Abstract
The anti-epidermal growth factor receptor antibody cetuximab (Erbitux, CTX) is currently used for the treatment of locally advanced squamous cell carcinoma of the head and neck (LA-SCCHN), as yet with modest effectiveness, prompting for the identification of response predictors to this treatment and for the targeting of additional pathways implicated in this disease. Within this scope, we investigated the effect of SRC/STAT pathway components on LA-SCCHN patient outcome. SRC, STAT1, STAT3, STAT5A, STAT5B, ANXA1, CAV1, IGFBP2, EPHA2, EPHB2, and MSN relative gene expression, as well as Stat protein activation, were assessed on LA-SCCHN tumor tissues from 35 patients treated with combined radiotherapy (RT) and CTX-based regimens. Stat1, Stat3, and Stat5 proteins were usually found activated in neoplastic nuclei (70.4%, 85.7%, and 70.8%, respectively). Activated Stat3 and Stat5 were associated with each other (P = .017) and with a CAV1high/MSNhigh/IGFBP2low profile. All patients with tumors expressing high STAT5A/EPHA2 experienced a complete response on RT-CTX-based treatments (12/15 complete responders, P < .0001) and a longer progression-free survival (P = .024). Few tumors expressed high ANXA1/CAV1/EPHA2 and low IGFBP2, a putative dasatinib response-related profile, whereas high ANXA1 was associated with shorter overall survival (P = .003). In conclusion, Stat activation is common in LA-SCCHN, where overexpression of STAT5A and EPHA2 may predict for response to RT-CTX treatments. The STAT5A/EPHA2 profile seems of particular interest for validation in larger cohorts and in multiple tumor types because markers for the positive selection of patients to benefit from CTX-containing treatments are currently lacking.
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) is a disease potentially curable with surgery and radiotherapy (RT) alone when diagnosed at early stages, as is the case for approximately one-third of patients. When diagnosed at advanced stages, however, SCCHN has a high risk for locoregional recurrence and metastasis on surgery and RT or tumors may be nonresectable. Treatment options for patients with locally advanced SCCHN (LA-SCCHN) include RT with concomitant administration of cisplatin or cetuximab ([CTX], Erbitux), a monoclonal antibody that blocks epidermal growth factor receptor (EGFR) activation, or both [1,2]. CTX in combination with RT has received US Food and Drug Administration approval for the treatment of patients with LA-SCCHN [3] based on clinical trial results showing that the drug reduces mortality and may control disease in collectively 50% of the cases [4], whereas it may also be effective in patients with recurrent and/or metastatic SCCHN who fail on platinum therapy [5]. EGFR targeting represents a successful proof-of-concept paradigm regarding the tumorigenic contribution of this receptor in SCCHN, as established in both clinical and experimental settings [6,7]. However, EGFR targeting still remains inefficient for a large proportion of SCCHN patients. Hence, resolving the mechanisms of intrinsic and acquired resistance to CTX in this type of tumors and identifying predictors of response to this drug are largely needed [7,8].
EGFR signaling is accomplished through three main pathways (RAS-MAPK, PI3K-AKT, and SRC-STAT), all of which function aberrantly in most carcinomas mostly on a ground of genetic alterations (mutations, gene amplifications/deletions) in the member molecules. In colorectal and non-small cell lung cancers (NSCLCs), EGFR-activating mutations are associated with response, whereas KRAS and BRAF mutations are associated with resistance to therapeutic EGFR antibodies and kinase inhibitors [9–11]. SCCHNs, however, do not in general carry KRAS and BRAF mutations [12] or classic activating EGFR mutations. Further, although the AKT pathway is usually activated in SCCHN [13], there are as yet no solid data to associate this parameter with disease outcome or treatment response. The third signaling pathway downstream of EGFR seems of particular interest in understanding tumorigenesis and response to anti-EGFR treatments in several carcinoma models. Three of the seven known signal transducers and activators of transcription (STATs) have mostly been studied in SCCHNs, namely, STAT1, STAT3, as well as STAT5A and STAT5B, which are genes closely neighbored to each other and to STAT3. STATs are transcription factors that may play oncogenic roles in the development of SCCHN [14], where Stat proteins may be activated by EGFR and/or SRC [15] or by non-EGFR pathways [16]. STATs act through receptor tyrosine kinases or cytokines [14], whereas their effects on gene transcription and cell function are described as distinct and nonredundant [17,18].
SRC family kinases (v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog, SFKs) and STATs have been associated with resistance to pharmaceutical EGFR targeting in SCCHN. In SCCHN cells in culture, a forced expression of dominant-mutant STAT5 conferred resistance to erlotinib, a small-molecule EGFR inhibitor [19]. Further, elevated levels of active EGFR, MAPK, AKT/PKB, and STAT3 were observed in a model of acquired resistance to CTX, whereas STAT3 was found activated in resistant but not in the parental CTX-sensitive cells [20]. In the same line, SFKs were overactivated in CTX-resistant NSCLC cells and could be blocked by the Src inhibitor dasatinib [21]. SCCHN cell lines also have overactivated SFKs, STAT3, and STAT5 and can be growth-inhibited by dasatinib [22]. The commonly observed activation of the SRC/STAT pathway in experimental systems of SCCHN, especially in relevance to CTX resistance, has served as the rationale for proposing the clinical testing of Src inhibitors in these tumors [6,8].
Dasatinib (BMS-354825) has been developed as a specific Src/Abl inhibitor [23] and is currently in clinical use in imatinib-resistant, Philadelphia chromosome-positive leukemias [24,25], whereas it is also being tested in phase 1 and 2 studies in solid tumors [26,27]. In an attempt to predict for response to this drug, profiling of dasatinib-resistant and dasatinib-sensitive cell lines has been undertaken and dasatinib response gene expression signatures have been described [28,29]. These signatures include Src-inducing and target proteins like the highly homologous closely neighbored caveolins (CAV1 and CAV2), annexin 1A (ANXA1), the ephrin receptors A2 and B2 (EPHA2 and EPHB2), moesin (MSN), insulin growth factor binding protein 2 (IGFBP2), and others. An expression signature of low IGFBP2 and high ANXA1, CAV1, and EPHA2 would predict sensitivity to dasatinib.
On the basis of the previously mentioned data, we investigated the effect of SRC/STAT-related gene expression and protein activation in LA-SCCHN tissues on patient outcome and response to CTX-based treatments. We also assessed the previously described dasatinib sensitivity profile because these patients might be candidates for receiving SRC inhibitors on failure of CTX in the near future. STAT1, STAT3, and STAT5 were expressed, and the proteins were activated in most LA-SCCHN, whereas the dasatinib-response-predicting profile was encountered in few tumors. The most interesting finding of this study was that complete response to CTX was associated with high STAT5A/EPHA2 messenger RNA (mRNA) expression in tumor tissues. If validated in larger cohorts, this profile may prove useful as a positive predictor for response to anti-EGFR antibodies not only in LA-SCCHN but also in other tumors where currently negative predictors are used for the exclusion of patients to receive this type of treatment.
Patients and Methods
A retrospective review of the medical records of 36 patients with newly diagnosed and histologically confirmed nonnasopharyngeal LA-SCCHN was undertaken. Twenty-three patients had been treated with concomitant chemoradiotherapy (CCRT) in five centers. Details about patient dose modifications, follow-up, and RT technique have been reported previously [30]. Further, 13 patients had been treated with CTX and RT only (CTX-RT). All adverse events were graded for this analysis according to the National Cancer Institute Common Terminology Criteria (version 3.0). The Radiation Therapy Oncology Group's criteria were used to assess RT-related toxicities.
After a median follow-up of 24.5 months, 11 deaths and 13 progressions were reported. Median survival has not yet been reached, but 1-year survival rate was 68%. Response to treatment was evaluated according to the RECIST criteria, as described in Fountzilas et al. [30]. Selected patient and tumor characteristics are shown in Table 1.
Table 1.
Selected LA-SCCHN Patient and Tumor Characteristics.
| N = 36 | CCRT (n = 23) | CTX-RT (n = 13) | ||||
| Age, years | ||||||
| Median (range) | 64 (40–82) | 65 (40–82) | 60 (40–81) | |||
| n | % | n | % | n | % | |
| <60 | 16 | 44 | 12 | 33 | 4 | 11 |
| =60 | 20 | 56 | 11 | 31 | 9 | 25 |
| Sex | ||||||
| Man | 30 | 83 | 18 | 50 | 12 | 33 |
| Woman | 6 | 17 | 5 | 14 | 1 | 3 |
| Alcohol | ||||||
| No | 15 | 42 | 10 | 28 | 5 | 14 |
| Yes | 21 | 58 | 13 | 36 | 8 | 22 |
| Smoking | ||||||
| No | 13 | 36 | 12 | 33 | 1 | 3 |
| Yes | 23 | 64 | 11 | 31 | 12 | 33 |
| Performance status | ||||||
| 0 | 31 | 86 | 19 | 53 | 12 | 33 |
| 1 | 4 | 11 | 3 | 8 | 1 | 3 |
| 2 | 1 | 3 | 1 | 3 | — | — |
| Primary tumor location | ||||||
| Oropharynx | 7 | 19 | 5 | 14 | 2 | 6 |
| Hypopharynx | 2 | 6 | 2 | 6 | — | — |
| Larynx | 15 | 42 | 6 | 17 | 9 | 25 |
| Oral cavity | 10 | 28 | 9 | 25 | 1 | 3 |
| Paranasal sinuses | 2 | 6 | 1 | 3 | 1 | 3 |
| Histology grade | ||||||
| Well differentiated | 1 | 3 | 1 | 3 | — | — |
| Moderately differentiated | 10 | 28 | 5 | 14 | 5 | 14 |
| Poorly differentiated | 22 | 61 | 15 | 42 | 7 | 19 |
| Undifferentiated | 1 | 3 | — | — | 1 | 3 |
| Unknown | 2 | 6 | 2 | 6 | — | — |
| Stage | ||||||
| II | 1 | 3 | — | — | 1 | 3 |
| II | 5 | 14 | 4 | 11 | 1 | 3 |
| IVA | 28 | 78 | 17 | 47 | 11 | 31 |
| IVB | 1 | 3 | 1 | 3 | — | — |
| Unknown | 1 | 3 | 1 | 3 | — | — |
Tissues and Processing
Formalin-fixed paraffin-embedded tumor tissue from 36 patients was used for protein and gene analysis. Hematoxylin and eosin-stained sections were assessed by two pathologists for tumor tissue adequacy and were marked for tissue microarray (TMA) construction and for macrodissection where necessary (cases with <70% neoplastic cells). TMAs (two cores per case, 1.5 mm in diameter) were constructed with a manual arrayer (Beecher Instruments, Sun Prairie, WI).
