yap grew more rapidly than vector-infected cells (data not shown), with a higher BrdU incorporation rate (Figure 7A). Furthermore, Yap significantly accelerated tumor onset and progression of p53−/−;myc liver progenitor cells (Figure 7C) and greatly increased tumor burden (myc;vector versus myc;Yap at day 40 [p < 0.005]). In contrast, Yap did not accelerate tumorigenesis together with activated Ras, although it did enhance Akt-driven tumorigenesis, particularly at later times (Figures 7D and 7E).
We also tested whether Yap was required for efficient tumor growth. Two distinct shRNAs were capable of suppressing Yap expression, and, interestingly, cells expressing either Yap shRNA downregulated cyclin E (Figure 7F). Despite the incomplete suppression of Yap, cells harboring the 9qA1 amplicon and expressing either Yap shRNA showed slower tumor progression compared to controls following injection into recipient mice (Figure 7G; p < 0.05 [0.013 (shYap 2)/0.018 (shYap 1)] at day 25 postinjection). Together, these data validate Yap as a potent oncogene.
cIAP1 and Yap Cooperate to Promote Tumorigenesis
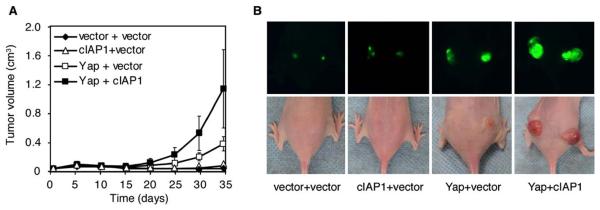
As prevailing view in cancer genomics is that focal genomic amplifications contain a key driver gene that is selected for during tumorigenesis, it was surprising to identify two oncogenes in the same focal amplicon. To determine whether cIAP1 and Yap might cooperate during tumorigenesis, p53−/−;myc liver progenitor cells were infected with either Yap and control vector or Yap plus cIAP1 and assayed for their ability to form tumors in vivo. Tumors arising from p53−/−;myc hepatoblasts coexpressing cIAP1 and Yap grew faster than those expressing either oncogene alone (Figure 8A; p < 0.005 and p < 0.05 [0.011] for cIAP1 + Yap versus cIAP1 or Yap alone, respectively). These effects were not merely additive: At time points when tumors expressing Yap alone were small and those harboring cIAP1 alone were barely detectable, tumors co-expressing Yap and cIAP1 were so large that the animals had to be sacrificed (Figures 8A and 8B). Other gene combinations did not have these effects; for example, coexpression of cIAP2 and cIAP1 had no further impact on tumorigenesis compared to cIAP1 alone, and the combination of Porimin with Yap appeared to even delay tumorigenesis (data not shown). Thus, our study establishes that two adjacent genes from the same focal amplification can cooperate during tumorigenesis.
Figure 8. cIAP1 and Yap Synergize to Drive Tumorigenesis.
(A) p53−/−;myc liver progenitor cells were infected with control vector, cIAP1, or Yap or coinfected with Yap + cIAP1 and then transplanted subcutaneously into nude mice (n = 6 per group). Error bars represent ±SD.
(B) GFP imaging of the tumors described in (A).
DISCUSSION
A New Mouse Model of Hepatocellular Carcinoma
Here we develop and utilize a genetically tractable mouse model of hepatocellular carcinoma that is based on the isolation and ex vivo genetic manipulation of mouse embryonic liver progenitor cells followed by their seeding into the livers of recipient mice. This model produces pathologies that resemble human liver carcinomas and has other features that make it amenable for studying liver cancer biology. First, the ability to manipulate liver progenitor cells—one presumed target of transformation in hepatocarcinogenesis—ex vivo allows the rapid production and analysis of tumors with complex genotypes without the cost and effort associated with genetic intercrossing of cancer-prone strains. Second, the fact that the system relies on transplantation of progenitor cells implies that the recipient mice could also have different genotypes, thereby facilitating studies of tumor/host interactions. Third, the model uses bipotential liver progenitors as the cancer-initiating cell, making it suitable for studying a potential “cell of origin” of different primary liver cancers. Finally, the ability to rapidly generate pathologically accurate liver tumors with different genotypes makes the model an ideal preclinical system for testing new drugs or drug combinations for HCC, a disease for which current therapies are ineffective.
