Abstract
Sunlight-induced C→T mutation hotspots occur most frequently at methylated CpG sites in tumor suppressor genes and thought to arise from trans-lesion synthesis past deaminated cyclobutane pyrimidine dimers (CPDs). While it is known that methylation enhances CPD formation in sunlight, little is known about the effect of methylation and sequence context on the deamination of 5-methyl C (mC) and its contribution to mutagenesis at these hotspots. Using an enzymatic method we have determined the yields and deamination rates of C and mC in CPDs and find that the frequency of UVB-induced CPDs correlates with the oxidation potential of the flanking bases. We also found that the deamination of TmC and mCT CPDs is about 25-fold faster when flanked by G’s than by A’s, C’s or T’s in duplex DNA and appears to involve catalysis by the O6 group of guanine. In contrast, the first deamination of either C or mC in ACmCG with a flanking G was much slower (t ½ >250 h) and rate limiting, while the second deamination was much faster. The observation that CmCG dimers deaminate very slowly but at the same time correlate with C→T mutation hotspots suggests that their repair must be slow enough to allow sufficient time for deamination. There are, however, a greater number of single C→T mutations than CC→TT mutations at CmCG sites even though the second deamination is very fast, which could reflect faster repair of doubly deaminated dimers.
Keywords: UV mutagenesis, p53 gene, DNA photoproduct, CC to TT mutation, flanking sequence effect
Introduction
In basal and squamous cell carcinomas, the p53 tumor suppressor gene exhibits a high percentage of C to T transition mutations at dipyrimidine sites.1–5 Previous studies of sunlight-induced mouse skin tumors indicate that 30–50% of these mutations are due to the formation of cyclobutane pyrimidine dimer (CPD) photoproducts (Fig. 1A).6,7 A comparison of sunlight-induced mutation spectra of cII and lacI transgenes as well as the p53 gene in human skin tumors reveal that 5-methylcytosine (mC) is involved in 25–40% of the mutations in all three systems.8 Most significantly, methylation at the C of PyCG increases CPD formation in sunlight 15-fold.9
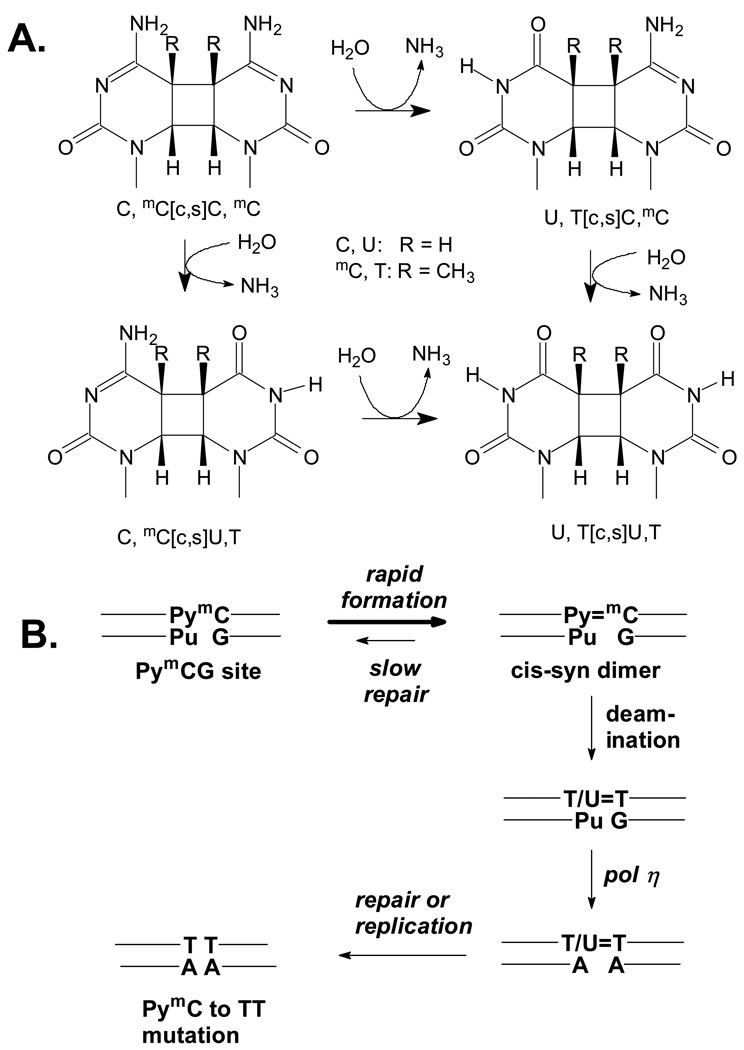
Figure 1.
Deamination of C-containing cis-syn cyclobutane pyrimidine dimers and their proposed role in UV-induced C to T mutations. A) Structures and deamination pathways. B) Deamination bypass mechanism for the origin of UV-induced C to T and CC to TT mutations.
The principal polymerase involved in synthesizing past CPDs is polymerase η. Both in vitro and in vivo studies have found, however, that T, U, C and mC-containing CPDs are not measurably mutagenic when replicated by pol η.10–14 Indeed, loss of functional pol η in Xeroderma pigmentosum variant (XPV) results in an increased risk of skin cancer due to the replication of photodimers by more error prone polymerases.15–17 While C and mC are very stable with a deamination half life of 30,000 years in duplex DNA,18 C and mC within CPD’s are more than a million-fold less stable and deaminate to U or T respectively (Fig. 1A), in hours to days due to loss of aromatic stabilization.14,19–27 It is the replication (bypass) of these deaminated products by a pol Y family polymerase that is thought to be the origin of UV-induced C to T mutations, a process otherwise known as the deamination-bypass mechanism (Fig. 1B).24–26,28–31 Little is known, however, about the deamination of C and mC within PymCG mutational hotspots.
Deamination of C and mC in cyclobutane pyrimidine dimers has been studied in dinucleotides and in single-strand DNA by chromatographic, mass-spectrometry and enzymatic analysis.14,19,20,22,23,27 Deamination has also been studied in vivo by genetic assays and by ligation mediated PCR.21,24–26 The reported deamination rates vary from hours to weeks depending on the substrate and organism with no clear explanation for the differences. In this manuscript, we describe a sensitive method for determining in vitro deamination rates of C and mC-containing cyclobutane dimers in duplex DNA using site-specifically radiolabeled nucleotides. While methylation only occurs at CG sites in vertebrates, such sequence restrictions do not apply in plants and fungi32 where mC-containing dimers could be formed in any sequence context. We therefore determinef the deamination rates of both NmC=PyN and NPy=mCN pyrimidine dimers where N is any base to better understand the structure-activity relationships in deamination of mC and it biological implications. We found that deamination of T=mC or mC=T cyclobutane dimers is at least 10-fold faster when flanked by G compared with A, C or T, and appears to involve the O6 carbonyl group of guanine. In contrast, deamination of C or mC in C=mCG, a known hotspot for C to T mutations in humans, is very slow and suggests the rate of repair of these dimers must also be slow to allow sufficient time for deamination.
Results and Discussion
Methodology
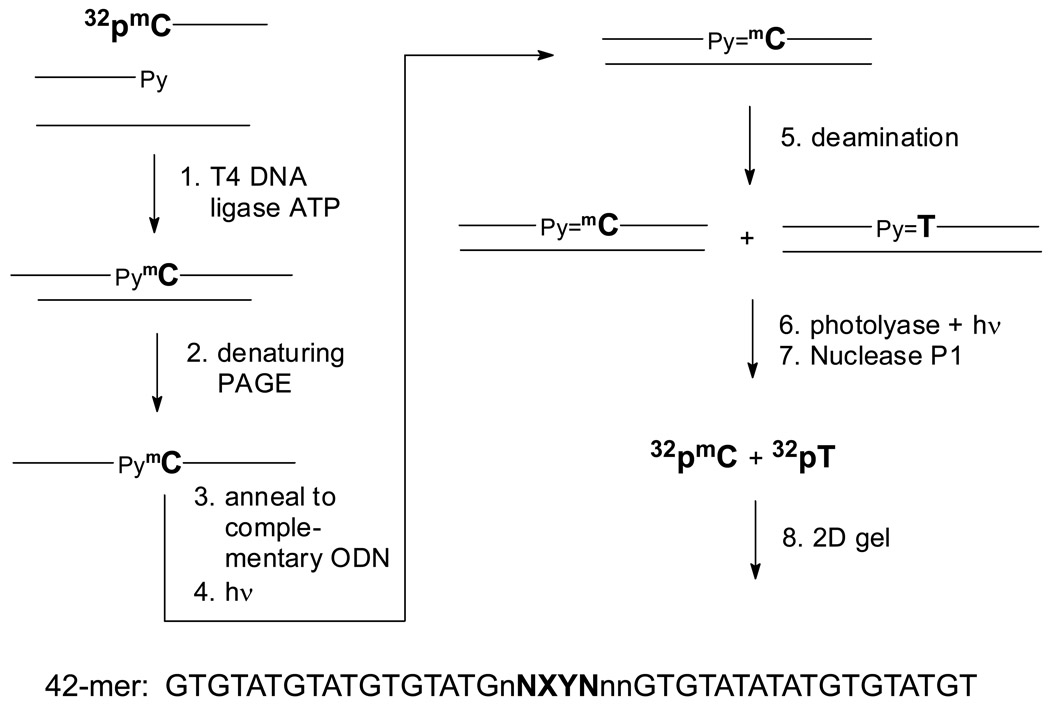
To determine the rate of formation and deamination of C and mC in cis-syn cyclobutane dimers we developed an assay that takes advantage of the specificity of E. coli photolyase for reverting this class of photoproduct. The idea was to 32P-label the C or mC of interest, and after photodimerization and deamination, specifically degrade the cis-syn photodimer along with undamaged DNA to mononucleotides with E. coli photolyase and nuclease P1 (Fig. 2). The extent of deamination could then be quantified by analysis of the amount of deaminated to undeaminated 32P-labeled nucleotide by gel electrophoresis (Fig. 3). To insure that the substrate was in the duplex form and to minimize end effects on deamination, we designed a 42-mer, containing two centrally located pyrimidines (X & Y) flanked by variable bases of interest (N & M) (Table 1). The other bases in the immediate vicinity (n) were chosen to minimize unwanted secondary structure formation in the single-strand that might result from the choice of N, X, Y and M.