Expression Profiling
RNA was extracted from whole or macrodissected formalin-fixed paraffin-embedded sections by using an experimental method based on proprietary magnetic beads from Siemens Healthcare Diagnostics (Cologne, Germany), as previously described [31]. The method involves DNase I treatment for the degradation of contaminating DNA. Reverse transcription was accomplished with random hexamers and Superscript III followed by excess RNA removal with RNase H (Invitrogen, Paisley, UK), according to the instructions of the manufacturer. Exon spanning TaqMan MGB assays (premade; Applied Biosystems, Biosolutions, Athens, Greece) were used to assess the relative expression of 12 genes in comparison to a housekeeping gene (β-glucuronidase [GUSB]). Target transcripts and assays are shown in Table 2. Except for the genes mentioned already, PIAS3 (protein inhibitor of activated STAT 3) was included in the target transcript panel because its low expression or absence had been reported in association with Stat3 activation in gliomas [32]. Samples were assessed twice in 20-µl reactions in separate runs along with no-template controls for 40 cycles under standard conditions in an ABI7500 real-time polymerase chain reaction (PCR) system and analyzed with the SDSv1.4 software (Applied Biosystems, Biosolutions) by keeping the reading threshold at 0.2 for all evaluations. Criteria for considering samples eligible for analysis were as follows: 1) for the identification of minimal quantities of amplifiable complementary DNA: GUSB CT values <36 and 2) for the evaluation of sample adequacy and PCR efficiency in consecutive runs: absolute difference of ΔCT (CTtarget - CTGUSB) values for the same sample was less than 0.5. By using these criteria, 31 of 36 samples were found eligible for further analysis. Relative expression of the target transcripts was assessed as the 2-ΔCT value (relative quantification value [RQ]) based on equal PCR efficiency for very short amplicons [33]. Mean RQs for each eligible sample were used for analysis.
Table 2.
Target Transcripts That Have Been Assessed for Relative Expression with FAM-TaqMan-MGB Assays.
| Target Transcript | Chromosomal Location | Assay ID | RNA Reference (GenBank) | Position | Amplicon Length (bp) |
| SRC | 20q12–q13 | Hs00178494_m1 | NM_198291.1, NM_005417.3 | ex 7–8 | 70 |
| STAT1 | 2q32.2 | Hs01014005_m1 | NM_139266.2, NM_007315.3 | ex 3–4 | 67 |
| STAT3 | 17q21.31 | Hs01047580_m1 | NM_213662.1, NM_139276.2, NM_003150.3 | ex 3–4 | 87 |
| STAT5A | 17q11.2 | Hs00234181_m1 | L41142.1 | ex 11–12 | 63 |
| STAT5B | 17q11.2 | Hs00560035_m1 | NM_012448.3 | ex 2–3 | 91 |
| PIAS3 | 1q21 | Hs00180666_m1 | NM_006099.3 | ex 7–8 | 102 |
| ANXA1 | 9q12-q21.2 | Hs00167549_m1 | NM_000700.1 | ex 8–9 | 66 |
| CAV1 | 7q31.1 | Hs00971716_m1 | NM_001753.3 | ex 2–3 | 66 |
| EPHA2 | 1p36 | Hs00171656_m1 | NM_004431.2 | ex 16–17 | 55 |
| EPHB2 | 1p36.1-p35 | Hs00362096_m1 | NM_017449.3, NM_004442.6 | ex 3–4 | 67 |
| ex 3–4 | 67 | ||||
| IGFBP2 | 2q33–q34 | Hs00167151_m1 | NM_000597.2 | ex 1–2 | 70 |
| MSN | Xq11.2-q12 | Hs01085677_g1 | NM_002444.2 | ex 10–11 | 61 |
| GUSB (reference transcript) | 7q21.11 | 4333767 | NM_000181.1 | ex 11–12 | 81 |
Immunohistochemistry
Activation of Stat1, Stat3, Stat5, and AKT/PKB was assessed with antibodies against phosphorylated sites in the corresponding proteins. TMA sections were incubated with monoclonal antibodies (Cell Signaling, Boston, MA) diluted 1:50 against Stat1-phospho-Tyr701, clone 58D6 (Stat1), Stat3-phospho-Tyr705, clone D3A7 (Stat3), Stat5-phospho- Tyr694, clone C11C5 (Stat5), AKT-phospho-Thr308, clone 244F9H2 (Akt-T308), and AKT-phospho-Ser473, clone 736E11 (Akt-S473). Antigen epitopes were retrieved with a citrate mixture, pH 6.2 to 4, whereas the Envision system and diaminobenzidine were used for visualizing the antibody-antigen complex (all reagents from DAKO, Glostrup, Denmark).
Immunohistochemistry (IHC) markers were evaluated as negative and positive by using a 5% positivity cutoff. Tumors were characterized as positive for activated Stat1, Stat3, and Stat5 when they exhibited greater than 5% positive nuclei; as positive for phosphorylated Akt-T308, when greater than 5% cytoplasmic staining was observed; and as positive for phosphorylated Akt-S473, when greater than 5% of cells exhibited cytoplasmic and/or nuclear staining (scoring modified from Mizoguchi et al. [34] and Yamashita et al. [35]).
Statistics
One of the major problems and challenges in studies dealing with relative gene expression is the reference system against which the obtained results are compared and analyzed. In this case, SCCHNs arise in a variety of epithelia with different functional properties, which makes it impossible to obtain one adequate reference tissue system for relative expression analysis. Ideally, normal counterparts of all tumors should be available for analysis, but this is practically not possible. Further, the commonly used pooled “normal” cell line template was considered inappropriate as well because transformation is required for normal epithelial cells to grow in culture, which per se abrogates normal control of survival and proliferation pathways. Hence, in this study, we considered it more pragmatic to analyze tumor samples only.
Categorical data were presented as counts and corresponding percentages, whereas continuous variables were summarized using medians and ranges. Correlations between the examined genes were calculated with the Spearman ρ correlation. To approach how high and low expression of the investigated genes associated with response to treatment, survival and progression-free survival (PFS), we used the extreme (upper and lower) quartile and median RQ values for each transcript target.
Comparisons between gene expression and clinicopathologic characteristics, as well as response to treatment were performed by using the χ2 and Fisher exact test where appropriate. The Mann-Whitney test was performed to compare the distribution of each transcript target according to the status (positive/negative) of each IHC marker.
Survival was measured from treatment initiation until death from any cause or date of last contact. PFS was measured from the time of treatment initiation to verified disease progression, death, or last contact. Both survival and PFS were estimated using the Kaplan-Meier product-limit method and comparisons were performed using the log-rank test. To estimate the hazard ratios of gene expression and protein activation data for survival and PFS, univariate Cox regression analyses were performed. Hazard ratios are presented along with the corresponding P values from the Wald test. Follow-up was last updated in March 2010. All tests were two-sided, and the level of significance was set at α = 0.05. Analysis was conducted by using the SPPS software for Windows, version 15 (SPSS Inc, Chicago, IL).
Results
SRC and SRC-Related Gene Expression in LA-SCCHNs
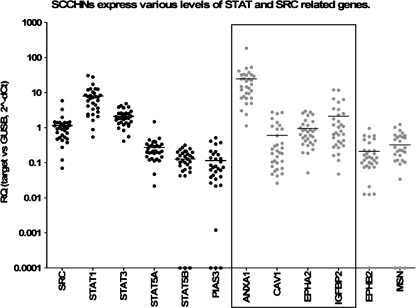
Relative expression results are shown in Table 3 and Figure 1. mRNA expression of SRC, STAT1, STAT3, and STAT5A, as well as of genes coding for proteins that are functionally related to Src and blocked by Src inhibitors, such as ANXA1, CAV1, EPHA2, and IGFBP2, was detectable in considerable although variable ratios versus GUSB in all LA-SCCHN samples eligible for relative quantification analysis (n = 31). In comparison, some of these tumors expressed very low to undetectable levels of PIAS3, a STAT3 inhibitor (n = 3); of STAT5B, the second STAT5 component (n = 3); of EPHB2, a receptor for ephrin-B family members (n = 1); and of MSN (moesin), a cytoskeleton stabilizer (n = 6). Among all STAT members, STAT1 was found to be expressed in relatively higher levels versus GUSB, whereas most LA-SCCHNs (20/31 [64.5%]) expressed ANXA1 >15-fold than this housekeeping gene.
