Abstract
Lactose is an interesting carbon source for the production of several bio-products by fermentation, primarily because it is the major component of cheese whey, the main by-product of dairy activities. However, the microorganism more widely used in industrial fermentation processes, the yeast Saccharomyces cerevisiae, does not have a lactose metabolization system. Therefore, several metabolic engineering approaches have been used to construct lactose-consuming S. cerevisiae strains, particularly involving the expression of the lactose genes of the phylogenetically related yeast Kluyveromyces lactis, but also the lactose genes from Escherichia coli and Aspergillus niger, as reviewed here. Due to the existing large amounts of whey, the production of bio-ethanol from lactose by engineered S. cerevisiae has been considered as a possible route for whey surplus. Emphasis is given in the present review on strain improvement for lactose-to-ethanol bioprocesses, namely flocculent yeast strains for continuous high-cell-density systems with enhanced ethanol productivity.
Key words: cheese whey, lactose, recombinant Saccharomyces cerevisiae, bio-ethanol, fermentation, metabolic engineering
Introduction
Lactose is a constituent of cheese whey, a major by-product of dairy industries. The lactose can constitute as much as 50 g·L−1. Whey streams could be used as an abundant and renewable raw material for microbial fermentations, with lactose providing the carbon source. In fact, whey disposal has been under consideration for several years, since it is highly polluting and generated in high amounts. Drying is one of the solutions that have been considered and has been implemented on an industrial scale. However, although the discharge problem is solved, no value is added. Whey protein concentrate (WPC) is nowadays one of the major products obtained from cheese whey.1 When producing WPC, high volumes of a lactose-rich permeate are also generated. This remains a major pollutant and a profitable use for this by-product must be found. The future trend for cheese factories is to move towards zero discharge, i.e., move away from high disposal costs and find more environmentally friendly and profitable applications for lactose.2 Lactose fermentation to bio-ethanol is one of the possibilities. The use of concentrated lactose solutions (up to 200 g·L−1) is important since it will permit high ethanol titres (up to 10–12% v/v) at the end of fermentation, therefore reducing considerably the ethanol distillation costs. However, natural lactose-fermenting microorganisms, such as the yeast Kluyveromyces marxianus, cannot ferment efficiently (i.e., rapidly and with high conversion yields) such high concentrations of lactose. Saccharomyces cerevisiae is the organism of choice for bioethanol production. However, this yeast is not able of metabolizing the sugar lactose. Thus, strain development programs through metabolic engineering of S. cerevisiae are required for the implementation of lactose-to-ethanol processes with increased productivity.
In 1991 Bailey3 proposed the emergence of a new discipline called “metabolic engineering”, which he defined as “the improvement of cellular activities by manipulations of enzymatic, transport and regulatory functions of the cell with the use of recombinant DNA technology.” Initially, metabolic engineering overlapped with applied molecular biology. Developments in genomics and high-throughput system biology tools enhanced the rapid characterization of cellular behavior, which led to a rapid expansion of metabolic engineering, where strain characterization is often the bottleneck in development programmes. Moreover, advanced genetic engineering techniques along with the sequencing of whole genomes of several organisms and developments in bioinformatics have speeded up the process of gene cloning and transformation.4 Metabolic engineering processes are categorized typically by the approach taken or the aim.4 These may be: (1) heterologous protein production (2) extension of substrate range (3) pathways leading to new products (4) pathways for degradation of xenobiotics (5) improvement of overall cellular physiology (6) elimination or reduction of by-product formation, and (7) improvement of yield or productivity. Metabolic engineering of S. cerevisiae strains for lactose fermentation fits in the substrate range category of metabolic engineering.
Lactose-Consuming Microorganisms
The number of microorganisms that can use lactose as a source of carbon and energy is limited, yet including bacteria, yeasts and filamentous fungi. Bacteria have evolved different strategies for the uptake and hydrolysis of lactose. The most effective implies the simultaneous phosphorylation and translocation of the sugar across the cell membrane, existing at least two alternative mechanisms for uptake (a lactose-proton symporter and a lactose-galactose antiporter). Once inside the bacterial cell, the phosphorylated lactose is hydrolyzed by a phospho-β-galactosidase (an enzyme that recognises phosphorylated lactose). When the uptake mechanism does not involve phosphorylation, lactose is cleaved intracellularly by a β-galactosidase.5
The regulation of lactose utilization by the lac operon in Escherichia coli has become a paradigm for prokaryotic gene regulation. The E. coli lacZ gene (encoding β-galactosidase) has been used commonly as a genetic and biotechnological tool, functioning as a reporter gene for protein expression.5 Moreover, the E. coli lacY gene (encoding lactose permease) was the first gene encoding a membrane transport protein to be cloned into a recombinant plasmid, overexpressed6 and sequenced.7 LacYp is a representative for the Major Facilitator Superfamily of transport proteins8 and its structure has been recently unveiled.9,10
Lactic acid bacteria (includes several genera such as Lactobacillus, Lactococcus, Leuconostoc, Pediococcus and Streptococcus) are among the most important lactose-consuming microorganisms, due to their occurrence in milk and dairy products. Besides their food-related significance, the importance of lactic acid bacteria in biotechnology is extended to the production of lactic acid, e.g., from whey fermentation.11
Filamentous fungi often utilize lactose at very low rates.12 In fungi there are two principal alternatives for the catabolism of lactose: (1) extracellular hydrolysis and subsequent uptake of the resulting glucose and galactose monomers and (2) uptake of the disaccharide and subsequent intracellular hydrolysis. Fungi species such as Aspergillus nidulans, Neurospora crassa or Fusarium graminearum follow the second strategy for lactose utilization, while others such as Hypocrea jecorina (Trichoderma reesei) and Aspergillus niger have the ability to secrete β-galactosidase that hydrolyzes lactose in the extracellular medium.12 The yeasts that assimilate lactose aerobically are widespread, but those that ferment lactose are rather rare,13 including e.g., Kluyveromyces lactis, K. marxianus and Candida pseudotropicalis.
Lactose metabolism in K. lactis and the GAL/LAC regulon.
Similarly to other microorganisms present in milk, K. lactis is adapted for the efficient utilization of lactose. The ability of this yeast to metabolize lactose results from the presence of lactose permease and β-galactosidase.14 The K. lactis LAC system is the best studied within the Kluyveromyces genus and is a good model for related species.