Figure 2.
Scheme for substrate preparation and analysis of the deamination rate of mC-containing cyclobutane dimers in duplex DNA. 1) 5'-32P-labeled 21-mer with a terminal mC is ligated to second 21-mer ODN in the presence of a 20-nt ligation scaffold. 2) The single-strand 42-mer ligation product is separated from unligated products by denaturing gel electrophoresis. 3) The double strand substrate is prepared by annealing the radiolabeled 42-mer substrate to the complementary 42-mer ODN. 4) The duplex is irradiated with 302 nm on ice to generate the CPD. 5) The irradiated product is deaminated for various times at 37°C. 6) The deamination mixture is treated with photolyase and light to photorevert the CPD. 7) Nuclease P1 degrades undamaged and photoreverted DNA to mononucleotides. 8) Two-dimensional acrylamide electrophoresis of the nuclease P1 products separates the non-deaminated mC from its deamination product T.
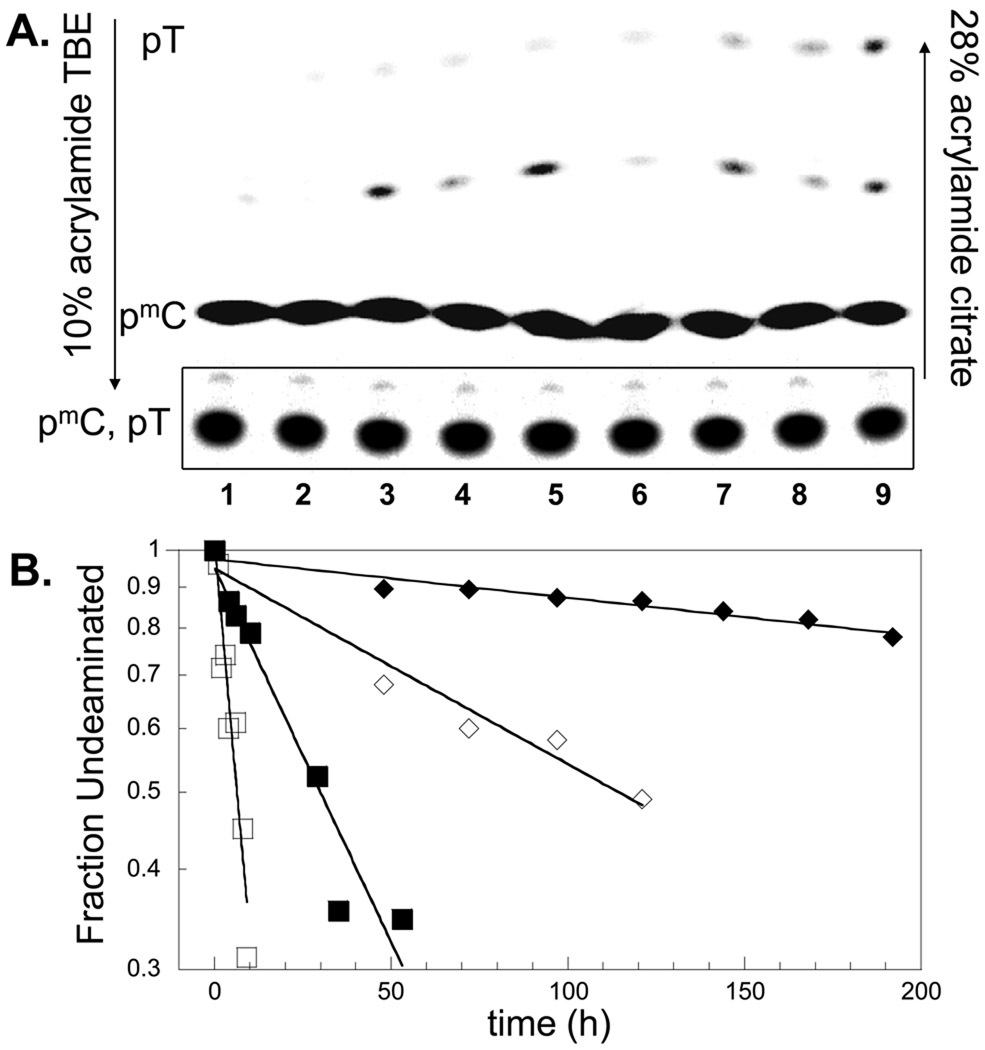
Figure 3.
2D gel electrophoresis and analysis of the deamination reaction. A) 2D acrylamide gel of the Nuclease P1 digestion products of duplex tGT=mCGtc that was deaminated for 0, 59, 120, 171, 218, 330, 456, 536 min at 37° C (Lanes 1–8, respectively) in 10 mM Tris/Mes/HCl pH 7.0 containing approximately 5 mM NaCl, and then photoreverted. Lane 9 contains the products following 16 h incubation at 67 °C at pH 6.3. The first dimension was carried out in the downward direction in 10% acrylamide with a 7 M urea, TBE buffer to give an unresolved mixture of the 32P-labeled mononucleotides pmdC and pT that migrate as a single intense band shown in the box. The second dimension was carried out in the upward direction towards the positive electrode in 28 % acrylamide/citrate. The second dimension separates pT (fastest migrating band) from pmdC (slowest migrating band). The bands in the middle (< 5%) are due to incomplete digestion products of P1 nuclease and non-photorevertible photoproduct-containing trinucleotides. B) Plots of the fraction undeaminated dimer vs time: data for duplex tGT=CGtc in 0 mM added NaCl (□) shown in panel A, and in 400 mM added NaCl (■) and for duplex tATmCAtc in 0 mM (◊), and 400 mM added NaCl (♦).
Table 1.
Cis-syn cyclobutane dimer photoproduct yield and deamination half life.
| entry | Abbrev. | Py=Py | Flanking N | sequencea | % yield | t1/2 (h) | |
|---|---|---|---|---|---|---|---|
| 1 | Double | GTmCG | T=mC | G--G | tGT=mCGtc | 5.4 | 5.8 ± 0.6 |
| 2 | Strand | GTCG | T=C | G--G | tGT=CGtc | 1 | 4.9 ± 0.8 |
| 3 | ITCI | T=mC | I--I | tIT=mCItc | 29 | 14.1 ± 0.8 | |
| 4 | aPTmCaP | T=mC | aP--aP | taPT=mCaPtc | 2.8 | 43 ± 2 | |
| 5 | ATmCA | T=mC | A--A | tAT=mCAtc | 17 | 175 ± 1 | |
| 6 | ATCA | T=C | A--A | tAT=CAtc | 7 | 46 ± 1.4 | |
| 7 | GmCTG | mC=T | G--G | tGmC=TGgc | 3.4 | 20.0 ± 0.5 | |
| 8 | AmCTA | mC=T | A--A | tAmC=TAgc | 7.8 | ~500 | |
| 9 | GTmCA | T=mC | G--A | tGT=mCAtc | 8.8 | 230 ± 18 | |
| 10 | ATmCG | T=mC | A--G | tAT=mCGtc | 12 | 7.1 ± 0.2 | |
| 11 | ATCG | T=C | A--G | tAT=CGtc | 5 | 5.3 ± 0.5 | |
| 12 | TTmCTT | T=mC/mC=T | T--T | tTTmCTtc | 15.7 | 385 ± 40 | |
| 13 | CCmCCC | C=mC/mC=C | C--C | tCCmCCcc | 6 | 300 ± 40 | |
| 14 | GTmCGGG | T=mC | G--GGG | tGT=mCGgg | 3.3 | 5.5 ± 0.5 | |
| 15 | GGGTmCG | T=mC | GGG--G | gGT=mCGtc | 3.9 | nd | |
| 16 | Single | GTmCG | TmC | G--G | tGT=mCGtc | 5.3 | 3.5 ± 0.1 |
| 17 | Strand | ATmCA | TmC | A--A | tAT=mCAtc | 7.8 | 4.2 ± 0.2 |
| 18 | GmCTG | mCT | G--G | tGmC=TGgc | 3.4 | 10.7 ± 0.6 | |
| 19 | AmCTA | mCT | A--A | tAmC=TAgc | 7.3 | 12.1 ± 0.4 | |
The sequence shown is embedded in the 42-mer single strand sequence: d(GTGTATGTATGTGTATGnNXYNnnGTGTATATATGTGTATGT). The double strand sequence is formed by annealing to its fully complementary strand. I: inosine, aP: 2-aminopurine, and mC: 5-methylcytosine.
To prepare the internally labeled 42-mer, an ODN containing a 5’-terminal mC was [γ-32P]-end-labeled and then ligated to a second ODN using a 20-mer scaffold. The ligated product was then separated from the ligation scaffold by denaturing gel electrophoresis in the presence of a six-fold excess of an ODN that was complementary to the scaffold to ensure that we only obtained the single-strand form of the 42-mer. The duplex form was then prepared by annealing the 32P-labeled 42-mer to the complementary 42-mer. Irradiation of single-strand or duplex 42-mer was performed at 302 nm (UVB light) for one hour at 4 °C and pH 8.3 to minimize the amount of deamination.14 Under these conditions, cis-syn dimers, (6–4) photoproducts and their Dewar isomers are the major products, whereas trans-syn and TA* photoproducts very minor. After irradiation, the samples were adjusted to pH 7 and allowed to deaminate at 37 °C for specific times and then the cis-syn dimers were specifically photoreverted with 100-fold excess of E. coli photolyase.33
After photoreversion, the DNA was degraded to 5’-mononucleotide phosphates with nuclease P1 to generate 32pmC/C and 32pT/U, which were separated by a 2D acrylamide gel system (Fig. 3A). All non-photorevertible dipyrimidine photoproducts are digested to trinucleotides, pPyPyN.34 Nuclease P1 interfered with migration of the nucleotides in the citrate gel and could not be used to separate 32pmC/C and 32pT/U directly by one single dimension. We therefore ran a 10% acrylamide gel containing TBE and 7 M urea to separate 32pmC/C and 32pT/U as a single intense band as a first dimension step (shown in rectangle). Because of the low percentage of acrylamide, this band also contained some incompletely digested di- and tri-nucleotides products as well as photoproduct-containing trinucleotides as discussed below.