Table 3.
mRNA Expression Characteristics of All Genes Examined: Relative Quantification* Was Performed for Individual LA-SCCHN Tumors.
| SRC | STAT1 | STAT3 | STAT5A | STAT5B | PIAS3 | ANXA1 | CAV1 | EPHA2 | EPHB2 | IGFBP2 | MSN | |
| Mean | 1.1390 | 7.9610 | 2.1000 | 0.2745 | 0.1265 | 0.1153 | 24.5500 | 0.5973 | 0.9563 | 0.2101 | 2.1290 | 0.3232 |
| SD | 1.0540 | 6.8930 | 1.0680 | 0.2478 | 0.0841 | 0.1298 | 32.1900 | 0.8089 | 0.8034 | 0.2182 | 3.0930 | 0.3181 |
| SE | 0.1862 | 1.2190 | 0.1889 | 0.0438 | 0.0149 | 0.0230 | 5.6910 | 0.1430 | 0.1420 | 0.0386 | 0.5468 | 0.0562 |
| Minimum | 0.0699 | 0.5404 | 0.4121 | 0.0217 | 0.0001 | 0.0001 | 1.1240 | 0.0260 | 0.0513 | 0.0001 | 0.0475 | 0.0001 |
| Maximum | 5.8320 | 30.5500 | 4.8200 | 1.4790 | 0.3169 | 0.5141 | 183.8000 | 2.7740 | 2.9340 | 0.9526 | 11.9700 | 1.2300 |
| 25th percentile | 0.5222 | 2.9450 | 1.3240 | 0.1465 | 0.0588 | 0.0284 | 7.1690 | 0.0830 | 0.3876 | 0.0764 | 0.3324 | 0.0943 |
| Median | 0.9758 | 7.0570 | 1.8700 | 0.2106 | 0.1202 | 0.0702 | 16.0100 | 0.2045 | 0.6922 | 0.1369 | 0.7533 | 0.2153 |
| 75th percentile | 1.3820 | 10.1400 | 2.5760 | 0.3306 | 0.1956 | 0.1608 | 31.6900 | 0.9123 | 1.2230 | 0.2235 | 2.4610 | 0.5378 |
Values were calculated with the 2-ΔCT method for target versus GUSB, where ΔCT = (CTtarget) - (CTGUSB).
Figure 1.
Relative expression of STAT and SRC-related genes in LA-SCCHN. The distribution of RQ values reflecting the presence of gene transcripts in comparison to those of GUSB (housekeeping gene) is presented in a logarithmic scale. Horizontal lines correspond to mean values per category; black dots, SRC/STAT genes; gray dots, genes producing SRC target proteins, some of which (box) are included in the proposed gene signature predicting for response to dasatinib.
STAT3 was expressed in parallel with STAT5B (P = .0019), which might suggest common regulatory events for these two neighbored genes that are located on the same strand at 17q21. By contrast, STAT5A, which is located between STAT3 and STAT5B but on the complementary strand, was expressed independently of these two genes and correlated with SRC expression (P = .0024). Thus, the two STAT5 genes did not seem to share common activators in LA-SCCHN.
ANXA1 and EPHA2 expression also correlated strongly with each other (P < .0001) but not with SRC or any STAT gene tested, providing evidence for separate transcription regulation of ANXA1/EPHA2 and SRC/STAT genes in LA-SCCHN.
MSN expression varied in parallel with CAV1 (P < .0001) and EPHB2 (P = .0191). MSN mRNA also correlated with the expression of STAT family members, namely, with STAT3 (P = .0020) and less with STAT5B (P = .0302) transcript levels. PIAS3 expression was positively related to STAT3 (P = .0166), whereas it was also strongly related to MSN (P < .0001) and to CAV1 (P = .0081). In all, the most significant above bivariate correlations suggest that 1) the negative regulatory loop for STAT3 inhibition through PIAS3 might be functional in LA-SCCHN, unlike to what seems to be the case in glioblastomas [32]; 2) transcription of MSN, STAT3, and its inhibitor PIAS3 may be regulated by common signaling; 3) transcriptional activation of CAV1 and SRC/STAT genes are not interrelated.
STAT1 and IGFBP2 expression did not correlate with any other mRNA target tested here. No significant negative correlations were observed.
Activation of Stat and Akt/PkB Proteins in LA-SCCHNs
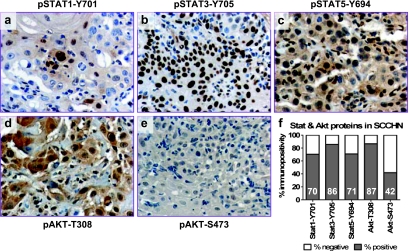
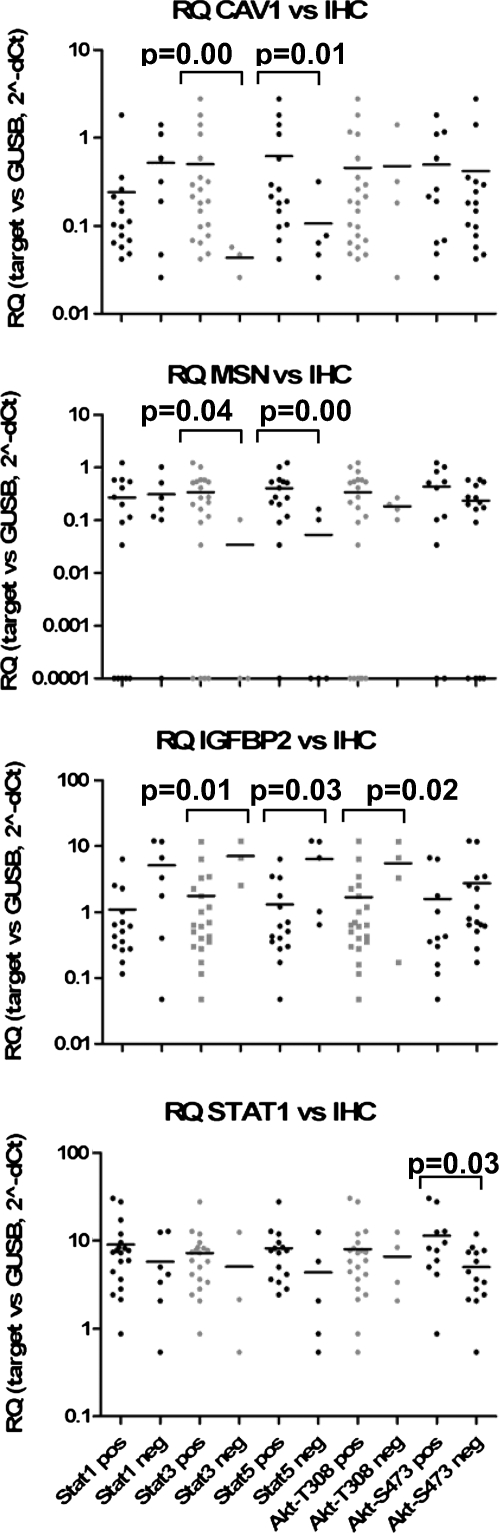
The three Stat proteins investigated in this study were found activated (phosphorylated) in most LA-SCCHN tumors, as indicated by the specific nuclear staining for each target (Figure 2, A–C). Stat3 phosphorylation was the most frequent event, usually coinciding with Stat5 phosphorylation (Fisher exact test, P = .017). Stat1-positive cells were usually confined to the most differentiated areas of the tumor (Figure 2A). No significant association was observed between each one of these activated proteins with the clinical and histopathologic parameters presented in Table 1. Activation of these proteins, as assessed by IHC, was not related to the level of the corresponding mRNA expression. Discordance between mRNA and protein expression for STAT3 and STAT5A has previously been reported [36,37], whereas activation of Stat proteins in SCCHN is induced by a number of signal transduction pathways that do not always depend on STAT mRNA expression (recently reviewed in Lai and Johnson [38]). Interestingly, however, tumors positive for Stat5 phosphorylation expressed relatively high CAV1 and MSN but relatively low IGFBP2 mRNA (Figure 3). Results in the same line were also obtained for Stat3 phosphorylation; however, because of the very small number of cases without activated Stat3, comparisons between positive and negative cases for this marker might represent statistical artifacts and should definitely be validated in larger studies.
Figure 2.
Immunohistochemical investigation of Stat and Akt/PKB activation in LA-SCCHN. Microphotographs A to E (all magnifications, x200): A typical case of a well-differentiated LA-SCCHN is shown (antibodies as indicated for each microphotograph). Stat1, Stat3, and Stat5 proteins seem activated, whereas Akt (Akt1-3) proteins are phosphorylated at Thr308 but not at Ser473. This profile was observed inmost LA-SCCHN, as collectively shown in the graph in F. Numbers within bars correspond to the rate of immunopositivity obtained with each antibody.
Figure 3.