Identification and Validation of cIAP1 and Yap as Human Oncogenes
We noted that amplifications of chromosome 9qA1 occurred at a high frequency in murine tumors derived from Myc-expressing cells, and at a lower frequency at the syntenic region of chromosome 11q22 in human liver cancers. Although the 11q22 amplicon has been observed in several tumor types, previous studies have not agreed on the likely driver gene(s), in part, because validation was lacking (Dai et al., 2003; Snijders et al., 2005; Bashyam et al., 2005; Imoto et al., 2001). However, through comparative oncogenomics and expression analyses, we pinpointed cIAP1 and Yap as the most likely driver genes in liver cancer; then, by returning to the mouse model, we rapidly demonstrated the oncogenic capacity of cIAP1 and Yap in the genetic setting where spontaneous amplification occurred. Despite the detection of the 11q22 amplicon in only 5%–10% of human tumor types examined, its association with common malignancies including lung, ovarian, esophageal, and liver carcinoma suggest that the overall contribution of cIAP1 and Yap to human cancer may be substantial.
The identification of cIAP1 as an oncogene is interesting in light of the controversial role of IAPs in modulating apoptosis in mammalian cells (Deveraux et al., 1998; Liston et al., 2003; Salvesen and Duckett, 2002). In Drosophila, the IAP genes are important cell death regulators, with loss-of-function mutants displaying profound defects in developmental and stress-induced apoptosis (Goyal et al., 2000). In mammalian cells, a large body of work shows that IAPs can suppress apoptosis (Lacasse et al., 1998), although most studies observe relatively modest effects despite overexpression in transient assays, and mice with knockout of single IAP genes do not display substantial apoptotic defects (Conze et al., 2005). Similarly, IAP expression has been associated with various cancer phenotypes (Wright and Duckett, 2005), but no IAP has been decisively linked to tumorigenesis in vivo.
cIAP1 promoted murine HCC in cooperation with Myc, an oncogene that drives proliferation but also promotes apoptosis through both p53-dependent and -independent mechanisms (Lowe et al., 2004). As in other cell types, Myc sensitizes hepatoblasts to diverse proapoptotic signals, and dampening of such signals is linked to tumorigenesis in other contexts. In our system, the effects of cIAP1 appeared more pronounced in vivo, suggesting that microenvironmental effects associated with tumor expansion may be critical for its prosurvival activities. Perhaps this explains the variable effects of cIAP1 observed in previous in vitro studies; alternatively, nonapoptotic cIAP1 activities may contribute to its oncogenic role. In any case, the antiapoptotic activity of cIAP1 would be unlikely to cooperate with Akt or Ras, which already transmit prosurvival signals (Lowe et al., 2004).
The finding that Yap is also an oncogene in the 9qA1/11q22 amplicon is surprising considering its ability to potentiate the proapoptotic functions of p73, a p53 family member (Basu et al., 2003). However, yorkie (yki), the Drosophila homolog of mammalian Yap, links warts/large tumor suppressor (wts/lats) signaling and transcriptional regulation in the Hippo (hpo) pathway (Huang et al., 2005). This pathway also includes the genes salvador (sav), mob as tumor suppressor (mats), NF2/Merlin, and expanded (Hamaratoglu et al., 2006; Edgar, 2006) and functions to precisely control tissue expansion by coordinately regulating cell proliferation and cell death. This is ultimately achieved through transcriptional regulation of cyclin E and the Drosophila inhibitor of apoptosis protein, dIAP.