The K. lactis lactose permease is a membrane protein of 587 amino acids encoded by the gene LAC12.15 The lactose uptake in K. lactis is mediated by a transport system inducible by lactose and galactose (the inducer is intracellular galactose).16 Uptake is mediated by a carrier and is saturated at high substrate concentrations. Dickson and Barr16 determined a Km of approximately 2.8 mM for this transport system, while Boze et al.17 reported a Km of 1.2–4 mM in a different strain. We have determined recently a Km of 1.0–1.8 mM for K. lactis CBS2359, and our results of lactose uptake by two recombinant S. cerevisiae strains expressing the K. lactis LAC12 gene are also consistent with such Km values.18 The transport of lactose in K. lactis is an active process, requiring an energy-generating system, which permits the intracellular accumulation of lactose against a concentration gradient.16,17 The transport is inhibited by the proton ionophore 2,4-dinitrophenol,16,17 and therefore it has been suggested that the transporter operates, at least in part, by a proton symport mechanism.16 In other Kluyveromyces species lactose uptake has also been described to proceed via a proton symport mechanism.19–21 Lac12p shows sequence similarity to the E. coli xylose and arabinose proton symporters15 and a significant sequence and structure homology with the S. cerevisiae maltose proton symporter Mal61p,22 but no significant sequence similarity with the lactose permease (lacY gene) of E. coli.15
The β-galactosidase (lactase) is encoded by the LAC4 gene23 and is described to be intracellular.18,24–26 This enzyme has a Km for lactose of 12–17 mM and its pH optimum is around 7.25
β-galactosidase hydrolyzes lactose into glucose and galactose. Intracellular glucose can enter glycolysis while galactose follows the Leloir pathway. In K. lactis, the metabolism of lactose and galactose are closely related. The regulatory circuit of the GAL/LAC regulon of K. lactis (Table 1) has been studied in detail (reviewed in refs. 24, 27 and 28) particularly in comparison with the GAL/MEL regulon of S. cerevisiae (Table 1), which is one of the most intensively studied and best understood genetic regulatory circuits in yeasts and a major model for the study of eukaryotic regulation (reviewed in refs. 29–31).
Table 1.
GAL/MEL genes of S. cerevisiae and GAL/LAC genes of K. lactis
| S. cerevisiae | K. lactis | ||
| Gene | Function | Gene | Function |
| Structural/Catabolic genes | Structural/Catabolic genes | ||
| MEL1 | α-Galactosidase | LAC12 | Lactose/galactose permease |
| GAL2 | Galactose permease | LAC4 | β-Galactosidase |
| GAL1a | Bifunctional galactokinase/sensor inducer | KlGAL1a | Bifunctional galactokinase/sensor inducer |
| GAL10 | Uridine diphosphoglucose 4-epimerase | KlGAL10 | Uridine diphosphoglucose 4-epimerase |
| GAL7 | Galactose-1-phosphate uridylyltransferase | KlGAL7 | Galactose-1-phosphate uridylyltransferase |
| GAL5b | Phosphoglucomutase | KlGAL5b | Phosphoglucomutase |
| Regulatory genes | Regulatory genes | ||
| GAL4 | Transcriptional activator | KlGAL4 (LAC9) | Transcriptional activator |
| GAL80 | Gal4p repressor | KlGAL80 | Gal4p repressor |
| GAL3 | Gal80p repressor (sensor/inducer) | ||
GAL1 has both catabolic (galactokinase) and regulatory (sensor/inducer) functions.
GAL5 is not specific of the GAL regulon, having a more generalized role in carbon metabolism.
S. cerevisiae cannot assimilate lactose, yet it can utilise galactose. Some Saccharomyces yeasts can also assimilate melibiose, which is hydrolyzed to glucose and galactose by a secretable α-galactosidase encoded by MEL1,32 and other genes of the MEL family.33 Galactose is taken up by a permease, encoded by the gene GAL2.34 Once inside the cell, catabolism of galactose proceeds through the highly evolutionarily conserved Leloir pathway, both in S. cerevisiae and in K. lactis.24,29
Despite the extensive degree of conservation in the group of genes involved in the utilization of galactose between the two yeasts, differences have arisen as a result of their evolution in different environments: S. cerevisiae has mainly adapted to glucose, whereas K. lactis has adapted to lactose. Therefore, the two yeasts have differences in the modes of regulation that have important consequences in their overall response to carbon sources and may account for major physiological differences between these yeasts.24,28
The induction of the GAL genes in both S. cerevisiae and K. lactis is determined by the interplay between three main GAL-specific regulatory proteins (Table 1): a transcriptional activator (Gal4p, also known as Lac9p in K. lactis), a repressor (Gal80p) and a ligand sensor (Gal3p in S. cerevisiae; Gal1p in K. lactis). This later activates GAL gene expression after binding galactose (the inducer) and ATP.24
Regulation of LAC12 and LAC4 expression in K. lactis is controlled by the same mechanisms that regulate GAL genes. LAC12 and LAC4 are transcribed divergently from an unusually large intergenic region, which works as promoter for the transcription of both genes. The LAC12-LAC4 intergenic region contains four functional UASG elements, which are binding sites for the trans-activator Lac9p. Two functional UASG elements are located in front of each of the genes at almost symmetrical positions. These elements cooperate in activating transcription of both genes.35
Metabolic Engineering of Lactose Consuming/Fermenting S. cerevisiae Cells
One of the first approaches to construct lactose-metabolizing S. cerevisiae cells consisted in the production of hybrids of S. cerevisiae and K. lactis or K. fragilis, using the protoplast fusion technique.36–38 The fusant strains were able to ferment lactose and produce more ethanol than the corresponding Kluyveromyces parental strain. More recently, the generation of hybrid strains of S. cerevisiae and K. lactis able to ferment lactose in sweet and salted whey has been reported.39,40 The genetic stability of the fusants is a concern when using this methodology.37
As aforementioned, some microorganisms are natural lactose consumers, with their lactose metabolization genes being potential candidates for cloning in S. cerevisiae cells in a direct metabolic engineering approach. Predominantly three lactose consumers have been used as sources of lactose genes: the bacteria E. coli, the yeast K. lactis and the filamentous fungi A. niger. Two different strategies can be devised: to clone both the lactose permease and β-galactosidase genes or to direct the β-galactosidase production to the extracellular medium.
Transfer of E. coli lactose genes.
Three different strategies have been designed to obtain lactose-consuming S. cerevisiae cells using E. coli lactose genes:
As the β-galactosidase from E. coli is cytosolic, lactose has to be transported to the cytoplasm to be hydrolised. Thus, the cloning of the lacZ gene alone is not enough to obtain recombinants able to utilize lactose. The functional expression of the lacY gene will also be required. Guarente and Ptashne41 have shown the functional expression of the lacZ gene under the regulation of a yeast promoter, which is used widely as a reporter gene. However, when cloning the lac operon in a multicopy plasmid in S. cerevisiae it was not possible to obtain transformants able to utilize lactose.42 The yeast transformants, although expressing β-galactosidase, were not able to grow on lactose due to the non-functionality of the E. coli transport system.