Relative expression of CAV1, MSN, IGFBP2, and STAT1 in association with Stat and Akt protein phosphorylation in LA-SCCHN. Tumors were categorized as positive/negative for phosphorylated Stat1-Y701, Stat3-Y705, Stat5-Y694, Akt-T308, and Akt-S473. RQ values are shown in a logarithmic scale. Horizontal lines correspond to mean values per category. Categories (IHC status, x axis) in the lower graph are valid for all graphs. Statistically significant associations of gene expression with Stat/Akt phosphorylation status are shown (Mann-Whitney P, exact significance, two-sided).
Akt/PKB was frequently found phosphorylated at Thr308 but less so at Ser473 (Figure 2, D–F). Immunopositive LA-SCCHNs for Akt-Ser473 showed increased STAT1 mRNA expression in comparison to Akt-Ser473-negative tumors (Figure 3). In the same line with Stat5 activation, tumors with phosphorylated Akt at Thr308 expressed lower levels of IGFBP2; however, this result was again limited by the very small number of A4kt-Thr308-negative cases.
STAT and SRC-Related Expression Profiles in Association with Treatment Response
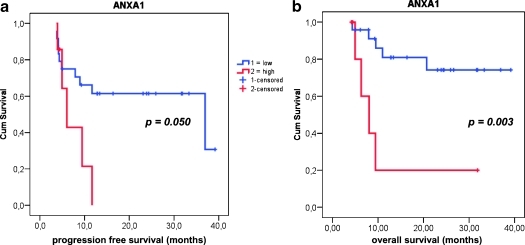
Patient follow-up data in association with the observed gene expression profiles are shown in Table 4. LA-SCCHN tumors expressing very high ANXA1 levels (upper quartile) were associated with significantly shorter survival (hazard ratio [HR] = 8.03, 95% confidence interval [CI] = 4.32–11.74, P = .0026) and marginally with earlier relapse of disease (HR = 6.03, 95% CI = 3.81–8.25, P = .0502; Figure 4). No further significant association was observed on overall and PFS for any other parameter tested, including IHC-determined activation of Stat and Akt proteins.
Table 4.
Gene Expression in Comparison to LA-SCCHN Patient Outcome and Response to CTX-Containing Treatments.
| Sample | Treatment | Overall Survival (mo) | Progression Free Survival (mo) | Response | SRC | STAT1 | STAT3 | STAT5A | STAT5B | PIAS3 | ANXA1 | CAV1 | EPHA2 | EPHAB2 | IGFBP2 | MSN |
| 1 | CCRT | 31.87* | 11.67 | CR | 433 | 7.255 | 2.155 | 0.215 | 0.101 | 0.127 | 41.098 | 0.098 | 1.159 | 0.369 | 0.515 | 0.588 |
| 2 | CCRT | 39.18* | 39.18† | CR | 0.479 | 5.938 | 0.412 | 0.022 | 0.043 | 0.001 | 9.331 | 0.048 | 0.329 | 0.098 | 0.116 | 0.000 |
| 3 | CCRT | 23.08* | 23.08† | CR | 0.070 | 10.346 | 1.423 | 0.112 | 0.000 | 0.061 | 29.081 | 0.110 | 2.581 | 0.116 | 1.197 | 0.112 |
| 4 | CCRT | 4.33 | 4.33 | CR | 1.375 | 2.377 | 1.829 | 0.496 | 0.042 | 0.067 | 2.369 | 0.906 | 0.421 | 0.021 | 0.323 | 0.230 |
| 5 | CCRT | 33.34* | 33.34† | CR | 3.301 | 6.859 | 4.213 | 0.448 | 0.271 | 0.040 | 4.938 | 0.247 | 0.185 | 0.225 | 0.796 | 0.458 |
| 6 | CCRT | 31.67* | 31.67† | CR | 1.184 | 17.268 | 3.848 | 1.479 | 0.160 | 0.043 | 20.966 | 0.113 | 0.602 | 0.139 | 0.269 | 0.115 |
| 7 | CCRT | 4.95 | 4.95 | CR | 1.252 | 2.809 | 2.556 | 0.199 | 0.237 | 0.107 | 52.527 | 0.042 | 2.757 | 0.013 | 0.619 | 0.091 |
| 8 | CCRT | 36.95* | 36.95† | CR | 0.967 | 7.738 | 1.654 | 0.247 | 0.071 | 0.055 | 33.174 | 0.068 | 1.597 | 0.092 | 0.304 | 0.304 |
| 9 | CCRT | 24.33* | 24.33† | CR | 1.154 | 2.413 | 2.509 | 0.389 | 0.317 | 0.039 | 32.111 | 0.296 | 2.249 | 0.073 | 3.456 | 0.219 |
| 10 | CCRT | 9.44 | 9.44 | ED | 0.376 | 2.135 | 2.118 | 0.114 | 0.057 | 0.023 | 49.351 | 0.057 | 0.981 | 0.182 | 2.521 | 0.000 |
| 11 | CCRT | 7.93 | 3.70 | PD | 1.402 | 7.408 | 2.439 | 0.126 | 0.064 | 0.137 | 3.031 | 0.357 | 0.550 | 0.586 | 2.280 | 0.580 |
| 12 | CCRT | 6.30 | 3.93 | PD | 5.832 | 5.808 | 2.583 | 0.291 | 0.103 | 0.000 | 37.453 | 0.078 | 0.787 | 0.000 | 0.649 | 0.000 |
| 13 | CCRT | 24.49* | 4.13 | PD | 1.138 | 8.369 | 1.376 | 0.309 | 0.113 | 0.022 | 31.341 | 0.184 | 0.633 | 0.166 | 0.173 | 0.201 |
| 14 | CCRT | 26.62* | 4.16 | PD | 1.456 | 3.613 | 1.253 | 0.316 | 0.000 | 0.000 | 24.218 | 0.104 | 0.912 | 0.013 | 0.279 | 0.000 |
| 15 | CCRT | 10.98 | 4.85 | PD | 0.920 | 4.426 | 3.665 | 0.335 | 0.210 | 0.085 | 24.251 | 0.184 | 0.751 | 0.012 | 0.641 | 0.174 |
| 16 | CCRT | 31.87* | 31.87† | SD | 0.898 | 30.548 | 2.918 | 0.197 | 0.089 | 0.213 | 17.113 | 1.163 | 0.826 | 0.478 | 0.160 | 0.837 |
| 17 | CCRT | 25.93* | 25.93† | CR | 0.119 | 7.749 | 0.962 | 0.211 | 0.139 | 0.074 | 14.591 | 0.149 | 0.868 | 0.075 | 0.711 | 0.275 |
| 18 | CCRT | 23.87* | 23.87† | CR | 0.535 | 0.540 | 1.208 | 0.201 | 0.053 | 0.034 | 11.259 | 0.047 | 0.051 | 0.074 | 11.967 | 0.000 |
| 19 | CCRT | 20.69 | 11.67 | CR | 0.274 | 0.880 | 0.554 | 0.146 | 0.000 | 0.000 | 6.945 | 0.064 | 0.307 | 0.220 | 1.028 | 0.000 |
| 20 | CTX-RT | 13.38* | 13.38† | CR | 1.385 | 12.510 | 1.112 | 0.286 | 0.199 | 0.130 | 6.974 | 0.026 | 0.480 | 0.099 | 6.639 | 0.103 |
| 21 | CTX-RT | 12.85* | 12.85† | CR | 0.700 | 5.007 | 1.487 | 0.390 | 0.251 | 0.026 | 13.251 | 0.191 | 0.335 | 0.081 | 0.405 | 0.118 |
| 22 | CTX-RT | 12.79* | 12.79† | CR | 1.456 | 27.819 | 1.729 | 0.404 | 0.127 | 169 | 22.362 | 1.813 | 2.385 | 0.453 | 0.430 | 0.540 |
| 23 | CTX-RT | 4.16* | 4.16† | CR | 1.173 | 8.468 | 2.136 | 0.149 | 0.107 | 0.079 | 31.801 | 2.498 | 1.485 | 0.181 | 1.489 | 0.617 |
| 24 | CTX-RT | 16.36* | 16.36† | CR | 0.518 | 3.352 | 2.045 | 0.210 | 0.185 | 0.514 | 9.626 | 1.405 | 0.583 | 0.117 | 3.297 | 0.267 |
| 25 | CTX-RT | 4.43* | 4.43† | CR | 1.410 | 1.619 | 3.880 | 0.192 | 0.212 | 0.412 | 83.801 | 2.693 | 2.934 | 0.953 | 5.318 | 0.703 |
| 26 | CTX-RT | 9.51 | 8.95 | PD | 0.984 | 8.185 | 4.820 | 0.185 | 0.178 | 0.375 | 1.124 | 0.217 | 0.378 | 0.595 | 6.316 | 1.230 |
| 27 | CTX-RT | 6.1* | 3.80 | PD | 0.726 | 9.540 | 1876 | 0.190 | 0.151 | 0.176 | 4.817 | 0.262 | 0.511 | 0.146 | 0.360 | 0.412 |
| 28 | CTX-RT | 8.03 | 6.03 | PD | 1.967 | 11.951 | 1.863 | 0.286 | 0.159 | 0.073 | 25.142 | 2.774 | 1.244 | 0.138 | 1.215 | 0.532 |
| 29 | CTX-RT | 9.28* | 9.28† | PR | 0.469 | 12.790 | 0.832 | 0.130 | 0.082 | 0.062 | 6.807 | 0.586 | 0.418 | 0.216 | 0.047 | 0.513 |
| 30 | CTX-RT | 7.9* | 7.9† | PR | 1.068 | 4.132 | 2.592 | 0.120 | 0.200 | 0.340 | 12.034 | 1.100 | 0.874 | 0.136 | 1.785 | 1.021 |
| 31 | CTX-RT | 5.05* | 5.05† | PR | 0.611 | 2.063 | 1.307 | 0.047 | 0.128 | 0.205 | 14.908 | 0.320 | 0.193 | 0.102 | 11.737 | 0.162 |
Relative quantification values were calculated with the 2-ΔCT method for target versus GUSB.