Interestingly, mammalian homologs exist for most Hippo pathway members (Chan et al., 2005; Tamaskovic et al., 2003), some of which have previously been linked to cancer development (e.g., Lats2 and NF2) (St John et al., 1999; Takahashi et al., 2005; McClatchey and Giovannini, 2005). In addition, Drosophila mutants can be rescued by the human homologs for Yap, LATS1, MATS1, and MSTS2. Interestingly, although Yap may have additional targets in mammalian cells, we note that mammalian cells and tumors with high Yap levels proliferate rapidly and have high levels of cyclin E. It is intriguing that cIAP1—a gene with structural and functional similarity to dIAP—resides in an adjacent chromosomal location in mice and humans and is coamplified with Yap in tumors. These data demonstrate a functional conservation and reveal the potential importance of the Hippo signaling network in hepatocellular carcinoma and perhaps other cancers. In further support of this possibility, Yap can also transform mammary epithelial cells in vitro (D. Haber and J. Brugge, personal communication).
Perhaps the most remarkable observation in our study was the demonstration that two genes embedded within the same focal amplification cooperate in cancer development. Thus, while cIAP1 and Yap can act independently as oncogenes, they synergize in transforming hepatoblasts and promoting tumorigenesis. Such codriver effects may not be restricted to the 11q22 amplicon, as suggested in a recent study of the 20q13.2 breast cancer amplicon, where there is universal overexpression of two genes: ZNF217, which is capable of immortalizing human mammary epithelial cells in culture, and prefoldin 4 (PFDN4) (Collins et al., 2001). These results underscore the complexity of cancer genomes and should force a reexamination of well-characterized amplification events for the presence of additional oncogenic lesions and potential drug targets.
Comparative Oncogenomics to Accelerate Cancer Genome Annotation and Target Identification
Our study took advantage of cross-species comparisons to help prioritize candidate oncogenes and then used a relevant mouse model to validate their role in tumorigenesis. Thus, our genome-wide analysis of human HCC identified a large number of genetic changes including the 11q22 amplicon, but the fact we detected a syntenic lesion on chromosome 9qA1 in a specific type of murine HCC provided a filter that allowed us to prioritize this lesion for further study. Importantly, our analysis of mouse tumors identified an appropriate setting to study the oncogenic potential of cIAP1 and Yap: a genetic background in which amplification of cIAP1 and Yap spontaneously occurred. A similar comparative genomic approach has been applied by Chin and colleagues to identify NEDD9 as a metastasis-promoting gene that allows escape from tumor cell dependence on Ras signaling (Kim et al., 2006 [this issue of Cell]).
Moving forward, our study suggests an integrative strategy to complement the Human Cancer Genome Project, whose goal is to catalog all of the mutations that contribute to human cancer (Garber, 2005). The advantages of using genomics to survey human tumors are obvious: These tumors harbor changes relevant to the human disease. However, spontaneous human cancers are heterogeneous and can display extreme genomic instability. Thus, it is often difficult to separate causative from passenger mutations or to identify the key gene (or genes) in a larger region of chromosomal gain or loss (Cox et al., 2005; Futreal et al., 2005). Even with a statistically good candidate, validation can be challenging—it is often necessary to survey many in vitro assays to link a particular lesion to cancer phenotypes. Furthermore, without in vivo validation, therapeutic development efforts are difficult to envision.
By integrating data from human cancer genomics with corresponding data from appropriate mouse models, it should be possible to expeditiously focus on the cancer lesions most likely to have translational potential. First, identifying common lesions through cross-species comparisons increases confidence that these lesions will prove important, a particularly relevant issue when studying relatively low-frequency events. In fact, owing to reduced selective pressure for multiple mutations and/or inherent differences in telomere biology (Artandi and Depinho, 2000), genetically defined murine tumors are often less complex than their corresponding human counterparts. Second, by incorporating murine tumors into the analysis, it is possible to increase the sample size and exploit synteny to exclude candidates outside the region of overlap. Third, genetically engineered mice, by definition, develop more defined cancers than humans. Thus, the identification of a particular lesion in murine cancers immediately provides information concerning the evolutionary context in which it arose and identifies a relevant setting in which to study its oncogenic potential. Finally, mouse models provide excellent settings to test the suitability of a cancer lesion as a therapeutic target and, at the same time, provide ideal preclinical models for subsequent drug testing. We believe that such integrative approaches will speed up the pace of discovery and help translate the promise of cancer genetics into improvements in cancer diagnosis, prognosis, and therapy.