An alternative approach involves the secretion of the E. coli β-galactosidase in S. cerevisiae cells. In E. coli, the attempt to direct β-galactosidase to the membrane using the signal sequence of a membranar protein (lamb) was ineffective.43 In S. cerevisiae, different signal sequences have been tried, namely from the SUC2,44 MFα45 and STA2,46 genes, but these attempts were also unsuccessful. With the STA2 signal sequence the authors were able to detect 76% of β-galactosidase activity in the periplasmatic space but no enzyme activity was detected in the culture medium. The fusion of glucoamylase residues with E. coli β-galactosidase was shown to facilitate its secretion although the secretion was not as efficient as with the glucoamylase gene.47 However, the authors do not mention if the recombinants were able to grow on lactose. Using the signal sequence of the membranar protein GgpI (the major yeast glycosylphosphatidylinositol-containing protein), it was possible to direct the E. coli β-galactosidase to the extracellular medium and for the first time, positive growth on lactose was observed.48
The third approach described in the literature deals with the spontaneous lysis of yeast cells overproducing the E. coli β-galactosidase enzyme.49 However, it is worth noting that cell lysis has a negative impact on downstream processing, which represents a disadvantage over the secretion approach. Porro et al.49 related the release of β-galactosidase activity in the culture medium by recombinant S. cerevisiae with the overexpression of the transcriptional activator GAL4, which induced partial lysis of the mother cells.50 Fermentation experiments with these transformants have shown that the release of β-galactosidase in the culture medium was enough to support growth in culture medium containing lactose as the sole carbon source and in whey-based culture medium. Ethanol production was observed in stationary phase with interesting yields (73–84% of the theoretical conversion yield) but unsatisfactory productivities (0.1–0.2 g·L−1·h−1) (Table 2). Interestingly, diauxic growth was not observed. The authors suggest that an excess of Gal4p may modify the regulatory pathways, leading to a change in cell wall composition, which in turn would be responsible for the lysis of older cells. As Gal4p is involved in glucose repression of galactose-utilizing genes, the excess of Gal4p could also be responsible for the simultaneous metabolization of glucose and galactose by the transformants.49 Compagno et al.51 reported the use of S. cerevisiae cells expressing the lacZ gene permeabilized by toluene in the bioconversion of lactose/whey to fructose diphosphate. Compagno et al.52 have crossed the S. diastaticus yeast strain JM2099 (having glucoamylase activities allowing a partial hydrolysis of starch) with the laboratory strain W303 to develop a strain able to grow simultaneously on starch and whey/lactose. Haploid cells able to grow on starch and bearing the appropriate mutation (i.e., leu2) have been isolated and transformed with the plasmid pM1 (previously used in the generation of lactose-utilizing yeast strains49). In this way, a yeast strain for the simultaneous utilization of lactose and starch has been developed.
Table 2.
Fermentation of lactose to ethanol by recombinant S. cerevisiae strains
| Characteristics of the strain | Cultivation conditions | Ethanol productivity (g·L·h−1) | Ethanol produced (g·L−1) | Ethanol yield (%)a | Lactose consumed (%)b | Reference |
| Autolytic cells expressing E. coli lacZ | Shake-flasks; Yeast Nitrogen Base/Lactose (2–6%) | 0.1–0.2 | 5–18 | 73–84 | >97 | 49 |
| Batch/Fed-batch; YP/Lactose (6%) + whey | 1.0 | 9 | 60–70 | 100 | 52 | |
| Expression of K. lactis LAC4 and LAC12 | Batch; Synthetic lactose (2.2%) medium | 0.3 | 4 | 34 | 100 | 55 |
| Continuous; Semi-synthetic lactose (5%) medium | 10–11 | 20 | 70–80 | >94 | 58 | |
| Batch; Cheese whey permeate (10% lactose) | 1.8 | 53 | >98 | 100 | 63 | |
| Continuous; Cheese whey permeate (5% lactose) | 9–10 | 20 | 70–80 | >98 | 63 | |
| Batch; Whey powder solution (15% lactose) | 0.46 | 55 | 70 | >98 | 66 | |
| Batch; Synthetic lactose (15%) medium | 1.5–2.0 | 63 | 78–84 | >98 | 68 | |
| Secretion of A. niger β-galactosidase | Anaerobic shake-flasks; Whey permeate (10% lactose) | 0.14–0.6 | 9.7 | 86 | 21 | 76 |
| Aerobic bioreactor; Synthetic lactose (10%) medium | 0.6 | 30 | 58 | 97 | 76 | |
| Batch; Semi-synthetic lactose (5%) medium | 1.0 | 25 | >80 | >90 | 78 | |
| Continuous; Semi-synthetic lactose (5%) medium | 9.0 | 20 | 74–83 | >90 | 81 | |
| Continuous; Semi-synthetic lactose (10%) medium | 3.4–7.0 | 32–48 | 70–90 | >75 | 81 |
Percentage of the theoretical yield, which is 0.538 g of ethanol produced per g of lactose consumed (i.e., 4 mol of ethanol produced per mol of lactose consumed).
Percentage of the initial lactose that was consumed during fermentation.
Transfer of K. lactis lactose genes.