The second dimension with citrate buffer 35 was used to separate 32pT/U from 32pmC/C, which appear as distinct and reproducible bands of high and low mobility respectively. Oligonucleotides in a citrate gel move primarily according to base composition, and are not very sensitive to size, with electrophoretic mobility increasing in the order C>A>>G>T. A number of intermediate bands of variable amount (<5 %) were also observed, which correspond to oligodeoxynucleotide dimers, trimers and possibly higher, with variable base composition that were incompletely digested by nuclease P1. Control experiments with unirradiated DNA suggested that the incompletely digested products may have arisen from a small fraction of labeled substrate that was in poor contact with NP1 and because each time point was digested separately, are of variable amount. Because NP1 cannot discriminate between undamaged DNA and photoreverted DNA, the calculation of the percentage of deaminated product is not affected by the presence of partially digested products.
Another contributor to the intermediate bands are trimers resulting from digestion of non-photorevertible DNA photoproducts. Experiments with and without photoreversal by photolyase showed that the cis-syn dimer-containing trinucleotide photoproducts also migrated with these bands but are completely eliminated by photoreversal conditions used in our experiment. Further experiments with T4 endo V to quantify cis-syn dimers in 5'-end labeled substrate also showed that under the conditions used, photolyase reverted >95% of TT and TmC cis-syn dimers. The incompletely digested products and non-revertible photoproducts do not interfere with quantification of the pT/U band as very little product was observed at the position of this band at zero deamination time, and any background activity at this time point was subtracted from the other lanes.
To determine the rate of deamination, the yield of cis-syn photodimer was first determined by heating the irradiated DNA to 67°C at pH 6.3 for 16 h to completely deaminate the dimer prior to the photolyase and nuclease P1 steps, and calculating the ratio of the pT/U band to the pmC/C + pT/U band. The half-lives were obtained from a plot of the natural log of the fraction undeaminated dimer versus time. Fig. 3B shows typical plots of the data from different 2-D gels from which the half-lives were calculated.
Deamination of mC in T=mC is faster than in mC=T
In our first experiments, we compared deamination rates of 5-methyl C within mC=T and T=mC dimers flanked either by A’s or G’s. Comparing the same flanking base, the 3'-mC in the dimer deaminates about 3–4 times faster than the 5'-mC in both single-strand and duplex DNA. (Table 1, Fig. 4). This contrasts with the deamination of C in the cis-syn dimers of the dinucleotides TpdC and dCpT, which have similar half-lives of 1.3 and 1.2 hours, respectively, at pH 7 at 50°C.22 This initially suggested that the slower deamination of the 5'-mC is due to steric interference by the 5'-flanking base, which might impede the attack of water on C4 of a 5'-C. In the crystal structure of a cis-syn thymine dimer flanked by A's (Fig. 5 A & B)36, the 3'-flanking base is moved greatly to the side, and would not pose the same degree of steric interference to the attack of water on C4 of a 3'-mC. Single-stranded DNA is also partially stacked and might confer a similar degree of steric interference. On the other hand, mC appears to deaminate almost 200 times faster in the dimer of the dinucleotide TpdmC (5.6 h, pH 7.4 50°C)27 than in the dimer of dmCpT (48 d, 1152 h, pH 7 25°C).23 Barring some experimental error, this large difference in rate suggests that the methyl group interferes more with approach of hydroxide from the 5'-side than the 3'-side due to the cyclobutane ring conformation, which must be different in the dinucleotide compared with single strand or duplex DNA. In support of this idea, the cyclobutane ring conformation of a TT dimer has been shown to be significantly different in duplex DNA compared with the dinucleotide.37
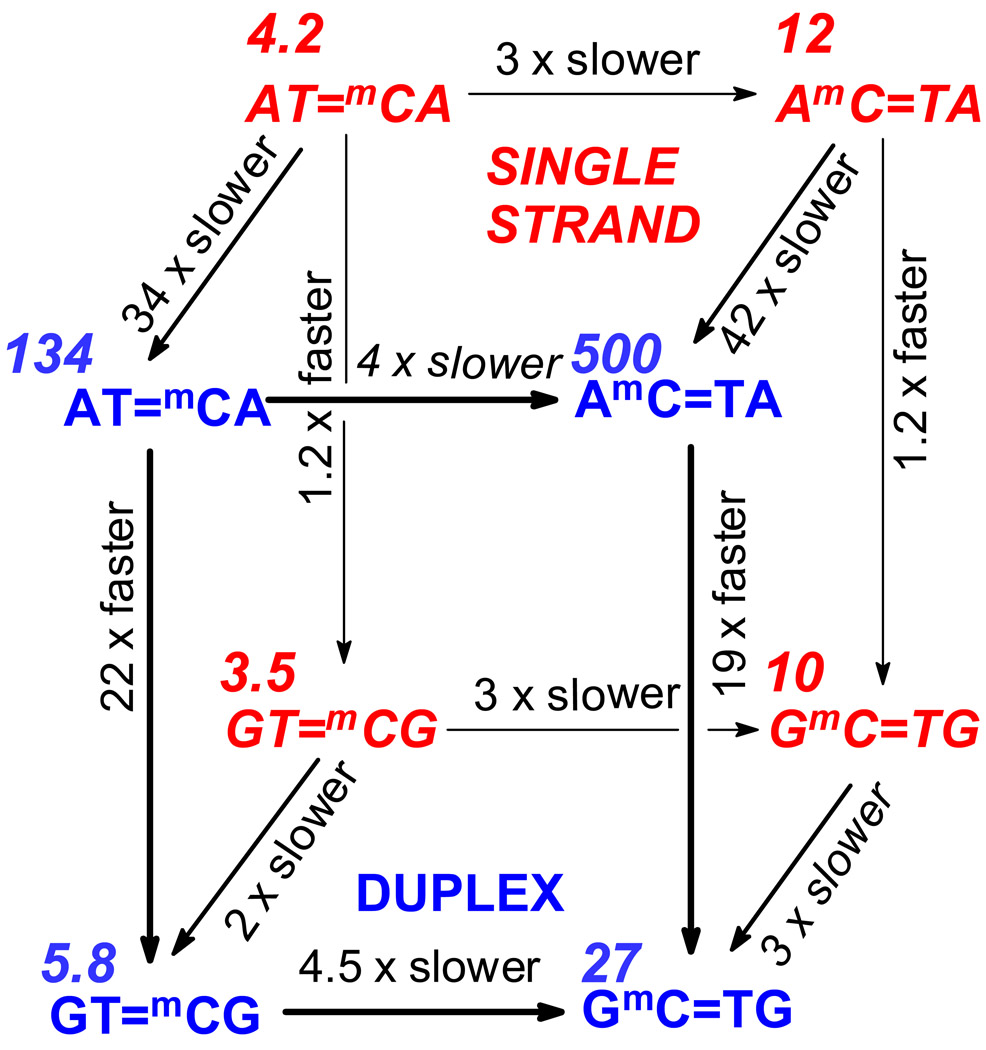
Figure 4.
Effect of sequence context and duplex formation on deamination rate. Sequences in italic and red on the back face of the cube refer to the single stranded form. Sequences in bold and blue on the front face of the cube refer to the double stranded form. Numbers in italics are the deamination half lives in hours.
Figure 5.
Influence of base stacking on nucleophilic attack and exciplex formation in duplex DNA. A) View down helix axis from 3'-end, and B) a side view of a AT=TA cyclobutane dimer (PDB File 1NE4)36 showing that the 5'-A can stack on top of the 5'-T of the dimer and block attack of water, whereas the 3'-A does not stack on the 3'-T and cannot block attack of water. C) View from 3'-end of a ATTA site in duplex DNA (PDB File 1D49) 49 showing significantly more base stacking between the 5'-A and the 5'-T than for the 3'-A with the 3'-T.
Deamination of mC=T and T=mC is fastest when the mC is flanked by G
The deamination half-lives of mC in a CPD flanked by either A or G are very similar in single-strand DNA with values of 3.5 and 4.2 h for GT=mCG and AT=mCA respectively, and 10.7 and 12.1 h for GmC=TG and AmC=TA (Table I, Fig. 4). In contrast, deamination of mC in duplex DNA was fastest when flanked by G’s. The t½ for mC was 22-times shorter for GT=mCG (5.8 h) than for AT=mCA (134 h), and approached that observed in single-strand DNA. Likewise, the t1/2 for deamination in GmC=TG (26.6 h) was 19-times shorter than for AmC=TA (>500 h).