CCRT indicates concomitant chemoradiotherapy; CR, complete response; CTX-RT, cetuximab + radiotherapy; ED, early death; mo, months; PD, progressive disease; PR, partial response; SD, stable disease.
Patients still alive.
Patients without events during the follow-up period.
Figure 4.
Association of low ANXA1 expression with favorable LA-SCCHN patient outcome. Although relative ANXA1 expression was only marginally related to progression free survival (PFS, shown in A), low ANXA1 RQ values were observed in tumors from patients with longer overall survival (B) (HR = 8.03, 95% CI = 4.32–11.74).
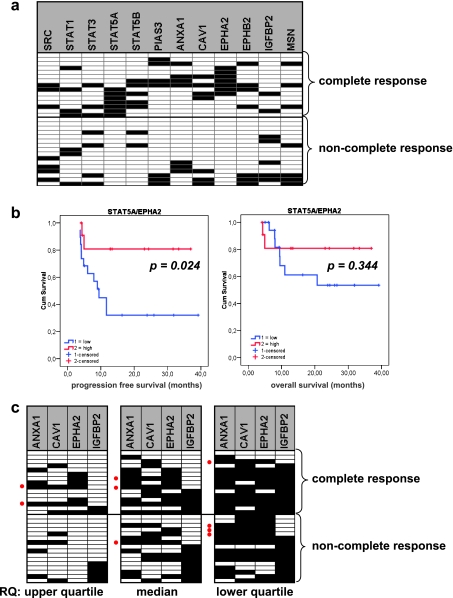
Of 37 patients, 35 were assessable for response to CTX-based treatments; 17 (47%) were complete responders, 6 (17%) showed partial response, 2 (6%) had stable disease, and 10 (28%) developed progressive disease while treated. Among patients with disease progression, there was one patient who discontinued treatment because of grade 3 mucositis and died a few months later. One patient was not assessable for response because he died early of disseminated disease, and for the rest, no information for response was available. mRNA profiling data were obtained in 31 of the above 35 cases (Table 4 and Figure 5A).
Figure 5.
STAT/SRC-related mRNA expression profiles were associated with response to treatment in LA-SCCHN. In A, a high expression of STAT5A and/or EPHA2 was associated with complete response to CTX-based treatments. Most tumors from patients who performed best on CTX-based treatments (12/15 [80%]) expressed high levels of STAT5A and/or high levels of EPHA2 (P < .0001; odds ratio = 58.67; 95% CI = 5.4–64.5; positive predictive value = 1, negative predictive value = 0.8) as assessed by using the upper RQ value quartile (75% percentile). This high STAT5A/EPHA2 pattern was further associated with prolonged PFS (HR = 9.44, 95% CI = 6.48–12.40) but not with patient overall survival, as shown in B. In C, the proposed dasatinib response-predictive signature with high ANXA1/CAV1/EPHA2 and low IGFBP2 was seldom encountered in LA-SCCHN (red dots). Cases are listed in the same order in the three panels. According to the definition of high and low (upper quartile, median, and lower quartile cutoffs for RQ values), different tumors seemed positive for this profile, some of which responded completely to CTX-based treatments. Panels in A and C: ▪ = high; □ = low RQ values.
All patients with tumors expressing very high levels of STAT5A and EPHA2 mRNA (RQ values in the upper quartile) exhibited a complete response on CTX-based treatments; by contrast, none of the tumors in the group of non-complete responders (0/16) expressed very high STAT5A and EPHA2 (P = .0002 each; Figure 5A). When assessing these two genes together at the same RQ value cutoff (upper quartile), 12 of 15 tumors in the complete responder group expressed very high STAT5A or EPHA2 mRNA or both (P < .0001). This expression pattern was further associated with prolonged PFS for LA-SCCHN patients (Figure 5B). This significant association of STAT5A and EPHA2 expression with best response to CTX-based treatment of LA-SCCHN was not maintained when assessing RQ values at lower cutoffs (median and lower quartile).
In addition, response to CTX-based treatment was negatively related to Stat1 protein phosphorylation (P = .009) because all patients bearing tumors without Stat1 activation were complete or partial responders (n = 8, almost half of the cases in the responders group). However, five of these tumors expressed very high STAT5A and two very high EPHA2 levels; hence, it is questionable whether the absence of Stat1 activation contributed to tumor response in these cases.
Because it had been proposed that SRC pathway targeting could be of benefit for patients with LA-SCCHN, especially those not responding to CTX treatments [6,21], it seemed rational to investigate whether the previously described dasatinib response-predictive signature for solid tumors [28] would be expressed in our cases. Four of the six genes in this signature were analyzed here (ANXA1, CAV1, EPHA2, and IGFBP2), and profiles were obtained for all RQ value cutoffs (upper and lower quartiles, and median [Figure 4B]). Among the 31 patients with available RNA data (15 complete responders, 16 non-complete responders), ratios of the four gene transcripts compatible with the described dasatinib response signature, i.e., high ANXA1/CAV1/EPHA2 - low IGFBP2, were obtained 1) for two responders, when assessing RQs with the upper quartile cutoff; 2) for three responders with the median cutoff; and 3) in four tumors with the low quartile cutoff. The latter four tumors corresponded to one complete responder, one patient with partial response and two patients with progressive disease. Except for one tumor that was identified as positive for this dasatinib response predictive signature with both the upper quartile and median cutoffs, all other tumors positive for this signature did not overlap among the three different cutoff result groups.
Discussion
This study shows that LA-SCCHN may express high levels of SRC, STAT1, STAT3, and STAT5A transcripts and that most of these tumors exhibit activated Stat1, Stat3, and Stat5 proteins. It seems that STAT3 and STAT5 are functionally more closely related to each other than to STAT1, perhaps in accordance with previous reports showing that STAT1 plays different roles than STAT3 in SCCHN[39], where STAT1 may function as a tumor suppressor and may be silenced [40]. Indeed, activated Stat1 was present in the best differentiated parts of the tumor, which might support a protective effect of this protein versus tumor progression [40,41]. However, STAT1 expression was the highest among all STATs examined, indicating that this gene is not silenced in SCCHN, in accordance with another previous report [42].
Overall, none of the STAT markers (mRNA or activated protein) was associated with disease outcome in our series; yet, activated Stat3 and Stat5 proteins were associated with CAV1, MSN, and IGFBP2 mRNA expression in LA-SCCHN. Although the number of cases examined in this study was small and the data obtained need validation in larger studies, the association of activated Stat5 with CAV1, MSN, and IGFBP2 expression is of interest especially because all these molecules represent SRC signaling targets, whereas SRC and STAT5A mRNA expression correlated significantly with each other. Caveolins participate in the function of the caveolae, structures involved in the internalization of inactive signaling molecules. CAV1 is generally considered as a tumor suppressor [43] and may play an antimetastatic role in SCCHN; its absence or low expression is associated with metastatic growth, whereas its restoration induces growth arrest and prevents metastatic spread of SCCHN cells in animal models [44]. Msn, a cytoskeletal protein, decreases with progression of carcinogenesis in the squamous oral cancer model [45], whereas another member of the same protein family, ezrin, has been associated with worse prognosis in SCCHN [46]. Decreased MSN expression [45] and altered subcellular localization [47] are also associated with increased metastatic potential of oral squamous cell tumors. Hence, CAV1 and MSN may be regarded as tumor suppressors in SCCHN. In comparison, IGFBP2 is usually regarded as an oncogene with a well-established role in promoting tumor growth and metastasis in various types of cancers [48–54], whereas IGFBP2 mRNA and protein expression have been associated with unfavorable tumor characteristics [55,56]. In addition, IGFBP2 regulates IGF binding to and activation of IGF1R, which is implicated in resistance to CTX and is considered as a therapeutic target in SCCHN [57]. On the basis of these data, because Stat3 and Stat5 phosphorylation was mostly observed in tumors with high CAV1 and MSN but low IGFBP2 expression, it does not seem likely that activation of these two proteins occurs in the tumor-promoting setting in LA-SCCHN. In line with this view, constitutive activation of Stat3 and Stat5 in nasopharyngeal carcinomas correlates with better prognosis [58]. In addition, when both Stat3 and Stat5 are activated in breast cancer cells, they are also associated with favorable prognostic parameters, such as decreased proliferation and increased chemosensitivity to taxanes and vinorelbine [59]. Individually, Stat5 protein expression has been described as a favorable prognostic factor in breast cancer [35], whereas nuclear Stat3 protein expression seems associated with a favorable outcome in SCCHNs [60]. The above evidence along with the data from this study indicates that the effect of Stat3 and Stat5 activation in SCCHN development, maintenance, and clinical/pharmacological behavior may be different in tissues than in cell culture systems.