The utilization of lactose by Kluyveromyces strains is based on a lactose transport system together with an intracellular β-galactosidase (see above). Hence, the same three strategies used with the E. coli lactose metabolizing genes have also been used with the Kluyveromyces LAC genes. However, as the lactose transport system from Kluyveromyces is eukaryotic, it is more prone to work in the phylogenetically related S. cerevisiae than the E. coli one. Indeed, using simultaneous expression of the lactose permease and intracellular β-galactosidase it was possible to obtain S. cerevisiae cells growing on lactose.53–56
Sreekrishna and Dickson53 were the first to construct Lac+ S. cerevisiae strains by transfer of the LAC12 and LAC4 genes of K. lactis. A 13 kb region of the K. lactis genome, comprising the two genes and their intergenic region, was used in the construction. Thus, transcriptional expression of the genes was controlled by the endogenous K. lactis promoter (see above). These authors have only obtained Lac+ transformants when using indirect selection (first selected for G418 resistance and then for growth on lactose). Moreover, they reported that the Lac+ transformants had integrated 15–25 tandem copies of the vector containing the LAC genes into a host chromosome. The transformants obtained presented a slow growth phenotype in lactose medium (doubling time in lactose minimal media of 6.7 h).53
Jeong et al.54 have constructed the plasmid SH096 by isolating the K. lactis lactose-utilizing genes, including LAC4 and LAC12, and cloning that DNA into a yeast integrative vector. The yeast strains transformed with this vector grew weakly on minimal lactose medium.54
More recently, Rubio-Teixeira et al.55 cloned the LAC4 and LAC12 genes in S. cerevisiae but their strategy involved placing LAC genes under the control of the CYC-GAL promoter (a galactose-inducible hybrid promoter) and targeting genomic integration to the ribosomal DNA region (RDN1 locus). The Lac+ transformants were selected in culture medium containing lactose as the sole carbon source. However, the transformants grew slowly in lactose while being stable mitotically. The transformants were then crossed with wild-type strains, yielding meiotic segregants with good growth and lactose assimilation capacity. Finally, two selected haploids were mated to generate a fast-growing Lac+ diploid strain. This strain exhibited a respiro-fermentative metabolism similar to that of K. lactis, with high biomass yield but low ethanol production55 (Table 2). The same approach was used to construct Lac+ baker's yeast57 with the ability to metabolize lactose. Growth of the new strain on cheese whey affected neither the quality of bread nor the yeast gassing power.57
In our laboratory, a flocculent S. cerevisiae Lac+ strain was constructed56 using the same plasmid (pKR1B-LAC4-1) employed by Sreekrishna and Dickson53 but with a different selection procedure. The plasmid KR1B-LAC4-1 was co-transformed with a linear fragment of the plasmid YAC4 (containing the URA3 gene) into an ura− strain (S. cerevisiae NCYC869-A3). Selection was done for ura− complementation in minimal medium plates containing galactose as carbon source. Xgal (5-bromo-4-chloro-3-indolyl- β-D-galactopyranoside) was included in the plates, allowing the identification of clones with β-galactosidase activity (blue colonies). Only four blue colonies were obtained (out of 1212) and only 2 kept a stable Lac+ phenotype. One of these transformants exhibited unusual morphology and pseudo-mycelium and therefore was rejected. The other (named S. cerevisiae NCYC869-A3/T1, or simply T1) was selected for more detailed characterization. Surprisingly, the recombinant strain metabolized the same amount of lactose (10 g·L−1) regardless of the initial lactose concentration and ethanol production was very low. Moreover, the doubling time of the recombinant strain in lactose minimal medium was 5 h.56 After an adaptation period, where the strain was maintained in periodically-refreshed liquid lactose medium, the performance of the recombinant strain in lactose culture medium was significantly improved and an ethanol conversion yield close to the theoretical value could be obtained. T1 kept the plasmid pKR1B-Lac4-1 in its autonomous form contrary to the Lac+ transformants obtained by Sreekrishna and Dickson.53 The recombinant T1 presented different flocculation behaviour from the host strain S. cerevisiae NYCY869-A3 although the recombinant strain was able to flocculate. The flocculation phenotype of the recombinant strain was more sensitive to environmental conditions than that of the host by losing its ability to flocculate more easily. After the adaptation period referred above, it was observed that this recombinant strain metabolized 50 g·L−1 lactose in less than 40 h, producing 16 g·L−1 ethanol.56
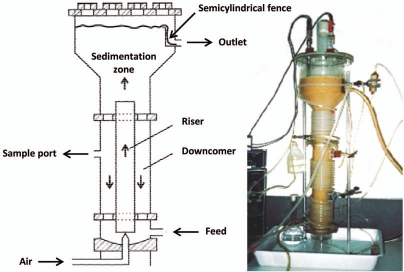
These preliminary results indicated that the recombinant strain could be used in a continuous high-cell-density fermentation system if the flocculation instability could be overcome.58 Continuous operation in a bioreactor with an appropriate design59 can be used to select for the most flocculating cells from a mixed culture, allowing for the possibility of accumulating a high biomass concentration in the bioreactor with the inherent advantages of operating as a continuous high-cell-density system.60 The choice of using an airlift bioreactor, which exhibits low shear stresses due to the absence of mechanical agitation,61 enabled the selection for the most flocculating yeast cells. The non-flocculent cells were washed-out from the bioreactor and the existence of a sedimentation zone in the top of the reactor coupled with the semicylindrical fence in the outlet region helped in the retention of the cells with higher sedimentation ability (Fig. 1). Selection of a 100% flocculent culture was achieved within just 13 days of continuous operation. For the continuously operating bioreactor, an ethanol productivity of 11 g·L−1·h−1 (corresponding to a feed lactose concentration of 50 g·L−1 and a dilution rate of 0.55 h−1) was obtained (Table 2), which is 7 times larger than the continuous conventional systems.58 The system stability was confirmed by keeping it in operation for 6 months.58 Also, the resistance of this system to nonflocculent contaminants was proved by artificially contaminating the bioreactor operating at 0.45 h−1 dilution rate with a 1 × 107 cells·mL−1 culture of recombinant E. coli expressing GFP (Green Fluorescent Protein).62
Figure 1.
Schematic representation and photography (credits: Lucília Domingues) of the airlift bioreactor.
When operating in continuous high-cell-density system using cheese whey permeate as substrate an ethanol productivity near 10 g·L−1·h−1 (corresponding to 0.45 h−1 dilution rate) was obtained63 (Table 2). While producing ethanol, the recombinant S. cerevisiae strain cleared the cheese whey permeate of most organic substances, allowing for a significant reduction in the pollutant load of cheese whey. The use of two-fold concentrated cheese whey permeate was also considered, resulting in a fermentation product with 5% (w/v) ethanol.63 However, it was not possible to operate continuously using the high-cell-density airlift bioreactor with concentrated cheese whey due to a deflocculating effect attributed to the salts concentration.63 A hydrodynamic and rheological analysis of the continuous airlift bioreactor operating at high-cell-density with the recombinant T1 strain was conducted.64 Measurements of liquid circulation velocity revealed a critical value of biomass concentration at which a dramatic deceleration of net liquid flow appeared with increasing biomass quantity. Rheological analysis demonstrated an exponential increase in viscosity of the yeast floc suspension at the same biomass concentration (around 73 g·L−1) corresponding to 42.8% v/v of solid fraction.64 A multi-route, non-structural kinetic model was developed for interpretation of ethanol fermentation of lactose using the recombinant flocculent T1 strain.65 In this model, the values of different metabolic pathways were calculated applying a modified Monod equation rate in which the growth rate is proportional to the concentration of a key enzyme controlling the single metabolic pathway. Three main metabolic routes for S. cerevisiae were considered: oxidation of lactose, reduction of lactose (producing ethanol), and oxidation of ethanol. A very good agreement between experimental data and simulated profiles of the main variables (lactose, ethanol, biomass and dissolved oxygen concentrations) was achieved.65
Unexpectedly, the strain lost its improved phenotype after storage at −80°C. Thus, another adaptation period was required for the already adapted culture of T1 that had been kept at −80°C.56 With the objective of obtaining a stable recombinant that could be used industrially, a long-term evolutionary engineering experiment was conducted and a stable evolved strain was obtained and named T1-E.66 We identified two molecular events that targeted the LAC construct in the evolved strain: a 1,593-bp deletion in the intergenic region (promoter) between LAC4 and LAC12 and a decrease of the plasmid copy number by about 10-fold compared to that in the original recombinant. Moreover, we have compared the transcriptomes of the original and the evolved recombinant strains growing in lactose, using cDNA microarrays. Microarray data revealed 173 genes whose expression levels differed more than 1.5-fold.67 About half of these genes were related to RNA-mediated transposition and the others were genes involved in DNA repair and recombination mechanisms, response to stress, chromatin remodeling, cell cycle control, mitosis regulation, glycolysis and alcoholic fermentation.67 The evolved T1-E strain retained improved lactose fermentation and enhanced flocculation phenotype even after −80°C storage. Strain T1-E was able to ferment efficiently high concentrations of lactose to ethanol, producing a maximum of 8% (v/v) ethanol from mineral medium with 150 g·L−1 lactose68 (Table 2). It was also capable of fermenting three-fold concentrated cheese whey, containing 150 g·L−1 lactose, yielding an ethanol titre of 7% (v/v)66 (Table 2).