To determine the effect of a flanking C or T on deamination, we determined the deamination rates of mC within TTmCTT and CCmCCC sequences in duplex DNA. Irradiation of these sequences produces a mixture of two dipyrimidine dimers with the mC located either in the 5’ or 3’ positions. Using the cis-syn dimer specific T4 endonuclease V we found that TT=mCT and TmC=TT were produced in a 2:1 ratio, and that CC=mCC and CmC=CC were produced in a 3:4 ratio. The observed deamination rate is therefore a composite of the rates of deamination for mC in the 5' and 3'-positions, which could differ by 4-fold based on what was observed for the deamination of mC flanked by A or G. The observed deamination half-life of 300 h for mC in dimers flanked by C's is about 2–3 times than the 107–160 h determined for deamination of the central two C's in CCCC of plasmid DNA in vitro25 as would be expected from the rate-retarding effect of methylation. The deamination half-life is also similar to the 160 h calculated from the % deamination data for CCC in codon 300 of the p53 gene in irradiated human cells.26 In that same study, the deamination half-life of six TCT sites in the p53 gene was also about 160 h and comparable to that of 385 h that we find for mC in a dimer flanked by T's. These deamination rates are very different, however, from that of 3.9 h reported for a C in TCT sequence tRNA suppressor gene in E. coli.21,38 This site in E. coli is actively transcribed, which would make the site more single-stranded and consequently, more prone to deamination. Since the deamination half-lives were very long for dimers flanked by T and C in our substrates and similar to those dimers flanked by A’s, we conclude that G is unique in its ability to accelerate the deamination of C. Only the G immediately flanking a mC had this effect with AT=mCG deaminating much more rapidly than GT=mCA (t ½’s of 6.6 and 170 h respectively) (Table 1).
O6 of guanine implicated in the deamination mechanism
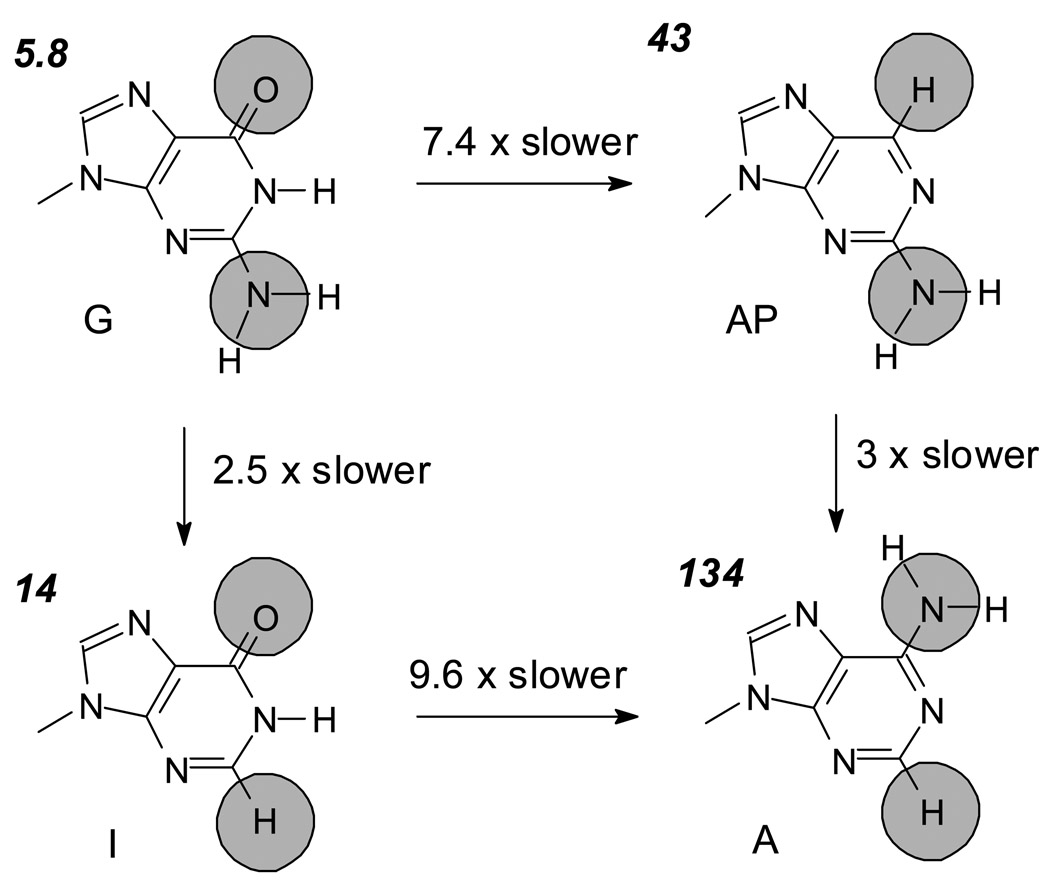
Spontaneous deamination of C or mC within a dinucleotide photoproduct is a general acid catalyzed reaction.22 The reaction is initiated by the protonation of cytosine at N3, which induces a positive charge at C4. Attack of hydroxide or water on C4 then produces a tetrahedral aminal intermediate that collapses to release ammonia or ammonium ion. To determine whether or not a particular functional group on G might be involved in the mechanism, we replaced the G’s flanking the photodimer with various analogs lacking specific functional groups of G (Fig. 6). When G was replaced by 2-aminopurine, which lacks the C6-carbonyl group but retains the C2-amino group, the deamination rate of mC in T=mC slowed by 8 fold. When G was replaced with inosine, which lacks the C2-amino group, but retains the carbonyl group, the rate only slowed 2 fold. Based on these results we conclude that O6 and possibly N7 of guanine are somehow involved in directing or activating water for attack at C4. Consecutive guanines as in GT=mCGGG did not enhance the effect (Table 1) and suggests there is no cooperative effect between adjacent guanines.
Figure 6.
Influence of individual functional groups of a 3'-G on the deamination rate of the 3'-C of the dimer in tGT=mCGtc. Values in italics are the half lives in hours at 37°C.
The O6 carbonyl group of G together with N7 may form a region of high negative electrostatic potential 39 that stabilizes the positively charged protonated C effectively raising the pKa to accelerate the reaction. This explanation is similar to the one used to explain why the cis-syn dimer of TpdC deaminates more quickly than the trans-syn-I dimer.22 In that case, it was found that the pKa of the protonated C in the trans-syn dimer was lower (2.9 vs 4.2), and attributed to a favorable alignment of the C4 carbonyl of T with the iminium ion of C located on the same side of the cyclobutane ring. In the trans-syn-I dimer, the O4 carbonyl of the T is on the opposite side of the cyclobutane ring and cannot interact as effectively. While the difference in rate is substantial at pH 4 (24-fold), it is much less at pH 7 (3-fold) than the 22-fold difference observed for deamination of mC flanked by G compared to A, C or T. G may come closer in the structure to achieve better alignment of its O4 carbonyl group (Fig. 5) compared with the O4 carbonyl of flanking T, while A and C possess an amino group with a less favorable dipole.
Effect of salt on deamination
We tested the effect of salt on deamination since mono and divalent cations are known to interact with both N7 and O6 of guanine by inner-sphere and water mediated interactions40 and stabilize the duplex form.41 Deamination was carried out in 0–400 mM added NaCl with duplex GT=mCG and AT=mCA (Fig. S1). At the highest NaCl concentration, the deamination rate slowed by a factor of 3.8 for the G sequence but also slowed by a similar factor of 3.3 for the sequence flanked by A’s. Surprisingly, the G flanked sequence deaminated 20-fold faster than the A flanked sequence over the entire range of salt concentrations. Consistent with the observation that both mono and divalent cations compete for the same sites in DNA,42 we found that the rate of deamination of GT=mCG decreased by a factor of 4.3 in 10 mM MgCl2. From these results, it appears that salt may slow down deamination by stabilizing the duplex form of the helix in a non-sequence specific manner, thereby inhibiting protonation of the C and addition of water. These results also indicate that while salt may not affect the relative rates of deamination between different sequences, deamination may be quite slow in vivo, where monovalent salt concentrations are about 150 mM.
Effect of methylation on deamination of C
Although it is known that C methylation greatly increases CPD formation in sunlight by about 15-fold,9 the effect of methylation on the deamination rate of photodimers in duplex DNA is not well established. We found that methylation of the C in GT=CG, AT=CG, and AT=CA slowed deamination down by factors of 1.2, 1.3 and 3.8 respectively (Table 1). It appears that the deamination rate of a 3'-C is more affected by methylation when flanked by an adjacent A than by a G. Deamination of a 5'-C flanked by A was also slowed down by a factor of 3.8 upon methylation (compare t1/2 of 500 h for AmCTA (Table 1) with 131 h for ACTG (Table 2)).
Table 2.
Formation and deamination of CmC dimers. 32P-labeled C in boldface. Mismatches are underlined.
| Entry | Abbrev. | sequence | % Yield | t1/2 |
|---|---|---|---|---|
| 1 | ACmCA | ACmCAt | 3.4 | nd |
| TG GTa | ||||
| 2 | GCmCG | GCmCGt | 1.4 | nd |
| CG GCa | ||||
| 3 | ACmCG | ACmCGt | 2.8 | 320 ± 12 |
| TG GCa | ||||
| 4 | ACmCG | ACmCGt | 3.0 | 256 ± 16 |
| TG GCa | ||||
| 5 | AUmCG | AUmCGt | 0.8 | 4.7 ± 1 |
| TG GCa | ||||
| 6 | ACTG | ACTGg | 3.9 | 28 ± 2.6 |
| TGGCc | ||||
| 7 | ACTG | ACTGg | 0.8 | 131 ± 12 |
| TGACc |
The NPyPyNn sequence shown is embedded in the 42-mer single strand sequence: d(GTGTATGTATGTGTATGTNXYNnCGTGTATATATGTGTATGT). The double strand sequence is formed by annealing to its fully complementary strand except were underlined.
Comparing the estimated deamination rate for C in the cis-syn dimer TpdC at 25°C at pH 7 (half life of 7.7 h)22 with methylated C at pH 7.4 (69 h)27 indicates that methylation of a 3'-C slows deamination by 9-fold. The effect appears to be even greater (172-fold) for 5'-C comparing the deamination rates of dCpT (6.7 h) and dmCpT (48 days). A possible explanation may involve the cyclobutane ring conformation as discussed above (see Deamination of mC in T=mC is faster than in mC=T). Our results with duplex DNA are more in accord with an in vitro PCR experiment with irradiated plasmid DNA that indicates little effect of methylation on deamination of CmCG at codon 248 or 282.26
Double deamination of C=mC to U=T
Both C residues in a C=C cyclobutane dimer can individually deaminate to ultimately yield U=U and thereby result in a CC→TT mutation after replication.25,26 To dissect the mechanism of the double deamination, we first tried to measure the deamination rates of C and mC in the sequence GC=mCG, but the yield of this product was not optimal (Table 2). We found that replacing the 5'-G with A increased the yield, and that both the C and the mC deaminate very slowly and at nearly the same rates (t ½ lives were 320 and 256 h respectively) (Table 2).