Activated Stat3 and Stat5 were not related to CTX-containing treatment response in our series. This finding may seem as contrasting to the previously reported experimental evidence on the role of these Stat proteins in promoting resistance to EGFR-inhibiting agents [19,20]. An explanation for this discrepancy might be that Stats may also be activated in an EGFR-independent paracrine manner in SCCHN [16], which may well be the case at the tissue level. As shown herein, LA-SCCHN expressing the highest levels of STAT5A mRNA responded best to CTX-based treatments in this study, whereas SCCHNs were found to express low to very low levels of STAT5B. With the IHC antibody available to assess Stat5 phosphorylation, it is impossible to distinguish whether Stat5a or Stat5b or both are phosphorylated in tissue sections; hence, the activation status of each one of these two proteins in our LA-SCCHN series remains unknown. Our finding on the association of STAT5A expression with CTX response may be related to the different actions of the two Stat5 components reported in several systems including SCCHN: Stat5a does not promote SCCHN growth, whereas Stat5b does [61,62], prompting for a distinction between these two molecules in the research setting and in data reporting.
Except for STAT5A, EPHA2 expression was also associated with best response to CTX-based treatments in our LA-SCCHN. EphA2 is generally considered as a pro-oncogenic molecule, but the function of this receptor and its ligand ephrin A1 in tumorigenesis and tumor progression is complex and seems to be dependent on cell type and microenvironment [63]. EphA2 has been correlated with poor prognosis in esophageal carcinomas [64], glioblastomas [65], and NSCLC [66], whereas EphA2 overexpression might be involved in the early development of SCCHN [67]. Interestingly, EPHA2 gene transcription may be upregulated by EGFR in squamous cell carcinoma cell lines because of EGF activation [68]. At the tissue level, we could observe a statistical trend for increased EPHA2 expression in LA-SCCHN with high EGFR protein score, as assessed by IHC [30] (data not shown). Thus, EPHA2 seems to be an EGFR target gene and may be considered as a surrogate marker for EGFR activation through EGF, which represents the optimal condition for CTX to exert its growth inhibitory properties [69]. Considering EPHA2 as a marker of EGF/EGFR activation might further explain why tumors expressing high levels of EPHA2 respond best to CTX-based treatments, as reported herein. Clearly, to use mRNA expression levels and profiles in drug response prediction, solid cutoffs and reference systems need to be established. As shown here, the STAT5A and EPHA2 mRNA profile seems promising as a marker predictive of response to CTX-based treatments in LA-SCCHN, when relative expression values in the upper quartile are considered (very high expression).
The threshold for “very high expression” remains to be defined in larger studies with reliable external reference systems. The same considerations on the evaluation of mRNA profiles apply to the proposed dasatinib response-predicting signature [28], which was tested here to identify LA-SCCHN that would probably benefit from dasatinib treatment on CTX failure. Except for EPHA2 that has been described as an SRC target in colon carcinogenesis [70] and as a dasatinib target in breast and prostate cancers [28,29], this profile further involves ANXA1, three caveolae-related genes, and IGFBP2. As shown here, according to the applied cutoff for relative expression values (upper/lower quartiles, median), only 7% to 13% of tumors expressed high ANXA1/EPHA2/CAV1 and low IGFBP2. Whereas it is not surprising that different patients were identified as possibly sensitive to dasatinib according to how the cutoffs were defined, it should be noted that the predictive value of this older one or of the new modified dasatinib predictive signatures remains unknown [71].
ANXA1, an inhibitor of phospholipase A2 and anti-inflammatory protein, is reported as downregulated in most types of cancer [72] and may serve as a differentiating factor in SCCHN development and histopathologic status [72–75]. In our series, however, overall survival of LA-SCCHN patients was adversely affected by high ANXA1 expression, in line with a recent report on high ANXA1 mRNA in association with poor survival in colon cancer [76], where this molecule was examined in the frame of inflammatory response in cancer. This unfavorable ANXA1 association was not related to treatment variations (CCRT vs CTX-RT). Of note, however, all treatment schemes for LA-SCCHN involve local irradiation, although response to this modality is not evaluated separately. In this context, ANXA1 has been found upregulated in radiation resistant NSCLC cells [77], whereas it may also protect breast cancer cells from heat-induced growth arrest [78]. Additional preclinical evidence is needed to elucidate whether ANXA1 affects survival of patients with LA-SCCHN and possibly other tumors as well, as an inflammatory molecule or by conferring tumor cell survival advantage on irradiation or by both mechanisms.
Finally, we observed that Akt/PKB is commonly activated in SCCHNs, mostly at T308, implying a preferential activation through the phosphoinositide-3-kinase/pyruvate dehydrogenase kinase (PI3K/PDK1) pathway than through the mechanistic target of rapamycin (mTOR) complex [79]. However, other than previously suggested for prediction of response to EGFR inhibitors [80], no clear association was observed between Akt phosphorylation and response to CTX-based treatments.
In summary, this exploratory study provides a global overview of the effect of STAT and SRC-related parameters in the behavior of LA-SCCHN. In this respect, activated Stat proteins are frequently observed in LA-SCCHN, but Stat3 and Stat5 activation seems associated with an antioncogenic gene expression profile. In the same tumors, an mRNA expression profile involving STAT5A and EPHA2 was associated with complete response to CTX-based treatments. If validated in larger cohorts and standardized for the evaluation of individual tumors, the high STAT5A/high EPHA2 profile may be applied for assessing LA-SCCHN patient treatment. The same profile might be worth investigating as a positive marker for predicting response to CTX in tumors lacking the established negative predictors in this context, namely, KRAS/BRAF mutations.
Footnotes
This study was supported by a Hellenic Cooperative Oncology Group research grant (HE R5A/06).
References
- 1.Licitra L, Felip E. Squamous cell carcinoma of the head and neck: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(suppl 4):121–122. doi: 10.1093/annonc/mdp149. [DOI] [PubMed] [Google Scholar]
- 2.Pivot X, Felip E. Squamous cell carcinoma of the head and neck: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2008;19(suppl 2):ii79–ii80. doi: 10.1093/annonc/mdn097. [DOI] [PubMed] [Google Scholar]
- 3.FDA, author. FDA approves first head & neck cancer treatment in 45 years data shows treatment with Erbitux extends survival. FDA News Release P06-34. 2006 [Google Scholar]
- 4.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al. Radiotherapy plus cetuximab for squamous cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 5.Vermorken JB, Herbst RS, Leon X, Amellal N, Baselga J. Overview of the efficacy of cetuximab in recurrent and/or metastatic squamous cell carcinoma of the head and neck in patients who previously failed platinum-based therapies. Cancer. 2008;112:2710–2719. doi: 10.1002/cncr.23442. [DOI] [PubMed] [Google Scholar]
- 6.Egloff AM, Grandis JR. Targeting epidermal growth factor receptor and SRC pathways in head and neck cancer. Semin Oncol. 2008;35:286–297. doi: 10.1053/j.seminoncol.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen EE. Role of epidermal growth factor receptor pathway-targeted therapy in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol. 2006;24:2659–2665. doi: 10.1200/JCO.2005.05.4577. [DOI] [PubMed] [Google Scholar]
- 8.Egloff AM, Grandis JR. Improving response rates to EGFR-targeted therapies for head and neck squamous cell carcinoma: candidate predictive biomarkers and combination treatment with Src inhibitors. J Oncol. 2009;2009:896407. doi: 10.1155/2009/896407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonomi PD, Buckingham L, Coon J. Selecting patients for treatment with epidermal growth factor tyrosine kinase inhibitors. Clin Cancer Res. 2007;13:s4606–s4612. doi: 10.1158/1078-0432.CCR-07-0332. [DOI] [PubMed] [Google Scholar]
- 10.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 11.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 12.Weber A, Langhanki L, Sommerer F, Markwarth A, Wittekind C, Tannapfel A. Mutations of the BRAF gene in squamous cell carcinoma of the head and neck. Oncogene. 2003;22:4757–4759. doi: 10.1038/sj.onc.1206705. [DOI] [PubMed] [Google Scholar]