Finally, the use of thermosensitive autolytic mutants has been reported in order to release Kluyveromyces β-galactosidase into the culture medium.69,70 Recombinant S. cerevisiae strains that are able of secreting K. lactis β-galactosidase have also been constructed.71,72 These approaches were used with the aim of developing a system for K. lactis β-galactosidase production and not for lactose bioconversion to ethanol. More recently, and with the same aim, a hybrid protein between K. lactis and A. niger β-galactosidase was constructed that increased the yield of the recombinant protein released to the growth medium.73
Transfer of A. niger lactose genes.
The filamentous fungi A. niger is an efficient producer of several secreted glycoproteins, some of which are used in industrial processes. Among these is β-galactosidase, mainly used to hydrolyze lactose in acid whey.74 The cloning of the lacA gene (coding for A. niger β-galactosidase) with its own signal sequence resulted in recombinant S. cerevisiae cells secreting β-galactosidase.75–78
Kumar et al.75 have obtained S. cerevisiae cells growing on lactose by transforming the cells with a yeast multicopy expression vector carrying the cDNA for A. niger secretory β-galactosidase under the control of ADH1 promoter and terminator. Ramakrishnan and Hartley76 studied the fermentation properties of the transformants and transformed polyploid distiller's yeast (Mauri) with the same vector. Diauxic growth patterns were not observed for the transformants growing on lactose while a typical biphasic growth was observed on a mixture of glucose and galactose under aerobic and anaerobic conditions. Rapid and complete lactose hydrolysis and higher ethanol (0.31 g per g of sugar) and biomass (0.24 g per g of sugar) production were observed with distiller's yeast grown under aerobic conditions.76 However, plasmid stability was low.
In our laboratory, flocculent S. cerevisiae strains secreting β-galactosidase were constructed77,78 using the vector developed by Kumar et al.75 Optimization of bioreactor operation together with culture conditions (lactose and yeast extract concentration) led to a 21-fold increase in the extracellular β-galactosidase produced when compared with preliminary shake-flask fermentations.79 To improve the genetic stability of the strains the lacA gene expression cassette was targeted to the δ-sequences in the genome.80 Even though our main goal was to produce heterologously A. niger β-galactosidase, we have observed that these strains produced ethanol from lactose/whey with close to theoretical yields in batch78 and in high-cell-density continuous fermentations81 with complete lactose utilisation (Table 2). The use of this strain in the dairy industry is very attractive for the simultaneous production of ethanol and β-galactosidase and the reduction of the organic load of the whey. The recombinant enzyme can be used for the generation of other products within the dairy industry (e.g., lactose-free products, hydrolyzed whey syrups).
Conclusions
For the conceivable future, there will be large surplus of whey and whey permeates worldwide. There is not one single solution to the problem of excess whey. The dairy industry should explore further the new possibilities for lactose as a raw material for processing in food and especially non-food industries.2 The development of processes and products for high volume markets will determine if a larger utilization of the lactose present in cheese whey is possible. Lactose-to-ethanol processes may be one of the solutions. Genetic engineering approaches have been used for the last 25 years with the aim of developing S. cerevisiae strains for such processes. Different strategies have been employed, as reviewed here, namely using the lactose metabolization genes from the bacteria E. coli, the yeast K. lactis and the filamentous fungi A. niger. When considering the metabolic engineering of S. cerevisiae cells for lactose-to-ethanol bioprocesses, the best results have been obtained with recombinants constructed with the K. lactis genes. Nevertheless, the direct cloning of LAC4 and LAC12 from K. lactis with its own promoter did not allow the direct selection of transformants in lactose plates and resulted in recombinants with a slow growth phenotype in lactose medium. When using other yeast promoters, direct selection of transformants was possible but the slow growth phenotype was still observed. This indicates that the cloning of the LAC4 and LAC12 genes per se is not enough to obtain a good lactose growth phenotype. Thus, the crossovers with wild-type strains or evolutionary engineering approaches were needed for the successful generation of efficient lactose-consuming recombinants. In the near future, further metabolic engineering efforts combined with improved bioprocess design will drive the development of more efficient fermentation processes for the conversion of concentrated whey to bioethanol. Furthermore, the simultaneous production of multiple commodities from whey (for instance β-galactosidase and ethanol) will improve the economics of whey fermentation processes.