The slow deamination of the mC flanked by G was surprising, since we had found that G greatly accelerates the deamination of TmC dimers. In AC=mCG, deamination of the C will lead to AU=mCG, whereas deamination of mC will lead to AC=TG, where the underlined bases are mispaired to G. Deamination of either C or mC was therefore expected to accelerate the deamination of the remaining base, since a base pair mismatch should enhance the accessibility of the C to protons and water through duplex destabilization. Indeed, we found that the deamination half lives of AC=TG and dAU=mCG with a G mismatch opposite the deaminated base were 28 and 4.7 h, respectively, and 11.4 and 54-fold faster than the corresponding sites in AC=mCG (Table 2). The U•G mismatch contributed very little to the 54-fold increase in the deamination rate of the 3'-mC as judged by only a 1.5-fold increase in the deamination rate of mismatched AU=mCG compared to matched AT=mCG (Table 1). The T•G mismatch, however, appeared to be the major factor for the 11.4 increase in the rate of deamination of the 5'-C, as judged from the 4.7-fold increase in rate of deamination of AC=TG compared to AC=TG. Thus it would appear that the 5'-C in AC=mCG exerts a much greater inhibitory effect on deamination of the 3'-mC than does the 3'-mC on the 5'-C. The slow overall deamination rate of AC=mCG can thus be explained by a rate limiting first deamination step that is of similar rate for both the 5'-C and the 3'-mC followed by a fast second deamination step.
Our results are consistent with the in vitro rate of deamination of CC dimers in a CCCC sequence in a plasmid DNA by a reversion assay, 25 A rate-limiting first deamination step with a half-life of 107–160 h was inferred since single C→T mutations plateaued at about 10%, while the frequency of CC→TT mutations increased to about 90% over time. A two step deamination mechanism with a rate limiting first deamination also fits the observed reversion frequencies induced by a CC dimer in a CCC sequence in E. coli.29 By fitting the mutation frequency to an analytical expression for two sequential first order steps the second deamination step was calculated to be 7.6 times faster than the first step. In contrast to the results of the in vitro deamination studies, the half-life for the first and second deamination steps were calculated to be 2.7 h and 21 min respectively, which is indicative of some other process, such as transcription, that would disrupt the duplex nature of the DNA.
The slower deamination of a CC dimer was first predicted by Lemaire and Ruzsicska from their studies of cis-syn and trans-syn-I TpdC dimers.22 As discussed above, (see O6 of guanine implicated in the deamination mechanism) these workers found that the trans-syn-I dimer deaminates slowler than the cis-syn dimer, which they attributed to a favorable dipole interaction between the O4 carbonyl of T with the imininum ion of the protonated C. On this basis, they argued that a cis-syn CC dimer should deaminate more slowly than a TC dimer because the amino group of the attached C would not have a favorable dipole interaction with the iminium ion of the protonated C. What is puzzling, however, is why mC in dimers of CCmCC which are flanked by C is deaminating at approximately the same rate as the dimer in the ACmCG, which should be accelerated by the 3'-flanking G (Table 1). Equally puzzling is why the mC in dimers of CCmCC, which should have an unfavorable dipole is deaminating at about the same rate as in dimers of TmCT. It may be that the complementary G-tract can slip and misalign with the dimer-containing C tract, thereby disrupting the duplex structure and accelerating deamination. It is also possible that a flanking C, unlike a C fixed in a particular position within the dimer, can help catalyze deamination by helping to direct attack of hydroxide on the protonated C by H-bonding with the C4 amino group of the flanking C.
Sequence and methylation effects on photoproduct yield
In preparing CPDs for deamination studies, we found that photoproduct yield at 302 nm was highly sensitive to flanking sequence (Table 1). We observed higher photoproduct yields when the dipyrimidines were flanked by A’s compared with G's and that this was independent of the position of the mC in the dipyrimidine sequence. A 17 % yield of CPD formation was observed for ATmCA compared to 5.4 % for GTmCG, and a 7.8 % yield of CPD formation for AmCTA compared to 3.4% for GmCTG. The yield was also high for an mC flanked by T's as judged by the combined yield of 15.7% for T=mC and mC=T CPDs in TTmCTT. The yield of CmC CPD’s were lowest (1.4%) when the sequence was flanked by G's in GCmCG, intermediate (2.9%) when flanked by an A and a G, higher (3.4%) when flanked by A's in ACmCA, and highest (6% combined yield) when flanked by C's in CCmCCC. Methylation did not increase the photoproduct yields with 302 nm light as much as the 15-fold increase reported for simulated sunlight, only increasing the yield for GT=CG, AT=CA, AT=CG and AC=TG by 5.4, 2.4, 2.4 and 8-fold, respectively (Tables 1 & 2).
Recent experiments have shown that a thymine dimer is produced in < 1 ps following UV excitation, which is faster than DNA can change conformation, suggesting that dimer yield is related to the proportion of photoreactive conformations.43 This proposal has been supported by molecular dynamics calculations correlating thymine dimer yield with conformation.44 Other factors can also enhance or diminish the yield of cyclobutane pyrimidine dimer formation by light, including photosensitization, and both direct and electron-transfer mediated photoreversal of the dimer. Because of the many potential factors, it is hard to attribute the observed dimer yield to any single cause. Some factors, however, are not expected to contribute greatly at 302 nm, such as direct photoreversal, due to the low absorptivity of pyrimidine dimers.22 Likewise base-mediated photoinduced electron transfer is also inefficient at this wavelength, except for the special case of a quadruplex-bearing DNAzyme.45 As for photosensitization, A is at least 10 fold more efficient in excited state energy-transfer than C, G, or T at 256 nm.46 If photosensitization represents a dominant mechanism at 302 nm, the yield of CPD's should be lower with flanking T’s compared with A's, which is not the case.
Correlation of oxidation potential with photoproduct yield
The observed yields are more consistent, however, with an electron-transfer mediated mechanism as revealed by CPD formation in the sequence NTmCN where N = G, A, and inosine (I). When G is replaced by A, which has a higher oxidation potential, the yield increased from 5.4% to 17%. When the A's are replaced with inosine (I), which has an even a higher oxidation potential,47 the yield further increased from 17% to 29%. We also found that consecutive G’s, which have a lower oxidation potential than a single G,48 decreased photoproduct yield. Dimer formation in ggGTmCG and GTmCGgg was lower (3.9 and 3.4% yields respectively) compared with GTmCG (5.4%). One possible explanation for these results is quenching by electron-transfer of the pyrimidine excited state by the flanking base. This would result in the formation of an exciplex that would deactivate by back electron transfer. When the flanking G's in GTmCG are replaced with 2-aminopurine (AP), which has a similar oxidation potential to A and therefore expected to increase the yield, the yield dropped from 5.4 % to 2.8 %. We attribute this drop in yield to competitive absorption of the 302 nm light by AP which has a red-shifted absorption maximum at about 320 nm.
In further support of an exciplex mechanism, we observe a lower photodimer yield when G is located in the 5’ position relative to the dipyrimidine site. When the 5'-A in the sequence ATmCA is replaced by G, the yield drops about 50% from 17% to 8.8%, but if the 3'-A is replaced by G, the yield only drops by about 30% to 12%. This is consistent with better stacking at Pu-Py sequences compared to Py-Pu sequences (Fig. 5C),49 that would facilitate electron transfer and exciplex formation. Interestingly, the effect of flanking A, C, G and T on the yield of photoproduct induced at 302 nm parallels the yield observed at 254 nm, which has been explained by a competition between dimer formation and photoinduced electron transfer-mediated reversion.50
Implications for the origin of sunlight-induced C to T mutations
In our study, we found that a 3'-flanking G has the unique ability to increase the deamination rate of mC in a TmC cyclobutane pyrimidine dimer, but not in a CmC dimer, which occurs about 50 times slower. If one accepts the deamination bypass mechanism as the principal pathway for C to T mutations, we would then expect that these mutations would occur more frequently at TmC sites than at CmCG sites as has been observed in a SupF system.30 On the other hand, CmCG and not TmCG are the major mutation hotspots in the p53 gene of skin cancers. Of 86 C → T mutations at 8 methylated CCG and 3 TCG sites in non-XP squamous and basal skin cancers, 81 (97%) were at CmCG sites (Table 3).51 The lower than expected frequency of TCG → TTG mutations can be explained, in part, by the very low frequency observed for this mutation at two of the these three TmCG sites in the p53 gene in all cancers, indicating that they are not as tumor promoting as those at the CmCG sites..
Table 3.
Analysis of mutation spectra of non-XP, squamous cell and basal cell carcinomas of the skin at 11 PymCG sites from the p53 database.a
| mCG codon | non-transcribed /transcribedb |
CC→TC | CC→CT | CC→TT | TC→TT | XPC |
|---|---|---|---|---|---|---|
| 152 | CCG/na | 8/na | ||||
| 158 | CCG/na | 1/na | 1/na | |||
| 196 | CCG/TCG | 13/0 | 2/0 | 2/0 | ||
| 213 | TCG/TCG | 3/0 | ||||
| 245 | na/CCG | na/2 | na/5 | na/2 | ||
| 248 | CCG/CCG | 12/9 | 9/1 | 6/0 | ||
| 282 | CCG/CCG | 9 | 13 | 6/0 | ||
| Total | 11 sites | 10 | 45 | 28 | 3 | 14 |
Data from: IARC TP53 Mutation Database (http://www-p53.iarc.fr/) (ref 38). Non-XP Basal cell carcinoma data from: Basal cell carcinoma NOS (C44._) data set restricted to the skin with XP germline mutations filtered out. Non-XP Squamous cell carcinoma data from: Squamous cell carcinoma NOS data set restricted to the skin and with XP germline mutations excluded.