- 13.Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, Meneses Garcia A, Raimondi AR, Jufe R, Itoiz M, Gao Y, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13:4964–4973. doi: 10.1158/1078-0432.CCR-07-1041. [DOI] [PubMed] [Google Scholar]
- 14.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 15.Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- 16.Squarize CH, Castilho RM, Sriuranpong V, Pinto DS, Jr, Gutkind JS. Molecular cross-talk between the NF-κB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia. 2006;8:733–746. doi: 10.1593/neo.06274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson CJ, Neoh K. The Stat family of transcription factors have diverse roles in mammary gland development. Semin Cell Dev Biol. 2008;19:401–406. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Fukuda S, Lee Y, Hangoc G, Cooper S, Spolski R, Leonard WJ, Broxmeyer HE. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J Exp Med. 2000;192:719–728. doi: 10.1084/jem.192.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koppikar P, Lui VW, Man D, Xi S, Chai RL, Nelson E, Tobey AB, Grandis JR. Constitutive activation of signal transducer and activator of transcription 5 contributes to tumor growth, epithelial-mesenchymal transition, and resistance to epidermal growth factor receptor targeting. Clin Cancer Res. 2008;14:7682–7690. doi: 10.1158/1078-0432.CCR-08-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benavente S, Huang S, Armstrong EA, Chi A, Hsu KT, Wheeler DL, Harari PM. Establishment and characterization of a model of acquired resistance to epidermal growth factor receptor targeting agents in human cancer cells. Clin Cancer Res. 2009;15:1585–1592. doi: 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler DL, Iida M, Kruser TJ, Nechrebecki MM, Dunn EF, Armstrong EA, Huang S, Harari PM. Epidermal growth factor receptor cooperates with Src family kinases in acquired resistance to cetuximab. Cancer Biol Ther. 2009;8:696–703. doi: 10.4161/cbt.8.8.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson FM, Saigal B, Talpaz M, Donato NJ. Dasatinib (BMS-354825) tyrosine kinase inhibitor suppresses invasion and induces cell cycle arrest and apoptosis of head and neck squamous cell carcinoma and non-small cell lung cancer cells. Clin Cancer Res. 2005;11:6924–6932. doi: 10.1158/1078-0432.CCR-05-0757. [DOI] [PubMed] [Google Scholar]
- 23.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, et al. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 24.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 25.Brave M, Goodman V, Kaminskas E, Farrell A, Timmer W, Pope S, Harapanhalli R, Saber H, Morse D, Bullock J, et al. Sprycel for chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia resistant to or intolerant of imatinib mesylate. Clin Cancer Res. 2008;14:352–359. doi: 10.1158/1078-0432.CCR-07-4175. [DOI] [PubMed] [Google Scholar]
- 26.Demetri GD, Lo Russo P, MacPherson IR, Wang D, Morgan JA, Brunton VG, Paliwal P, Agrawal S, Voi M, Evans TR. Phase I dose-escalation and pharmacokinetic study of dasatinib in patients with advanced solid tumors. Clin Cancer Res. 2009;15:6232–6240. doi: 10.1158/1078-0432.CCR-09-0224. [DOI] [PubMed] [Google Scholar]
- 27.Yu EY, Wilding G, Posadas E, Gross M, Culine S, Massard C, Morris MJ, Hudes G, Calabro F, Cheng S, et al. Phase II study of dasatinib in patients with metastatic castration-resistant prostate cancer. Clin Cancer Res. 2009;15:7421–7428. doi: 10.1158/1078-0432.CCR-09-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang F, Reeves K, Han X, Fairchild C, Platero S, Wong TW, Lee F, Shaw P, Clark E. Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res. 2007;67:2226–2238. doi: 10.1158/0008-5472.CAN-06-3633. [DOI] [PubMed] [Google Scholar]
- 29.Wang XD, Reeves K, Luo FR, Xu LA, Lee F, Clark E, Huang F. Identification of candidate predictive and surrogate molecular markers for dasatinib in prostate cancer: rationale for patient selection and efficacy monitoring. Genome Biol. 2007;8:R255. doi: 10.1186/gb-2007-8-11-r255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fountzilas G, Kalogera-Fountzila A, Lambaki S, Wirtz RM, Nikolaou A, Karayannopoulou G, Bobos M, Kotoula V, Murray S, Lambropoulos A, et al. MMP9 but not EGFR, MET, ERCC1, P16, and P-53 is associated with response to concomitant radiotherapy, cetuximab, and weekly cisplatin in patients with locally advanced head and neck cancer. J Oncol. 2009;2009:305908. doi: 10.1155/2009/305908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koutras AK, Kalogeras KT, Dimopoulos MA, Wirtz RM, Dafni U, Briasoulis E, Pectasides D, Gogas H, Christodoulou C, Aravantinos G, et al. Evaluation of the prognostic and predictive value of HER family mRNA expression in high-risk early breast cancer: a Hellenic Cooperative Oncology Group (HeCOG) study. Br J Cancer. 2008;99:1775–1785. doi: 10.1038/sj.bjc.6604769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brantley EC, Nabors LB, Gillespie GY, Choi YH, Palmer CA, Harrison K, Roarty K, Benveniste EN. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res. 2008;14:4694–4704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Mizoguchi M, Betensky RA, Batchelor TT, Bernay DC, Louis DN, Nutt CL. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol. 2006;65:1181–1188. doi: 10.1097/01.jnen.0000248549.14962.b2. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita H, Nishio M, Ando Y, Zhang Z, Hamaguchi M, Mita K, Kobayashi S, Fujii Y, Iwase H. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr Relat Cancer. 2006;13:885–893. doi: 10.1677/erc.1.01095. [DOI] [PubMed] [Google Scholar]
- 36.Lassmann S, Schuster I, Walch A, Gobel H, Jutting U, Makowiec F, Hopt U, Werner M. STAT3 mRNA and protein expression in colorectal cancer: effects on STAT3-inducible targets linked to cell survival and proliferation. J Clin Pathol. 2007;60:173–179. doi: 10.1136/jcp.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohankumar KM, Perry JK, Kannan N, Kohno K, Gluckman PD, Emerald BS, Lobie PE. Transcriptional activation of signal transducer and activator of transcription (STAT) 3 and STAT5B partially mediate homeobox A1-stimulated oncogenic transformation of the immortalized human mammary epithelial cell. Endocrinology. 2008;149:2219–2229. doi: 10.1210/en.2007-1320. [DOI] [PubMed] [Google Scholar]
- 38.Lai SY, Johnson FM. Defining the role of the JAK-STAT pathway in head and neck and thoracic malignancies: implications for future therapeutic approaches. Drug Resist Updat. 2010;13:67–78. doi: 10.1016/j.drup.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Investig. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xi S, Dyer KF, Kimak M, Zhang Q, Gooding WE, Chaillet JR, Chai RL, Ferrell RE, Zamboni B, Hunt J, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J Natl Cancer Inst. 2006;98:181–189. doi: 10.1093/jnci/djj020. [DOI] [PubMed] [Google Scholar]
- 41.Arany I, Chen SH, Megyesi JK, Adler-Storthz K, Chen Z, Rajaraman S, Ember IA, Tyring SK, Brysk MM. Differentiation-dependent expression of signal transducers and activators of transcription (STATs) might modify responses to growth factors in the cancers of the head and neck. Cancer Lett. 2003;199:83–89. doi: 10.1016/s0304-3835(03)00345-8. [DOI] [PubMed] [Google Scholar]
- 42.Shaw RJ, Hall GL, Lowe D, Liloglou T, Field JK, Sloan P, Risk JM. The role of pyrosequencing in head and neck cancer epigenetics: correlation of quantitative methylation data with gene expression. Arch Otolaryngol Head Neck Surg. 2008;134:251–256. doi: 10.1001/archoto.2007.50. [DOI] [PubMed] [Google Scholar]
- 43.Engelman JA, Zhang XL, Galbiati F, Lisanti MP. Chromosomal localization, genomic organization, and developmental expression of the murine caveolin gene family (Cav-1, -2, and -3). Cav-1 and Cav-2 genes map to a known tumor suppressor locus (6-A2/7q31) FEBS Lett. 1998;429:330–336. doi: 10.1016/s0014-5793(98)00619-x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Su L, Muller S, Tighiouart M, Xu Z, Zhang X, Shin HJ, Hunt J, Sun SY, Shin DM, et al. Restoration of caveolin-1 expression suppresses growth and metastasis of head and neck squamous cell carcinoma. Br J Cancer. 2008;99:1684–1694. doi: 10.1038/sj.bjc.6604735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belbin TJ, Singh B, Smith RV, Socci ND, Wreesmann VB, Sanchez-Carbayo M, Masterson J, Patel S, Cordon-Cardo C, Prystowsky MB, et al. Molecular profiling of tumor progression in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2005;131:10–18. doi: 10.1001/archotol.131.1.10. [DOI] [PubMed] [Google Scholar]
- 46.Madan R, Brandwein-Gensler M, Schlecht NF, Elias K, Gorbovitsky E, Belbin TJ, Mahmood R, Breining D, Qian H, Childs G, et al. Differential tissue and subcellular expression of ERM proteins in normal and malignant tissues: cytoplasmic ezrin expression has prognostic significance for head and neck squamous cell carcinoma. Head Neck. 2006;28:1018–1027. doi: 10.1002/hed.20435. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi H, Sagara J, Kurita H, Morifuji M, Ohishi M, Kurashina K, Taniguchi S. Clinical significance of cellular distribution of moesin in patients with oral squamous cell carcinoma. Clin Cancer Res. 2004;10:572–580. doi: 10.1158/1078-0432.ccr-1323-03. [DOI] [PubMed] [Google Scholar]