EXPERIMENTAL PROCEDURES
Generation of Genetically Defined Liver Carcinomas, Tumor Retransplantation, Analysis, and Immunohistochemistry
Isolation, culturing, and retroviral infection of purified hepatoblasts as well as surgical procedures, retrorsine treatment of mice, tumor monitoring, and tumor retransplantation are described in the Supplemental Experimental Procedures and Zender et al. (2006). All retroviruses were based on MSCV vectors containing human cDNAs encoding for myc, H-RasV12, and Akt or the murine cDNAs encoding Yap and myc-tagged cIAP1. Short-hairpin RNAs against cIAP1, cIAP2, or Yap were expressed from the LTR promoter of MSCV retroviruses. Tumor volume (cm3) was calculated as length × width × height. Paraffin-embedded liver tumor sections were stained with hematoxylin and eosin according to standard protocols or with α-GFP (Abcam 290). Standard Proteinase K antigen retrieval was used. Human hepatocellular carcinomas were analyzed using antibodies against Ck8 (RDI), cIAP1 (Silke et al., 2005), cIAP2 (sc-7944, Santa Cruz), YAP1 (sc-15407, Santa Cruz), and Porimin (IMG472, Imgenex).
Immunoblotting
Fresh tumor tissue or cell pellets were lysed in RIPA buffer using a tissue homogenizer. Equal amounts of protein (16 mg) were separated on 10% SDS-polyacrylamide gels and transferred to PVDF membranes. The blots were probed with antibodies against cIAP1 (Silke et al., 2005), cIAP1/2 (gift from P. Liston; 1:2000), YAP1 (sc-15407, Santa Cruz; 1:200), Porimin (IMG472, Imgenex; 1:300), cyclin E (#06-459, Up-state; 1:500), tubulin (B-5-1-2, Sigma; 1:5000), vimentin (Abcam; 1:1000), cytokeratin 19 (Biocare Medical; 1:1000), albumin (Biogenesis; 1:5000), or AFP (Dako; 1:1000).
Cell Proliferation Assay and Cell Death ELISA
Cells plated on gelatin-coated coverslips were incubated with 5-bromo-2′-deoxyuridine (BrdU; 100 mg/ml; Sigma) for 1 hr. Nuclei incorporating BrdU were visualized by immunolabeling using anti-BrdU antibody (Pharmingen; 1:400) as previously described (Narita et al., 2003). DNA was visualized by DAPI (1 μg/ml) after permeabilization with 0.2% Triton X-100/PBS. Cells were grown in various concentrations of serum, and apoptosis was measured using the Cell Death Detection ELISAPLUS kit (Roche).