Both the transcribed and non-transcribed strands contain a mCG sequence. If the strand contains a PymCG sequence capable of dimer-mediated C→T mutations, the sequence is given. If the strand does not contain a PymCG sequence, the abbreviation na for not applicable is given.
The high frequency of C→T mutations at CmCG in the p53 gene of skin cancers also suggests that dimers at these sites must be repaired slowly enough to allow deamination to occur prior to pol η replication. In support of this notion, it has been found that only 20% of dimers in CHO cells were repaired in 6 h, and only 25–50% after 24 h.52 The initial rapid removal of 20% of the dimers is likely due to transcription coupled repair, which has been shown to remove 70% of CPDs within 24 h.53 Transcription-coupled repair is known to preferentially remove dimers from the transcribed strand of the p53 gene, 54 which would explain why 71/84 mutations at the 8 CmCG sites are in the non-transcribed strand. It has also been found that 25–32% of cis-syn dimers at two CmCG sites in exon 8 of the mouse p53 gene remain after 48 hours.7 Thus it is likely that some fraction of dimers may persist for hundreds of hours in untranscribed regions as well as in untranscribed strands, giving them sufficient time to deaminate. Transcription, however, might also accelerate deamination in non-transcribed strands due to disruption of the duplex structure.
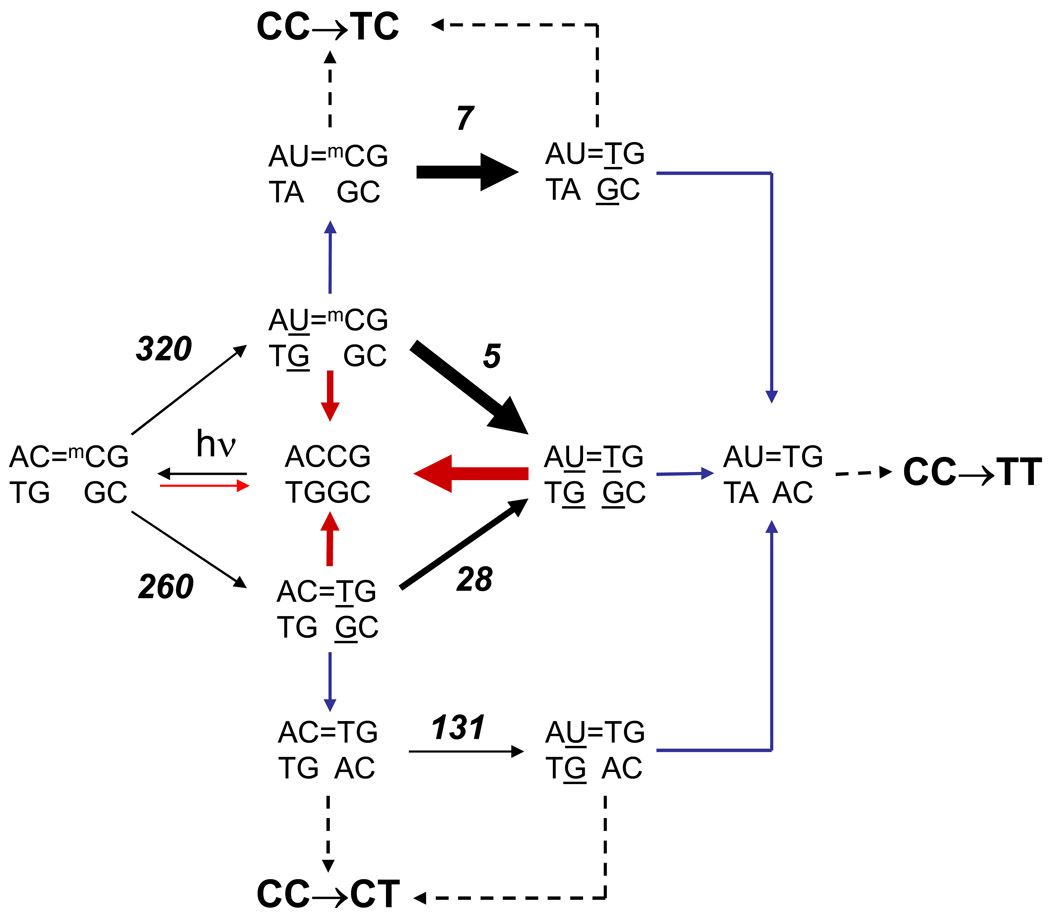
While slow repair of C=mC dimers may explain the high frequency of C→T mutations at CmCG sites, it does not explain the much higher percentage of single C→T mutations than tandem CC→TT mutations at these sites. Of 83 C→T and CC→TT mutations at the 8 CmCG sites, 8 are CC → TC and 45 are CC→CT single base mutations (66%), whereas only 28 are CC → TT tandem mutations (34%). Since the second deamination step in tandem deamination is much faster than the first, the doubly deaminated product is calculated from the kinetic data (Table 2) to predominate after only about 5% of the dimer has deaminated (Fig. S2). Unless replication of the dimers only takes place during this first 5% of reaction, an alternative explanation for the high percentage of single C→T mutations is needed. One possibility is that excision repair of the doubly deaminated dimer (U=T•GG) is faster than the singly deaminated dimers (U=mC•GG and C=T•GG), which would suppress CC→TT mutations as shown in the scheme in Fig. 7. We have previously found that a T=T•GA mismatched dimer destabilizes the DNA duplex by 0.7 kcal compared to a matched T=T•AA dimer,55 suggesting that a double mismatch would be even more destabilizing, and hence more easily detected by the excision repair system. Indeed, a doubly mismatched dimer, T=T•GG, is repaired faster than the singly mismatched dimer T=T•GA, which is repaired at a similar rate to T=T•AG, both of which are repaired much faster than T=T•AA.56,57
Figure 7.
Proposed scheme showing the possible deamination/bypass pathways leading to C→T and CC→TT pathways at an ACmCG site. Black arrows refer to chemical steps, red arrows to excision repair steps, blue arrows to pol η bypass steps, and dotted arrows to either excision repair, or replication of the undamaged strand. The thickness of the arrows represents the relative rates of deamination (experimentally determined) or repair (proposed). Values in italics are the experimentally determined half lives in hours at 37°C.
In further support of our scheme (Fig. 7), the frequency of CC→TT mutations is much higher in skin cancers from XPC patients than in non-XP patients (76% vs 8%) and occurs almost exclusively in the non-transcribed strand.58,59 At the 8 CmCG hotspots, only CC → TT mutations were observed. Sufferers of XPC lack the DNA damage recognition component of nucleotide excision repair, and thus C=C dimers in the non-transcribed strand have enough time to completely deaminate and cause CC → TT mutations. Our scheme also would predict that CC→CT mutations would predominate over CC→TC mutations because while both U=mC and C=T dimers are produced at the same rate, U=mC is more rapidly depleted due to the accelerating effect of G on deamination. In accord with this, 45/53 of the C→T mutations at the 8 CmC hotspots are CC→CT mutations. Because these CC→CT mutations correspond to missense mutations, it is possible that they predominate because of some selective survival advantage conferred by the mutant protein, and not because of selective deamination
Conclusion
Sunlight produces cis-syn dimers at PymCG sites, which correlate with C to T and CC to TT mutational hotspots found in basal and squamous cell carcinomas. Our results are consistent with a mutagenic pathway that involves spontaneous deamination of 5-methylcytosine to T in a CPD followed by error-free trans-lesion synthesis by pol η resulting in insertion of A opposite the T. We have discovered that deamination of mC in a CPD in duplex DNA is highly sequence dependent and is fastest for a TmCG site, and slowest for a CmCG site. The observation that a C=mCG dimer deaminates the slowest, but is at the same time a hotspot for C to T and CC to TT mutations in skin cancers, suggests that repair must be slow enough at these sites to allow sufficient time for deamination. The higher incidence of C→T than CC→TT mutations at CmC sites, in spite of a very rapid second deamination step, might be explained by more rapid repair of the doubly deaminated dimer than the singly deaminated dimer. Our deamination data provides a baseline of comparison for future studies aimed at dissecting the effects of DNA structure, dynamics, and protein interactions on photoproduct formation, deamination, repair, and bypass, which ultimately govern the overall mutation frequency.
Experimental Procedures
Materials
Oligonucleotides were from IDT. [γ-32P]ATP was from Amersham. Acrylamide was from EMD. N’N’ methylene bisacrylamide, Tris base, boric acid, Mes, EDTA, citrate, formamide, SDS, xylene cyanol, NaCl, MgCl2, ZnCl2, DTT and P1 nuclease were from SIGMA. T4 polynucleotide kinase was from NEB. T4 DNA ligase was from Promega. E. Coli photolyase was a gift from Chris Selby and Aziz Sancar, and T4 endo V from R. Stephen Lloyd.