- 48.Dunlap SM, Celestino J, Wang H, Jiang R, Holland EC, Fuller GN, Zhang W. Insulin-like growth factor binding protein 2 promotes glioma development and progression. Proc Natl Acad Sci USA. 2007;104:11736–11741. doi: 10.1073/pnas.0703145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukushima T, Tezuka T, Shimomura T, Nakano S, Kataoka H. Silencing of insulin-like growth factor-binding protein-2 in human glioblastoma cells reduces both invasiveness and expression of progression-associated gene CD24. J Biol Chem. 2007;282:18634–18644. doi: 10.1074/jbc.M609567200. [DOI] [PubMed] [Google Scholar]
- 50.Lee EJ, Mircean C, Shmulevich I, Wang H, Liu J, Niemisto A, Kavanagh JJ, Lee JH, Zhang W. Insulin-like growth factor binding protein 2 promotes ovarian cancer cell invasion. Mol Cancer. 2005;4:7. doi: 10.1186/1476-4598-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin JL, Baxter RC. Expression of insulin-like growth factor binding protein-2 by MCF-7 breast cancer cells is regulated through the phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway. Endocrinology. 2007;148:2532–2541. doi: 10.1210/en.2006-1335. [DOI] [PubMed] [Google Scholar]
- 52.Miyake H, Hara I, Yamanaka K, Muramaki M, Gleave M, Eto H. Introduction of insulin-like growth factor binding protein-2 gene into human bladder cancer cells enhances their metastatic potential. Oncol Rep. 2005;13:341–345. [PubMed] [Google Scholar]
- 53.Miyako K, Cobb LJ, Francis M, Huang A, Peng B, Pintar JE, Ariga H, Cohen P. PAPA-1 is a nuclear binding partner of IGFBP-2 and modulates its growth-promoting actions. Mol Endocrinol. 2009;23:169–175. doi: 10.1210/me.2008-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore MG, Wetterau LA, Francis MJ, Peehl DM, Cohen P. Novel stimulatory role for insulin-like growth factor binding protein-2 in prostate cancer cells. Int J Cancer. 2003;105:14–19. doi: 10.1002/ijc.11015. [DOI] [PubMed] [Google Scholar]
- 55.Marucci G, Morandi L, Magrini E, Farnedi A, Franceschi E, Miglio R, Calo D, Pession A, Foschini MP, Eusebi V. Gene expression profiling in glioblastoma and immunohistochemical evaluation of IGFBP-2 and CDC20. Virchows Arch. 2008;453:599–609. doi: 10.1007/s00428-008-0685-7. [DOI] [PubMed] [Google Scholar]
- 56.So AI, Levitt RJ, Eigl B, Fazli L, Muramaki M, Leung S, Cheang MC, Nielsen TO, Gleave M, Pollak M. Insulin-like growth factor binding protein-2 is a novel therapeutic target associated with breast cancer. Clin Cancer Res. 2008;14:6944–6954. doi: 10.1158/1078-0432.CCR-08-0408. [DOI] [PubMed] [Google Scholar]
- 57.Barnes CJ, Ohshiro K, Rayala SK, El-Naggar AK, Kumar R. Insulin-like growth factor receptor as a therapeutic target in head and neck cancer. Clin Cancer Res. 2007;13:4291–4299. doi: 10.1158/1078-0432.CCR-06-2040. [DOI] [PubMed] [Google Scholar]
- 58.Hsiao JR, Jin YT, Tsai ST, Shiau AL, Wu CL, Su WC. Constitutive activation of STAT3 and STAT5 is present in the majority of nasopharyngeal carcinoma and correlates with better prognosis. Br J Cancer. 2003;89:344–349. doi: 10.1038/sj.bjc.6601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, Frank DA. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009;7:966–976. doi: 10.1158/1541-7786.MCR-08-0238. [DOI] [PubMed] [Google Scholar]
- 60.Pectasides E, Egloff AM, Sasaki C, Kountourakis P, Burtness B, Fountzilas G, Dafni U, Zaramboukas T, Rampias T, Rimm D, et al. Nuclear localization of signal transducer and activator of transcription 3 in head and neck squamous cell carcinoma is associated with a better prognosis. Clin Cancer Res. 2010;16:2427–2434. doi: 10.1158/1078-0432.CCR-09-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xi S, Zhang Q, Gooding WE, Smithgall TE, Grandis JR. Constitutive activation of Stat5b contributes to carcinogenesis in vivo. Cancer Res. 2003;63:6763–6771. [PubMed] [Google Scholar]
- 62.Leong PL, Xi S, Drenning SD, Dyer KF, Wentzel AL, Lerner EC, Smithgall TE, Grandis JR. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene. 2002;21:2846–2853. doi: 10.1038/sj.onc.1205385. [DOI] [PubMed] [Google Scholar]
- 63.Wykosky J, Debinski W. The EphA2 receptor and ephrinA1 ligand in solid tumors: function and therapeutic targeting. Mol Cancer Res. 2008;6:1795–1806. doi: 10.1158/1541-7786.MCR-08-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyazaki T, Kato H, Fukuchi M, Nakajima M, Kuwano H. EphA2 overexpression correlates with poor prognosis in esophageal squamous cell carcinoma. Int J Cancer. 2003;103:657–663. doi: 10.1002/ijc.10860. [DOI] [PubMed] [Google Scholar]
- 65.Wang LF, Fokas E, Bieker M, Rose F, Rexin P, Zhu Y, Pagenstecher A, Engenhart-Cabillic R, An HX. Increased expression of EphA2 correlates with adverse outcome in primary and recurrent glioblastoma multiforme patients. Oncol Rep. 2008;19:151–156. [PubMed] [Google Scholar]
- 66.Brannan JM, Dong W, Prudkin L, Behrens C, Lotan R, Bekele BN, Wistuba I, Johnson FM. Expression of the receptor tyrosine kinase EphA2 is increased in smokers and predicts poor survival in non-small cell lung cancer. Clin Cancer Res. 2009;15:4423–4430. doi: 10.1158/1078-0432.CCR-09-0473. [DOI] [PubMed] [Google Scholar]
- 67.Rivera RS, Gunduz M, Nagatsuka H, Gunduz E, Cengiz B, Fukushima K, Beder LB, Pehlivan D, Yamanaka N, Shimizu K, et al. Involvement of EphA2 in head and neck squamous cell carcinoma: mRNA expression, loss of heterozygosity and immunohistochemical studies. Oncol Rep. 2008;19:1079–1084. [PubMed] [Google Scholar]
- 68.Larsen AB, Pedersen MW, Stockhausen MT, Grandal MV, van Deurs B, Poulsen HS. Activation of the EGFR gene target EphA2 inhibits epidermal growth factor-induced cancer cell motility. Mol Cancer Res. 2007;5:283–293. doi: 10.1158/1541-7786.MCR-06-0321. [DOI] [PubMed] [Google Scholar]
- 69.Burtness B. The role of cetuximab in the treatment of squamous cell cancer of the head and neck. Expert Opin Biol Ther. 2005;5:1085–1093. doi: 10.1517/14712598.5.8.1085. [DOI] [PubMed] [Google Scholar]
- 70.Leroy C, Fialin C, Sirvent A, Simon V, Urbach S, Poncet J, Robert B, Jouin P, Roche S. Quantitative phosphoproteomics reveals a cluster of tyrosine kinases that mediates SRC invasive activity in advanced colon carcinoma cells. Cancer Res. 2009;69:2279–2286. doi: 10.1158/0008-5472.CAN-08-2354. [DOI] [PubMed] [Google Scholar]
- 71.Moulder S, Yan K, Huang F, Hess KR, Liedtke C, Lin F, Hatzis C, Hortobagyi GN, Symmans WF, Pusztai L. Development of candidate genomic markers to select breast cancer patients for dasatinib therapy. Mol Cancer Ther. 2010;9:1120–1127. doi: 10.1158/1535-7163.MCT-09-1117. [DOI] [PubMed] [Google Scholar]
- 72.Lim LH, Pervaiz S. Annexin 1: the new face of an old molecule. FASEB J. 2007;21:968–975. doi: 10.1096/fj.06-7464rev. [DOI] [PubMed] [Google Scholar]
- 73.Garcia Pedrero JM, Fernandez MP, Morgan RO, Herrero Zapatero A, Gonzalez MV, SuarezNieto C, Rodrigo JP. Annexin A1 down-regulation in head and neck cancer is associated with epithelial differentiation status. Am J Pathol. 2004;164:73–79. doi: 10.1016/S0002-9440(10)63098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alves VA, Nonogaki S, Cury PM, Wunsch-Filho V, de Carvalho MB, Michaluart-Junior P, Moyses RA, Curioni OA, Figueiredo DL, Scapulatempo-Neto C, et al. Annexin A1 subcellular expression in laryngeal squamous cell carcinoma. Histopathology. 2008;53:715–727. doi: 10.1111/j.1365-2559.2008.03186.x. [DOI] [PubMed] [Google Scholar]
- 75.Nomura H, Uzawa K, Yamano Y, Fushimi K, Nakashima D, Kouzu Y, Kasamatsu A, Ogawara K, Shiiba M, Bukawa H, et al. Down-regulation of plasma membranous Annexin A1 protein expression in premalignant and malignant lesions of the oral cavity: correlation with epithelial differentiation. J Cancer Res Clin Oncol. 2009;135:943–949. doi: 10.1007/s00432-008-0530-z. [DOI] [PubMed] [Google Scholar]
- 76.Schetter AJ, Nguyen GH, Bowman ED, Mathe EA, Yuen ST, Hawkes JE, Croce CM, Leung SY, Harris CC. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878–5887. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu QY, Gao Y, Liu Y, Yang WZ, Xu XY. Identification of differential gene expression profiles of radioresistant lung cancer cell line established by fractionated ionizing radiation in vitro. Chin Med J (Engl) 2008;121:1830–1837. [PubMed] [Google Scholar]
- 78.Nair S, Hande MP, Lim LH. Annexin-1 protects MCF7 breast cancer cells against heat-induced growth arrest and DNA damage. Cancer Lett. 2010;294:111–117. doi: 10.1016/j.canlet.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 79.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 80.Pernas FG, Allen CT, Winters ME, Yan B, Friedman J, Dabir B, Saigal K, Mundinger GS, Xu X, Morris JC, et al. Proteomic signatures of epidermal growth factor receptor and survival signal pathways correspond to gefitinib sensitivity in head and neck cancer. Clin Cancer Res. 2009;15:2361–2372. doi: 10.1158/1078-0432.CCR-08-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]