Acknowledgements
The financial support of Fundação para a Ciência e a Tecnologia (FCT), Portugal, is acknowledged: project ProBioethanol PTDC/BIO/66151/2006, grant SFRH/BPD/44328/2008 to P.M.R. Guimarães. The authors gratefully acknowledge Russell Paterson for English proof read of the manuscript and useful comments.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/10619
References
- 1.Smithers GW. Whey and whey proteins—From ‘gutter-to-gold’. Int Dairy J. 2008;18:695–704. [Google Scholar]
- 2.Lifran EV, Hourigan JA, Sleigh RW, Johnson RL. New wheys for lactose. Food Australia. 2000;52:120–125. [Google Scholar]
- 3.Bailey JE. Toward a science of metabolic engineering. Science. 1991;252:1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen J. Metabolic engineering. Appl Microbiol Biotechnol. 2001;55:263–283. doi: 10.1007/s002530000511. [DOI] [PubMed] [Google Scholar]
- 5.Adam AC, Rubio-Texeira M, Polaina J. Lactose: the milk sugar from a biotechnological perspective. Crit Rev Food Sci Nutr. 2004;44:553–557. doi: 10.1080/10408690490931411. [DOI] [PubMed] [Google Scholar]
- 6.Teather RM, Muller-Hill B, Abrutsch U, Aichele G, Overath P. Amplification of the lactose carrier protein in Escherichia coli using a plasmid vector. Mol Gen Genet. 1978;159:239–248. doi: 10.1007/BF00268260. [DOI] [PubMed] [Google Scholar]
- 7.Büchel DE, Gronenborn B, Muller-Hill B. Sequence of the lactose permease gene. Nature. 1980;283:541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- 8.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 10.Kaback HR. Structure and mechanism of the lactose permease. C R Biol. 2005;328:557–567. doi: 10.1016/j.crvi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Panesar PS, Kennedy JF, Gandhi DN, Bunko K. Bioutilisation of whey for lactic acid production. Food Chem. 2007;105:1–14. [Google Scholar]
- 12.Seiboth B, Pakdaman BS, Hartl L, Kubicek CP. Lactose metabolism in filamentous fungi; how to deal with an unknown substrate. Fungal Biology Reviews. 2007;21:42–48. [Google Scholar]
- 13.Fukuhara H. Kluyveromyces lactis—a retrospective. FEMS Yeast Res. 2006;6:323–324. doi: 10.1111/j.1567-1364.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 14.Rubio-Texeira M. Endless versatility in the biotechnological applications of Kluyveromyces LAC genes. Biotechnol Adv. 2006;24:212–225. doi: 10.1016/j.biotechadv.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Chang YD, Dickson RC. Primary structure of the lactose permease gene from the yeast Kluyveromyces lactis. Presence of an unusual transcript structure. J Biol Chem. 1988;263:16696–16703. [PubMed] [Google Scholar]
- 16.Dickson RC, Barr K. Characterization of lactose transport in Kluyveromyces lactis. J Bacteriol. 1983;154:1245–1251. doi: 10.1128/jb.154.3.1245-1251.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boze H, Moulin G, Galzy P. Galactose and lactose transport in Kluyveromyces lactis. Folia Microbiol. 1987;32:107–111. doi: 10.1007/BF02883236. [DOI] [PubMed] [Google Scholar]
- 18.Guimarães PMR, Multanen JP, Domingues L, Teixeira JA, Londesborough J. Stimulation of zero-trans rates of lactose and maltose uptake into yeasts by preincubation with hexose to increase the adenylate energy charge. Appl Environ Microbiol. 2008;74:3076–3084. doi: 10.1128/AEM.00188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnett JA, Sims AP. The requirement of oxygen for the active transport of sugars into yeasts. J Gen Microbiol. 1982;128:2303–2312. [Google Scholar]
- 20.Van den Broek PJA, Van Steveninck J. Kinetic analysis of H+/methyl β-D-thiogalactoside symport in Saccharomyces fragilis. Biochim Biophys Acta. 1982;693:213–220. doi: 10.1016/0005-2736(82)90489-8. [DOI] [PubMed] [Google Scholar]
- 21.Carvalho-Silva M, Spencer-Martins I. Modes of lactose uptake in the yeast species Kluyveromyces marxianus. Antonie Van Leeuwenhoek. 1990;57:77–81. doi: 10.1007/BF00403158. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Q, Michels CA. The maltose permease encoded by the MAL61 gene of Saccharomyces cerevisiae exhibits both sequence and structural homology to other sugar transporters. Genetics. 1989;123:477–484. doi: 10.1093/genetics/123.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poch O, L'Hote H, Dallery V, Debeaux F, Fleer R, Sodoyer R. Sequence of the Kluyveromyces lactis β-galactosidase: comparison with prokaryotic enzymes and secondary structure analysis. Gene. 1992;118:55–63. doi: 10.1016/0378-1119(92)90248-n. [DOI] [PubMed] [Google Scholar]
- 24.Rubio-Texeira M. A comparative analysis of the GAL genetic switch between not-so-distant cousins: Saccharomyces cerevisiae versus Kluyveromyces lactis. FEMS Yeast Res. 2005;5:1115–1128. doi: 10.1016/j.femsyr.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Dickson RC, Dickson LR, Markin JS. Purification and properties of an inducible β-galactosidase isolated from the yeast Kluyveromyces lactis. J Bacteriol. 1979;137:51–61. doi: 10.1128/jb.137.1.51-61.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheetz RM, Dickson RC. Mutations affecting synthesis of β-galactosidase activity in the yeast Kluyveromyces lactis. Genetics. 1980;95:877–890. doi: 10.1093/genetics/95.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaffrath R, Breunig KD. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet Biol. 2000;30:173–190. doi: 10.1006/fgbi.2000.1221. [DOI] [PubMed] [Google Scholar]
- 28.Breunig KD, Bolotin-Fukuhara M, Bianchi MM, Bourgarel D, Falcone C, Ferrero I, et al. Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microbiol Technol. 2000;26:771–780. doi: 10.1016/s0141-0229(00)00170-8. [DOI] [PubMed] [Google Scholar]
- 29.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 31.Bhat PJ, Murthy TVS. Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction. Mol Microbiol. 2001;40:1059–1066. doi: 10.1046/j.1365-2958.2001.02421.x. [DOI] [PubMed] [Google Scholar]
- 32.Sumner-Smith M, Bozzato RP, Skipper N, Davies RW, Hopper JE. Analysis of the inducible MEL1 gene of Saccharomyces carlsbergensis and its secreted product α-galactosidase (melibiase) Gene. 1985;36:333–340. doi: 10.1016/0378-1119(85)90188-x. [DOI] [PubMed] [Google Scholar]