Preparation of site-specifically 32P-labeled substrates
In a typical experiment, 7 µg of a gel purified 21-mer oligodeoxynucleotide containing a 5’-mC was end-labeled with 2 µl of T4 polynucleotide kinase in a 40 µl reaction mix containing 50–100 µC [γ-32P]ATP. The reaction was allowed to proceed at 37 °C for 1 h and then terminated by heating to 100 °C for 40 min. The 5'-32P-labeled labeled 21-mer oligonucleotide was ligated to a second 21-mer ODN using a short complementary ligation scaffold of 20 nucleotides. Both the second ODN and the scaffold were added in 2-fold molar excess over the 32P-end labeled ODN in a volume of 144 µl. This mixture was heated to 80 °C for 5 min and then allowed to slowly cool down to room temperature before adding 16 µl of 10× ligation buffer supplied by the vendor and 4 µl T4 DNA ligase. The reaction was allowed to proceed overnight at 23 °C. The next day, the reaction mixture was heated to 100 °C for 10 min to inactivate the ligase and the sample then divided into two 80 µl aliquots. A 20-mer ODN (42 µg) complementary to the scaffold was added to one aliquot (6 fold molar excess over the scaffold in that aliquot). This was done in order to minimize any possible re-annealing of the scaffold and ligated DNA in subsequent heating and acrylamide gel electrophoresis steps. No additions were made to the second aliquot. It is important to purify the ligated product to ensure that all radiolabeled nucleotides resulting from subsequent NP1 digestion arise only from the ligated sequence. Therefore, each aliquot was evaporated to dryness at 43 °C, resuspended in 30 µl formamide containing xylene cyanol dye marker and then heated to 100 °C for 10 min before loading a 1 mm thick acrylamide slab containing TBE and 7M urea. The excess scaffold and its complementary sequence are easily separated by the acrylamide gel giving close to 100% yield of single-strand ligated 42-mer product. The second sample, without the complementary scaffold sequence, targeted for duplex DNA production also ran primarily as the single-strand ligated product. In each case, phosphorimaging was used to locate the single-strand product. A 1 × 6 cm acrylamide rectangle containing each internally labeled ligated product was cut out and crushed with a glass rod in a 50 ml plastic centrifuge tube. The internally labeled DNA was eluted in 5 ml water after 2 days at room temperature with rapid agitation by a magnetic stir bar. After elution, the sample was centrifuged to pellet any polyacrylamide and the supernatant containing the DNA was then evaporated to 600 µl. The DNA was pelleted by ethanol precipitation with 0.5 M NaCl and 3 volumes of ethanol. The sample was re-precipitated and then washed with 95% ethanol. The DNA pellet was air dried in a 37 °C oven for 30 min before re-suspension in 200 µl of 10 mM Tris-HCl, pH 8.3. The 42-mer ODN (28 µg) complementary to the 42-mer ligation product, was added to the sample targeted for duplex deamination studies and heated to 100 °C followed by slowly cooling to room temperature.
Irradiation and deamination
Each sample (about 200 µl) was irradiated at 302 nm for 1 h at 4 °C on a model TR-302 Spectroline transilluminator light box in a 4 °C cold room at pH 8.3. The samples were spotted in 100 µl aliquot droplets onto a single layer of clear plastic cut from a Ziploc bag. After irradiation, the separate 100 µl aliquots were pooled and the volume brought up to 200 µl with the addition of water. We observed about a 30% loss in volume due to some evaporation. Mes buffer pH 6.0 was added to each sample to bring the pH to 7.0 which introduced about 5 mM NaCl and stored frozen. Deamination was carried out by quickly heating a frozen sample (~200 µl) to 37 °C after which 10 µl aliquots were removed at time intervals and quick-frozen on dry ice. The collected deamination samples were stored frozen at −70 °C until the photolyase step. After all the time points were collected the remaining sample was brought to pH 6.3 and incubated at 67 °C for 16 h for complete deamination. At 67 °C the 42 basepair duplex dissociates into single-strands, which deaminate rapidly at the lower pH. After the 16 h incubation, the pH was brought back up to pH 7 by the addition of Tris buffer.
Photoreversion and degradation
NaCl and DTT (50 mM and 1 mM respectively in a total volume of 0.35 µl) were added to each 10 µl deamination sample. This was followed by the addition of 0.2 µl (~1 µg) photolyase. This mixture was incubated at 23 °C for 5 min in the dark before incubation under a black light at 23 °C for 60 min. Each microfuge tube (1.5 ml size) was placed on its side so that the light source was roughly 1.3 cm from the photolyase reaction mixture at the bottom of the tube. A maximum of 20 tubes could be placed under the 32 cm length bulb leaving 4 cm at the ends of the bulb vacant. After photoreversion, ZnCl2 (1 mM) was added to each sample in addition to enough Mes buffer to lower the pH to 6.2. P1 nuclease (7 µg in 1 µl volume) was then added to each sample and the reaction allowed to proceed for 1 h at 23°C. Samples after the reaction were stored at −20°C.
Two-dimensional acrylamide electrophoresis and analysis
Sonicated-denatured salmon sperm DNA (5 µl of 10 mg/ml) was added to each sample follwed by 18 µl of formamide containing 0.1 % SDS and xylene cyanol dye marker. An 5 µl aliquot of each sample (without heating) were loaded onto a 1 mm thick, 30.5 × 35 cm 10% acrylamide gel containing 7 M urea and TBE buffer. The wells were 0.5 cm wide and two wells were left empty between each sample. The xylene cyanol dye was allowed to migrate 10 cm before phosphorimaging. A long razor blade tool was used to cut a 4 × 25 cm rectangle in the gel that included the mononucleotide band. The rest of the gel was removed leaving that piece of gel attached to the glass plate. Spacers and glass plate were then put in position to pour the 2nd dimension. The 2nd dimension was a 28 % citrate acrylamide gel containing 7 M urea, which was poured and allowed to polymerize vertically. Pouring the gel vertically provides much better contact with the top of the 1st dimension of the TBE gel. The gel was poured in three steps. First, 10 ml of the citrate gel was poured and allowed to polymerize at the bottom of the slab, providing a plug. We found that sealing the gel form with agarose alone was not effective in stopping leaks when pouring the gel in a vertical position. Second, the gel form was filled with 50 mM citrate and allowed to stand for 30 min and then poured off. We found that TBE inhibits polymerization and results in poor contact with the 1st dimension gel. Third, the rest of the citrate gel could be poured at this point. Large amounts of polymerization reagents were required for the citrate gel in order to obtain good contact with the 1st dimension gel (for 150 ml acrylamide we used 3.25 mg FeSO4, 130 mg ascorbate and 410 µl of 3% H2O2). At these high concentrations, the gel would polymerize in about 3 min. The 2nd dimension gel was run for 4 h, at 400 Watts. Phosphorimaging was used to quantify the 32P-T (or 32P-dU) and 32P-mC bands in the 2nd dimension citrate gel, and the fraction of deaminated band relative to the undeaminated band, F(t), corresponding to pT/U/(pT/U+pC), was calculated for each time point. The fraction of pT/U in the sample that had been heated at 16 h at pH 6.3 to completely deaminate the dimer was taken to correspond to F(∞) and the yield of the cis-syn photodimer. The deamination half lives were obtained from the slope of a linear least squares fit to a plot of the natural log of the fraction of undeaminated product, ln[(F(∞)-F(t))/F(∞)], vs time.
Supplementary Material
Acknowledgments
We thank Chris Selby and Aziz Sancar for a generous gift of photolyase, R. Stephen Lloyd for T4 endonuclease V, and NIH Grant CA40463 for support.
Abbreviations used
- aP
2-aminopurine
- CPD
cyclobutane pyrimidine dimer
- I
inosine
- mC
5-methylcytosine
- ODN
oligodeoxynucleotide
- Pu
purine
- Py
pyrimidine
- TBE
tris borate EDTA
- XPV
xeroderma pigmentosum variant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at
References
- 1.Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziegler A, Leffell DJ, Kunala S, Sharma HW, Gailani M, Simon JA, Halperin AJ, Baden HP, Shapiro PE, Bale AE, Brash DE. Mutation hotspots due to sunlight in the p53 gene on nonmelanoma skin cancers. Proc. Natl. Acad. Sci. U.S.A. 1993;90:4216–4220. doi: 10.1073/pnas.90.9.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumaz N, van Kranen HJ, de Vries A, Berg RJ, Wester PW, van Kreijl CF, Sarasin A, Daya-Grosjean L, de Gruijl FR. The role of UV-B light in skin carcinogenesis through the analysis of p53 mutations in squamous cell carcinomas of hairless mice. Carcinogenesis. 1997;18:897–904. doi: 10.1093/carcin/18.5.897. [DOI] [PubMed] [Google Scholar]
- 4.Ananthaswamy HN, Fourtanier A, Evans RL, Tison S, Medaisko C, Ullrich SE, Kripke ML. p53 Mutations in hairless SKH-hr1 mouse skin tumors induced by a solar simulator. Photochem. Photobiol. 1998;67:227–232. doi: 10.1562/0031-8655(1998)067<0227:mihshm>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Queille S, Seite S, Tison S, Medaisko C, Drougard C, Fourtanier A, Sarasin A, Daya-Grosjean L. p53 mutations in cutaneous lesions induced in the hairless mouse by a solar ultraviolet light simulator. Mol. Carcinog. 1998;22:167–174. doi: 10.1002/(sici)1098-2744(199807)22:3<167::aid-mc4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Drouin R, Therrien JP. UVB-induced cyclobutane pyrimidine dimer frequency correlates with skin cancer mutational hotspots in p53. Photochem. Photobiol. 1997;66:719–726. doi: 10.1111/j.1751-1097.1997.tb03213.x. [DOI] [PubMed] [Google Scholar]