Representational Oligonucleotide Microarray Analysis
Human tumor samples were obtained from the NCI-sponsored Cooperative Human Tissue Network or the tissue bank of the University of Hong Kong. Genomic DNA was isolated from human or mouse tumors using the PureGene DNA Isolation Kit (Gentra). Hybridizations were carried out on 85K arrays (NimbleGen) (Lucito et al., 2003; B. Lakshmi, I.M. Hall, C. Egan, J. Alexander, J. Healy, L.Z., W.X., M.S.S., S.W.L., M.W., and R.L., unpublished data). The genome position was determined from the UCSC GoldenPath browser (freezes April 2003 for human and February 2003 for mouse). Focal gains or losses were defined as spanning <5 Mb.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed on a PRISM 7700 sequence detector (Applied Biosystems, Foster City, CA, USA). Quantification of genomic copy number was based on standard curves derived from serial dilutions of normal human genomic DNA (Invitrogen). For quantitation of mRNA expression, mouse tumors were freshly homogenized in Trizol (GIBCO). RNA was isolated and treated with RNase-free DNase (QIAGEN) and purified over QIAGEN RNAeasy columns. Total RNA was converted to cDNA using TaqMan reverse transcription reagents (Applied Biosystems) and used in qPCR reactions with incorporation of Sybr Green PCR Master Mix (Applied Biosystems) done in triplicate using gene-specific primers. Quantification of mRNA expression of human tumor samples was performed using TaqMan probes (Applied Biosystems). Samples were normalized to the level of β-actin. Primer sequences are listed in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. C. Duckett and P. Liston for providing cDNA constructs and antibodies. We thank K. Nguyen, A. Brady, L. Bianco, and M. Jiao for excellent technical assistance and E. Hernando for assistance with histology. We also thank B. Stillman for critical reading of the manuscript and E. Cepero and other members of the Lowe lab for advice and discussions. This work was generously supported by the German Research Foundation; Alan and Edith Seligson; the Miracle Foundation; the Breast Cancer Research Foundation; Long Islanders Against Breast Cancer; the West Islip Breast Cancer Foundation; Long Island Breast Cancer (1 in 9); an Elizabeth McFarland Group breast cancer research grant; Breast Cancer Help Inc.; and grants CA078544, CA13106, CA87497, and CA105388 from the National Institutes of Health. M.W. is an American Cancer Society Research Professor. G.J.H. and S.W.L. are Howard Hughes Medical Institute investigators.
Footnotes
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, four figures, and three tables and can be found with this article online at http://www.cell.com/cgi/content/full/125/7/1253/DC1/.
REFERENCES
- Aguilar F, Harris CC, Sun T, Hollstein M, Cerutti P. Geographic variation of p53 mutational profile in nonmalignant human liver. Science. 1994;264:1317–1319. doi: 10.1126/science.8191284. [DOI] [PubMed] [Google Scholar]
- Alison MR, Lovell MJ. Liver cancer: the role of stem cells. Cell Prolif. 2005;38:407–421. doi: 10.1111/j.1365-2184.2005.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Depinho RA. Mice without telomerase: what can they teach us about human cancer? Nat. Med. 2000;6:852–855. doi: 10.1038/78595. [DOI] [PubMed] [Google Scholar]
- Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A, Pollack JR. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14–3-3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J. Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Collins C, Volik S, Kowbel D, Ginzinger D, Ylstra B, Cloutier T, Hawkins T, Predki P, Martin C, Wernick M, et al. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 2001;11:1034–1042. doi: 10.1101/gr.174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze DB, Albert L, Ferrick DA, Goeddel DV, Yeh WC, Mak T, Ashwell JD. Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol. Cell. Biol. 2005;25:3348–3356. doi: 10.1128/MCB.25.8.3348-3356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C, Bignell G, Greenman C, Stabenau A, Warren W, Stephens P, Davies H, Watt S, Teague J, Edkins S, et al. A survey of homozygous deletions in human cancer genomes. Proc. Natl. Acad. Sci. USA. 2005;102:4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, Wu YZ, Rush LJ, Ross P, Molina JR, et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum. Mol. Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–2604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Wooster R, Stratton MR. Somatic mutations in human cancer: insights from resequencing the protein kinase gene family. Cold Spring Harb. Symp. Quant. Biol. 2005;70:1–8. doi: 10.1101/sqb.2005.70.015. [DOI] [PubMed] [Google Scholar]