- 33.Naumov GI, Naumova ES, Louis EJ. Genetic mapping of the α-galactosidase MEL gene family on right and left telomeres of Saccharomyces cerevisiae. Yeast. 1995;11:481–483. doi: 10.1002/yea.320110512. [DOI] [PubMed] [Google Scholar]
- 34.Nehlin JO, Carlberg M, Ronne H. Yeast galactose permease is related to yeast and mammalian glucose transporters. Gene. 1989;85:313–319. doi: 10.1016/0378-1119(89)90423-x. [DOI] [PubMed] [Google Scholar]
- 35.Gödecke A, Zachariae W, Arvanitidis A, Breunig KD. Coregulation of the Kluyveromyces lactis lactose permease and β-galactosidase genes is achieved by interaction of multiple LAC9 binding sites in a 2.6 kbp divergent promoter. Nucleic Acids Res. 1991;19:5351–5358. doi: 10.1093/nar/19.19.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taya M, Honda H, Kobayashi T. Lactose-utilizing hybrid strain derived from Saccharomyces cerevisiae and Kluyveromyces lactis by protoplast fusion. Agric Biol Chem. 1984;48:2239–2243. [Google Scholar]
- 37.Farahnak F, Seki T, Ryu DD, Ogrydziak D. Construction of lactose-assimilating and high ethanol producing yeasts by protoplast fusion. Appl Environ Microbiol. 1986;51:362–367. doi: 10.1128/aem.51.2.362-367.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu YW, Jang HW, Lee HS. Enhancement of ethanol tolerance of lactose assimilating yeast strain by protoplast fusion. J Microbiol Biotechnol. 1991;1:151–156. [Google Scholar]
- 39.Tahoun MK, El-Nemr TM, Shata OH. Ethanol from lactose in salted cheese whey by recombinant Saccharomyces cerevisiae. Zeitschrift Fur Lebensmittel-Untersuchung Und-Forschung A—Food Research and Technology. 1999;208:60–64. (Ger). [Google Scholar]
- 40.Tahoun MK, El-Nemr TM, Shata OH. A recombinant Saccharomyces cerevisiae strain for efficient conversion of lactose in salted and unsalted cheese whey into ethanol. Nahrung. 2002;46:321–326. doi: 10.1002/1521-3803(20020901)46:5<321::AID-FOOD321>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 41.Guarente L, Ptashne M. Fusion of Escherichia coli LacZ to the cytochrome C gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadaban MJ, Martinezarias A, Shapira SK, Chou J. β-Galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 43.Moreno F, Fowler AV, Hall M, Silhavy TJ, Zabin I, Schwartz M. A signal sequence is not sufficient to lead β-galactosidase out of the cytoplasm. Nature. 1980;286:356–359. doi: 10.1038/286356a0. [DOI] [PubMed] [Google Scholar]
- 44.Das RC, Shultz JL, Lehman DJ. α-Factor leader sequence directed transport of Escherichia coli β-galactosidase in the secretory pathway of Saccharomyces cerevisiae. Mol Gen Genet. 1989;218:240–248. doi: 10.1007/BF00331274. [DOI] [PubMed] [Google Scholar]
- 45.Emr SD, Schauer I, Hansen W, Esmon P, Schekman R. Invertase β-galactosidase hybrid proteins fail to be transported from the endoplasmic reticulum in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2347–2355. doi: 10.1128/mcb.4.11.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanoni M, Porro D, Martegani E, Alberghina L. Secretion of Escherichia coli β-galactosidase in Saccharomyces cerevisiae using the signal sequence from the glucoamylase-encoding STA2 gene. Biochem Biophys Res Com. 1989;164:1331–1338. doi: 10.1016/0006-291x(89)91815-9. [DOI] [PubMed] [Google Scholar]
- 47.Venturini M, Morrione A, Pisarra P, Martegani E, Vanoni M. In Saccharomyces cerevisiae a short amino acid sequence facilitates excretion in the growth medium of periplasmic proteins. Mol Microbiol. 1997;23:997–1007. doi: 10.1046/j.1365-2958.1997.2841649.x. [DOI] [PubMed] [Google Scholar]
- 48.Pignatelli R, Vai M, Alberghina L, Popolo L. Expression and secretion of β-galactosidase in Saccharomyces cerevisiae using the signal sequences of GgpI, the major yeast glycosylphosphatidylinositol-containing protein. Biotechnol Appl Biochem. 1998;27:81–88. doi: 10.1111/j.1470-8744.1998.tb01378.x. [DOI] [PubMed] [Google Scholar]
- 49.Porro D, Martegani E, Ranzi BM, Alberghina L. Lactose/whey utilization and ethanol production by transformed Saccharomyces cerevisiae cells. Biotechnol Bioeng. 1992;39:799–805. doi: 10.1002/bit.260390802. [DOI] [PubMed] [Google Scholar]
- 50.Martegani E, Brambilla L, Porro D, Ranzi BM, Alberghina L. Alteration of cell population structure due to cell lysis in Saccharomyces cerevisiae cells overexpressing the GAL4 gene. Yeast. 1993;9:575–582. doi: 10.1002/yea.320090603. [DOI] [PubMed] [Google Scholar]
- 51.Compagno C, Tura A, Ranzi BM, Martegani E. Bioconversion of lactose whey to fructose diphosphate with recombinant Saccharomyces cerevisiae cells. Biotechnol Bioeng. 1993;42:398–400. doi: 10.1002/bit.260420319. [DOI] [PubMed] [Google Scholar]
- 52.Compagno C, Porro D, Smeraldi C, Ranzi BM. Fermentation of whey and starch by transformed Saccharomyces cerevisiae cells. Appl Microbiol Biotechnol. 1995;43:822–825. doi: 10.1007/BF02431914. [DOI] [PubMed] [Google Scholar]
- 53.Sreekrishna K, Dickson RC. Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc Natl Acad Sci USA. 1985;82:7909–7913. doi: 10.1073/pnas.82.23.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong YS, Vieth WR, Matsuura T. Fermentation of lactose to ethanol with recombinant yeast in an immobilized yeast membrane bioreactor. Biotechnol Bioeng. 1991;37:587–590. doi: 10.1002/bit.260370614. [DOI] [PubMed] [Google Scholar]
- 55.Rubio-Texeira M, Castrillo JI, Adam AC, Ugalde UO, Polaina J. Highly efficient assimilation of lactose by a metabolically engineered strain of Saccharomyces cerevisiae. Yeast. 1998;14:827–837. doi: 10.1002/(SICI)1097-0061(19980630)14:9<827::AID-YEA281>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 56.Domingues L, Teixeira JA, Lima N. Construction of a flocculent Saccharomyces cerevisiae fermenting lactose. Appl Microbiol Biotechnol. 1999;51:621–626. doi: 10.1007/s002530051441. [DOI] [PubMed] [Google Scholar]