- 7.You YH, Szabo PE, Pfeifer GP. Cyclobutane pyrimidine dimers form preferentially at the major p53 mutational hotspot in UVB-induced mouse skin tumors. Carcinogenesis. 2000;21:2113–2117. doi: 10.1093/carcin/21.11.2113. [DOI] [PubMed] [Google Scholar]
- 8.You YH, Pfeifer GP. Similarities in sunlight-induced mutational spectra of CpG-methylated transgenes and the p53 gene in skin cancer point to an important role of 5-methylcytosine residues in solar UV mutagenesis. J. Mol. Biol. 2001;305:389–399. doi: 10.1006/jmbi.2000.4322. [DOI] [PubMed] [Google Scholar]
- 9.Tommasi S, Denissenko MF, Pfeifer GP. Sunlight induces pyrimidine dimers preferentially at 5-methylcytosine bases. Cancer. Res. 1997;57:4727–4730. [PubMed] [Google Scholar]
- 10.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 11.Yu SL, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase eta for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 13.Takasawa K, Masutani C, Hanaoka F, Iwai S. Chemical synthesis and translesion replication of a cis-syn cyclobutane thymine-uracil dimer. Nucleic Acids Res. 2004;32:1738–1745. doi: 10.1093/nar/gkh342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vu B, Cannistraro VJ, Sun L, Taylor JS. DNA synthesis past a 5-methylC-containing cis-syn-cyclobutane pyrimidine dimer by yeast pol eta is highly nonmutagenic. Biochemistry. 2006;45:9327–9335. doi: 10.1021/bi0602009. [DOI] [PubMed] [Google Scholar]
- 15.Masutani C, Kusumoto R, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA Polymerase eta. Nature. 1999;399:700. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 16.Wang YC, Maher VM, Mitchell DL, McCormick JJ. Evidence from mutation spectra that the uv hypermutability of xeroderma-pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 18.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 19.Setlow RB, Carrier WL. Pyrimidine dimers in ultraviolet-irradiated DNA's. J. Mol. Biol. 1966;17:237–254. doi: 10.1016/s0022-2836(66)80105-5. [DOI] [PubMed] [Google Scholar]
- 20.Liu F-T, Yang NC. Photochemistry of cytosine derivatives. 1. Photochemistry of thymidylyl-(3'→5')-deoxycytidine. Biochemistry. 1978;17:4865–4876. doi: 10.1021/bi00616a003. [DOI] [PubMed] [Google Scholar]
- 21.Fix D, Bockrath R. Thermal resistance to photoreactivation of specific mutations potentiated in E. coli B/r ung by ultraviolet light. Mol. Gen. Genet. 1981;182:7–11. doi: 10.1007/BF00422759. [DOI] [PubMed] [Google Scholar]
- 22.Lemaire DGE, Ruzsicska BP. Kinetic analysis of the deamination reactions of cyclobutane dimers of dTpdC and dCpdT. Biochemistry. 1993;32:2525–2533. doi: 10.1021/bi00061a009. [DOI] [PubMed] [Google Scholar]
- 23.Douki T, Cadet J. Formation of cyclobutane dimers and (6-4) photoproducts upon far-UV photolysis of 5-methylcytosine-containing dinucleoside monophosphates. Biochemistry. 1994;33:11942–11950. doi: 10.1021/bi00205a033. [DOI] [PubMed] [Google Scholar]
- 24.Barak Y, Cohen-Fix O, Livneh Z. Deamination of cytosine-containing pyrimidine photodimers in UV-irradiated DNA. J. Biol. Chem. 1995;270:24174–24179. doi: 10.1074/jbc.270.41.24174. [DOI] [PubMed] [Google Scholar]
- 25.Peng W, Shaw BR. Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC->TT transitions. Biochemistry. 1996;35:10172–10181. doi: 10.1021/bi960001x. [DOI] [PubMed] [Google Scholar]
- 26.Tu Y, Dammann R, Pfeifer GP. Sequence and time-dependent deamination of cytosine bases in UVB- induced cyclobutane pyrimidine dimers in vivo. J. Mol. Biol. 1998;284:297–311. doi: 10.1006/jmbi.1998.2176. [DOI] [PubMed] [Google Scholar]
- 27.Celewicz L, Mayer M, Shetlar MD. The Photochemistry of Thymidylyl-(3'-5')- 5-methyl-2'-deoxycytidine in Aqueous Solution. Photochem. Photobiol. 2005;81:404–418. doi: 10.1562/2004-06-15-ra-201.1. [DOI] [PubMed] [Google Scholar]
- 28.Jiang N, Taylor J-S. In vivo evidence that uv-induced C[to]T mutations at dipyrimdine sites could result from the replicative bypass of cis-syn cyclobutane dimers or their deamination products. Biochemistry. 1993;32:472–481. doi: 10.1021/bi00053a011. [DOI] [PubMed] [Google Scholar]
- 29.Burger A, Fix D, Liu H, Hays J, Bockrath R. In vivo deamination of cytosine-containing cyclobutane pyrimidine dimers in E. coli: a feasible part of UV-mutagenesis. Mutat. Res. 2003;522:145–156. doi: 10.1016/s0027-5107(02)00310-x. [DOI] [PubMed] [Google Scholar]
- 30.Lee D-H, Pfeifer GP. Deamination of 5-Methylcytosines within Cyclobutane Pyrimidine Dimers Is an Important Component of UVB Mutagenesis. J. Biol. Chem. 2003;278:10314–10321. doi: 10.1074/jbc.M212696200. [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer GP, You Y-H, Besaratinia A. Mutations induced by ultraviolet light. Mutat. Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 33.Sancar GB, Sancar A. Purification and characterization of DNA photolyases. Methods Enzymol. 2006;408:121–156. doi: 10.1016/S0076-6879(06)08009-8. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Taylor JS, Gross ML. Nuclease P1 Digestion Combined with Tandem Mass Spectrometry for the Structure Determination of DNA Photoproducts. Chem. Res. Toxicol. 1999;12:1077–1082. doi: 10.1021/tx9900831. [DOI] [PubMed] [Google Scholar]
- 35.Gould H, Matthews HR. Separation methods for nucleic acid and oligonucleotides. In: Work TS, Work E, editors. Laboratory Techniques in Biochemistry and Molecular Biology. Amsterdam: North Holland Publishing Company; 1976. [Google Scholar]
- 36.Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor J-S, Garrett DS, Brockie IR, Svoboda DL, Telser J. 1H NMR assignment and melting temperature study of cis-syn and trans-syn thymine dimer containing duplexes of d(CGTATTATGC)•(GCATAATACG) Biochemistry. 1990;29:8858–8866. doi: 10.1021/bi00489a049. [DOI] [PubMed] [Google Scholar]
- 38.Fix D. Thermal resistance of UV-mutagenesis to photoreactivation in E. coli B/r uvrA ung: estimate of activation energy and further analysis. Mol. Gen. Genet. 1986;204:452–456. doi: 10.1007/BF00331023. [DOI] [PubMed] [Google Scholar]
- 39.Bonnaccorsi R, Scrocco E, Tomasi J, Pullman A. Ab initio molecular electrostatic potentials. Guanine compared to adenine. Theoretica Chimica Acta. 1975;36:339–344. [Google Scholar]
- 40.Hud NV, Polak M. DNA-cation interactions: The major and minor grooves are flexible ionophores. Curr. Opin. Struct. Biol. 2001;11:293–301. doi: 10.1016/s0959-440x(00)00205-0. [DOI] [PubMed] [Google Scholar]
- 41.Nakano S, Fujimoto M, Hara H, Sugimoto N. Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai Y, Greenfeld M, Travers KJ, Chu VB, Lipfert J, Doniach S, Herschlag D. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J. Am. Chem. Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreier WJ, Schrader TE, Koller FO, Gilch P, Crespo-Hernandez CE, Swaminathan VN, Carell T, Zinth W, Kohler B. Thymine Dimerization in DNA Is an Ultrafast Photoreaction. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Law YK, Azadi J, Crespo-Hernandez CE, Olmon E, Kohler B. Predicting thymine dimerization yields from molecular dynamics simulations. Biophys. J. 2008;94:3590–3600. doi: 10.1529/biophysj.107.118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chinnapen DJ, Sen D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:65–69. doi: 10.1073/pnas.0305943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu DG, Nordlund TM. Sequence dependence of energy transfer in DNA oligonucleotides. Biophys. J. 2000;78:1042–1058. doi: 10.1016/S0006-3495(00)76663-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crespo-Hernandez CE, Close DM, Gorb L, Leszczynski J. Determination of Redox Potentials for the Watson-Crick Base Pairs, DNA Nucleosides, and Relevant Nucleoside Analogues. J. Phys. Chem. B. 2007;111:5386–5395. doi: 10.1021/jp0684224. [DOI] [PubMed] [Google Scholar]
- 48.Saito I, Nakamura T, Nakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. Mapping of the Hot Spots for DNA Damage by One-Electron Oxidation: Efficacy of GG Doublets and GGG Triplets as a Trap in Long-Range Hole Migration. J. Am. Chem. Soc. 1998;120:12686–12687. [Google Scholar]
- 49.Quintana JR, Grzeskowiak K, Yanagi K, Dickerson RE. Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G. J. Mol. Biol. 1992;225:379–395. doi: 10.1016/0022-2836(92)90928-d. [DOI] [PubMed] [Google Scholar]
- 50.Holman MR, Ito T, Rokita SE. Self-repair of thymine dimer in duplex DNA. J. Am. Chem. Soc. 2007;129:6–7. doi: 10.1021/ja0668365. [DOI] [PubMed] [Google Scholar]
- 51.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 52.Perdiz D, Grof P, Mezzina M, Nikaido O, Moustacchi E, Sage E. Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J. Biol. Chem. 2000;275:26732–26742. doi: 10.1074/jbc.M001450200. [DOI] [PubMed] [Google Scholar]
- 53.Vreeswijk MP, van Hoffen A, Westland BE, Vrieling H, van Zeeland AA, Mullenders LH. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J. Biol. Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 54.Ford JM, Lommel L, Hanawalt PC. Preferential repair of ultraviolet light-induced DNA damage in the transcribed strand of the human p53 gene. Mol. Carcinog. 1994;10:105–109. doi: 10.1002/mc.2940100208. [DOI] [PubMed] [Google Scholar]
- 55.Jing Y, Kao JF-L, Taylor J-S. Thermodynamic and base-pairing studies of matched and mismatched DNA dodecamer duplexes containing cis-syn, (6-4) and Dewar photoproducts of TT. Nucleic Acids Res. 1998;26:3845–3853. doi: 10.1093/nar/26.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol. Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giglia G, Dumaz N, Drougard C, Avril MF, Daya-Grosjean L, Sarasin A. p53 mutations in skin and internal tumors of xeroderma pigmentosum patients belonging to the complementation group C. Cancer Res. 1998;58:4402–4409. [PubMed] [Google Scholar]
- 59.Giglia-Mari G, Sarasin A. TP53 mutations in human skin cancers. Hum. Mutat. 2003;21:217–228. doi: 10.1002/humu.10179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.