- Garber K. Human Cancer Genome Project moving forward despite some doubts in community. J. Natl. Cancer Inst. 2005;97:1322–1324. doi: 10.1093/jnci/dji324. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Fu T, Nelson JA, Superina RA, Soriano HE. Liver repopulation after cell transplantation in mice treated with retrorsine and carbon tetrachloride. Transplantation. 2002;73:1818–1824. doi: 10.1097/00007890-200206150-00020. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, Huang M, Wang Y, Wu MC, Huang JF, et al. Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene. 2004;23:298–302. doi: 10.1038/sj.onc.1206483. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Imoto I, Yang ZQ, Pimkhaokham A, Tsuda H, Shimada Y, Imamura M, Ohki M, Inazawa J. Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res. 2001;61:6629–6634. [PubMed] [Google Scholar]
- Kallioniemi OP, Kallioniemi A, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization: a rapid new method for detecting and mapping DNA amplification in tumors. Semin. Cancer Biol. 1993;4:41–46. [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik J-H, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. this issue. [DOI] [PubMed] [Google Scholar]
- Lacasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17:3247–3259. doi: 10.1038/sj.onc.1202569. [DOI] [PubMed] [Google Scholar]
- Laconi E, Oren R, Mukhopadhyay DK, Hurston E, Laconi S, Pani P, Dabeva MD, Shafritz DA. Long-term, near-total liver replacement by transplantation of isolated hepatocytes in rats treated with retrorsine. Am. J. Pathol. 1998;153:319–329. doi: 10.1016/S0002-9440(10)65574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Ma F, Zhang C, Prasad KV, Freeman GJ, Schlossman SF. Molecular cloning of Porimin, a cell lung cancer to gefitinib cell death. Proc. Natl. Acad. Sci. USA. 2001;98:9778–9783. doi: 10.1073/pnas.171322898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis—the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- Murakami H, Sanderson ND, Nagy P, Marino PA, Merlino G, Thorgeirsson SS. Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor alpha in hepatic oncogenesis. Cancer Res. 1993;53:1719–1723. [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Nitou M, Sugiyama Y, Ishikawa K, Shiojiri N. Purification of fetal mouse hepatoblasts by magnetic beads coated with monoclonal anti-e-cadherin antibodies and their in vitro culture. Exp. Cell Res. 2002;279:330–343. doi: 10.1006/excr.2002.5615. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int. J. Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- Peng SY, Lai PL, Hsu HC. Amplification of the c-myc gene in human hepatocellular carcinoma: biologic significance. J. Formos. Med. Assoc. 1993;92:866–870. [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Sandgren EP, Quaife CJ, Pinkert CA, Palmiter RD, Brinster RL. Oncogene-induced liver neoplasia in transgenic mice. Oncogene. 1989;4:715–724. [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat. Genet. Suppl. 2005;37:S38–S45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, Huang DC, Vaux DL. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc. Natl. Acad. Sci. USA. 2005;102:16182–16187. doi: 10.1073/pnas.0502828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brown-stein DG, Parlow AF, McGrath J, Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- Suzuki K, Hayashi N, Yamada Y, Yoshihara H, Miyamoto Y, Ito Y, Ito T, Katayama K, Sasaki Y, Ito A. Expression of the c-met protooncogene in human hepatocellular carcinoma. Hepatology. 1994;20:1231–1236. [PubMed] [Google Scholar]
- Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Tamaskovic R, Bichsel SJ, Hemmings BA. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 2003;546:73–80. doi: 10.1016/s0014-5793(03)00474-5. [DOI] [PubMed] [Google Scholar]
- Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CW, Duckett CS. Reawakening the cellular death program in neoplasia through the therapeutic blockade of IAP function. J. Clin. Invest. 2005;115:2673–2678. doi: 10.1172/JCI26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Cordon Cardo C, Hannon GJ, Lucito R, Powers S, Flemming P, Spector MS, Lowe SW. Generation and analysis of genetically-defined liver carcinomas derived from bi-potential liver progenitors. Cold Spring Harb. Symp. Quant. Biol. 2006 doi: 10.1101/sqb.2005.70.059. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.