- 57.Adam AC, Prieto JA, Rubio-Texeira M, Polaina J. Construction of a lactose-assimilating strain of baker's yeast. Yeast. 1999;15:1299–1305. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1299::AID-YEA454>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 58.Domingues L, Dantas MM, Lima N, Teixeira JA. Continuous ethanol fermentation of lactose by a recombinant flocculating Saccharomyces cerevisiae strain. Biotechnol Bioeng. 1999;64:692–697. doi: 10.1002/(sici)1097-0290(19990920)64:6<692::aid-bit8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 59.Teixeira JA, Mota M, Goma G. Continuous ethanol production by a flocculating strain of Kluyveromyces marxianus: bioreactor performance. Bioprocess Eng. 1990;5:123–127. [Google Scholar]
- 60.Domingues L, Lima N, Teixeira JA. An integrated approach for cheese whey lactose valorisation. Recent Res Devel Biotech Bioeng. 2003;5:65–78. [Google Scholar]
- 61.Domingues L, Vicente AA, Lima N, Teixeira JA. Applications of yeast flocculation in biotechnological processes. Biotechnol Bioprocess Eng. 2000;5:288–305. [Google Scholar]
- 62.Domingues L, Lima N, Teixeira JA. Contamination of a high-cell-density continuous bioreactor. Biotechnol Bioeng. 2000;68:584–587. doi: 10.1002/(sici)1097-0290(20000605)68:5<584::aid-bit14>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 63.Domingues L, Lima N, Teixeira JA. Alcohol production from cheese whey permeate using genetically modified flocculent yeast cells. Biotechnol Bioeng. 2001;72:507–514. doi: 10.1002/1097-0290(20010305)72:5<507::aid-bit1014>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 64.Klein J, Maia J, Vicente AA, Domingues L, Teixeira JA, Jurašcík M. Relationships between hydrodynamics and rheology of flocculating yeast suspensions in a high-cell-density airlift bioreactor. Biotechnol Bioeng. 2005;89:393–399. doi: 10.1002/bit.20335. [DOI] [PubMed] [Google Scholar]
- 65.Jurašcík M, Guimarães P, Klein J, Domingues L, Teixeira J, Markoš J. Kinetics of lactose fermentation using a recombinant Saccharomyces cerevisiae strain. Biotechnol Bioeng. 2006;94:1147–1154. doi: 10.1002/bit.20941. [DOI] [PubMed] [Google Scholar]
- 66.Guimarães PMR, François J, Parrou JL, Teixeira JA, Domingues L. Adaptive evolution of a lactose-consuming Saccharomyces cerevisiae recombinant. Appl Environ Microbiol. 2008;74:1748–1756. doi: 10.1128/AEM.00186-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guimarães PMR, Le Berre V, Sokol S, François J, Teixeira JA, Domingues L. Comparative transcriptome analysis between original and evolved recombinant lactose-consuming Saccharomyces cerevisiae strains. Biotechnol J. 2008;3:1591–1597. doi: 10.1002/biot.200800111. [DOI] [PubMed] [Google Scholar]
- 68.Guimarães PMR, Teixeira JA, Domingues L. Fermentation of high concentrations of lactose to ethanol by engineered flocculent Saccharomyces cerevisiae. Biotechnol Lett. 2008;30:1953–1958. doi: 10.1007/s10529-008-9779-1. [DOI] [PubMed] [Google Scholar]
- 69.Becerra M, Cerdan E, Siso MIG. Heterologous Kluyveromyces lactis β-galactosidase production and release by Saccharomyces cerevisiae osmotic-remedial thermosensitive autolytic mutants. Biochim Biophys Acta. 1997;1335:235–241. doi: 10.1016/s0304-4165(97)00048-2. [DOI] [PubMed] [Google Scholar]
- 70.Becerra M, Rodriguez-Belmonte E, Esperanza Cerdan M, Gonzalez Siso MI. Engineered autolytic yeast strains secreting Kluyveromyces lactis β-galactosidase for production of heterologous proteins in lactose media. J Biotechnol. 2004;109:131–137. doi: 10.1016/j.jbiotec.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 71.Becerra M, Prado SD, Cerdán E, Siso MIG. Heterologous Kluyveromyces lactis β-galactosidase secretion by Saccharomyces cerevisiae super-secreting mutants. Biotechnol Lett. 2001;23:33–40. [Google Scholar]
- 72.Becerra M, Prado SD, Siso MI, Cerdan ME. New secretory strategies for Kluyveromyces lactis β-galactosidase. Protein Eng. 2001;14:379–386. doi: 10.1093/protein/14.5.379. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez AP, Leiro RF, Trillo MC, Cerdan ME, Siso MI, Becerra M. Secretion and properties of a hybrid Kluyveromyces lactis-Aspergillus niger β-galactosidase. Microb Cell Fact. 2006;5:41. doi: 10.1186/1475-2859-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holsinger VH, Kligerman AE. Applications of lactase in dairy foods and other foods containing lactose. Food Technol. 1991;45:92. [Google Scholar]
- 75.Kumar V, Ramakrishnan S, Teeri TT, Knowles JKC, Hartley BS. Saccharomyces cerevisiae cells secreting an Aspergillus niger β-galactosidase grow on whey permeate. Bio-Technology. 1992;10:82–85. doi: 10.1038/nbt0192-82. [DOI] [PubMed] [Google Scholar]
- 76.Ramakrishnan S, Hartley BS. Fermentation of lactose by yeast cells secreting recombinant fungal lactase. Appl Environ Microbiol. 1993;59:4230–4235. doi: 10.1128/aem.59.12.4230-4235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Domingues L, Onnela ML, Teixeira JA, Lima N, Penttila M. Construction of a flocculent brewer's yeast strain secreting Aspergillus niger β-galactosidase. Appl Microbiol Biotechnol. 2000;54:97–103. doi: 10.1007/s002530000358. [DOI] [PubMed] [Google Scholar]
- 78.Domingues L, Teixeira JA, Penttila M, Lima N. Construction of a flocculent Saccharomyces cerevisiae strain secreting high levels of Aspergillus niger β-galactosidase. Appl Microbiol Biotechnol. 2002;58:645–650. doi: 10.1007/s00253-002-0948-1. [DOI] [PubMed] [Google Scholar]
- 79.Domingues L, Oliveira C, Castro I, Lima N, Teixeira JA. Production of β-galactosidase from recombinant Saccharomyces cerevisiae grown on lactose. J Chem Technol Biotechnol. 2004;79:809–815. [Google Scholar]
- 80.Oliveira C, Teixeira JA, Lima N, Da Silva NA, Domingues L. Development of stable flocculent Saccharomyces cerevisiae strain for continuous Aspergillus niger β-galactosidase production. J Biosci Bioeng. 2007;103:318–324. doi: 10.1263/jbb.103.318. [DOI] [PubMed] [Google Scholar]
- 81.Domingues L, Lima N, Teixeira JA. Aspergillus niger β-galactosidase production by yeast in a continuous high cell density reactor. Process Biochem. 2005;40:1151–1154. [Google Scholar]