Abstract
In this study, the effect of homologous multiple copies of the ask gene, which encodes aspartokinase catalyzing the first step of the aspartate pathway, on cephamycin C biosynthesis in S. clavuligerus NRRL 3585 and its hom mutant was investigated. The intracellular pool levels of aspartate pathway amino acids accorded well with the Ask activity levels in TB3585 and AK39. When compared with the control strain carrying vector alone without any gene insert, amplification of the ask gene in the wild strain resulted in a maximum of 3.1- and 3.3-fold increase in specific, 1.7- and 1.9-fold increase in volumetric cephamycin C production when grown in trypticase soy broth (TSB) and a modified chemically defined medium (mCDM), respectively. However, expression of multicopy ask gene in a hom-deleted background significantly decreased cephamycin C yields when the cells were grown in either TSB or mCDM, most probably due to physiological disturbance resulting from enzyme overexpression and high copy number plasmid burden in an auxotrophic host, respectively.
Key words: aspartate pathway, aspartokinase, cephamycin C, homologous expression, Streptomyces clavuligerus
Introduction
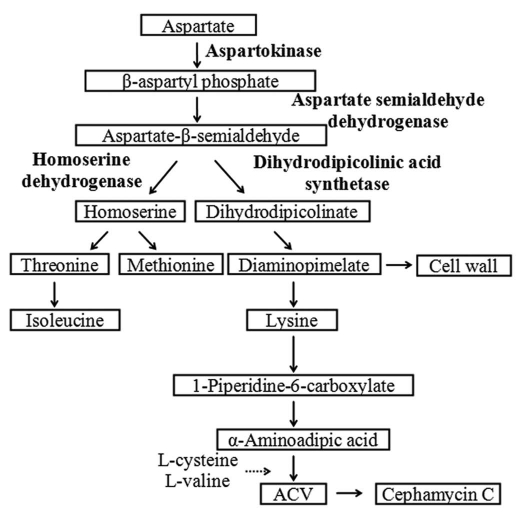
Streptomyces clavuligerus is an actinomycete well known for its ability to produce a variety of β-lactam antibiotics including cephamycin C and structurally related β-lactam compounds, the clavams. The initial step in the biosynthesis of β-lactams is the non-ribosomal condensation of L-α-aminoadipate (L-α-AAA), L-cysteine and L-valine, to form the tripeptide α-aminoadipyl-cysteinyl-valine.1 In actinomycetes, α-AAA is a catabolic product of L-lysine produced from the lysine branch of the aspartate pathway2 and as shown in Figure 1, its availability is critical regarding precursor flux from the aspartate pathway to β-lactam biosynthesis.3,4
Table 1.
Bacterial strains and plasmids used in this study
| Strains & plasmids | Description | Source or reference |
| Strains | ||
| S. clavuligerus | ||
| NRRL 3585 | Wild type, cephamycin C and clavulanic acid producer | Higgins and Kastner32 |
| AK39 | Null mutant of S. clavuligerus NRRL 3585, hom::kan | Yilmaz et al.17 |
| TB3585 | S. clavuligerus NRRL 3585 carrying pTB486, ThioR | This study |
| BA39 | S. clavuligerus AK39 carrying pTB486, ThioR, KanR | This study |
| TBV | S. clavuligerus NRRL 3585 carrying pIJ486, ThioR | This study |
| BAV | S. clavuligerus AK39 carrying pIJ486, ThioR, KanR | This study |
| E. coli | ||
| ESS | β-lactam supersensitive E. coli strain | Aharonowitz and Demain18 |
| ET12567 | F−, dam 13::Tn9 dcm-6 hsdM hsdR lacYI | Prof. K. Chater, John Innes Centre, Colney, Norwich, UK |
| Plasmids | ||
| pGEM-T | AmpR, lacZ′ | Promega |
| pBluescript II KS (+) | pBluescript II KS (+) | Stratagene |
| pTBKS | pBluescript II KS carrying 1,506 bp S. clavuligerus ask gene | This study |
| pIJ486 | Streptomyces plasmid vector, pIJ101 replicon, ThioR | Prof. K. Chater, John Innes Centre, Colney, Norwich, UK |
| pNST102 | pBluescript II KS carrying 3,500 bp S. clavuligerus ask-asd cluster | Tunca et al.11 |
| pTB486 | pIJ486 carrying 1,506 bp S. clavuligerus ask gene | This study |
Different gene organization and regulatory mechanisms controlling the metabolic flux through the aspartate pathway have been identified in bacteria.5 The ask (encoding the aspartokinase; EC 2.7.2.4) and asd (aspartate semialdehyde dehydrogenase; EC 1.2.1.11) genes are clustered in an operon in mycobacteria, corynebacteria and bacilli6–9 as well as in Amycolatopsis10 and S. clavuligerus,11 but not in Streptomyces akiyoshiensis.12 The regulation of the aspartate pathway involves the multiplicity of the aspartokinase, first enzyme of the pathway, in certain bacteria like Escherichia coli and bacilli.13,14 In other organisms, including actinomycetes, only one aspartokinase has been described. It is feedback-regulated by either lysine as in Amycolatopsis mediterranei15 or by the concerted action of lysine and threonine as in S. clavuligerus and A. lactamdurans.10,16
The formation of L-homoserine from aspartate-β-semialdehyde (ASA) by the homoserine dehydrogenase (Hsd; EC 1.1.1.3) is the first step of the branch of the aspartate pathway leading to L-threonine, L-isoleucine and L-methionine. Recently, the hom gene coding for this enzyme was cloned and characterized from S. clavuligerus and a hom mutant of this organism was constructed by cassette mutagenesis. The disruption of hom resulted in a two-fold increase in cephamycin C specific production in chemically defined medium.17 The present study is aimed at the expression of multiple copies of the homologous ask gene in the wild strain of S. clavuligerus NRRL3585 and its hom-disrupted mutant, and to determine the effects of ask amplification on cephamycin C production.
Results and Discussion
In this study, two different ask transformants were constructed, S. clavuligerus TB3585 and S. clavuligerus BA39, containing multiple copies of the ask gene in the wild-type strain and in the hom mutant S. clavuligerus AK39, respectively. In order to obtain control strains the plasmid pIJ486 was introduced into wild-type and AK39 and the transformants were named TBV and BAV, respectively (Table 1).
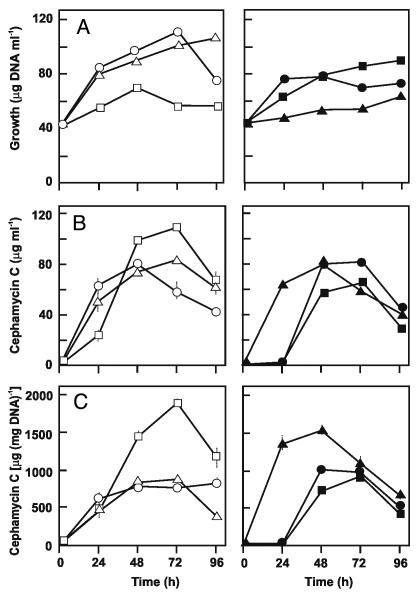
Cephamycin C volumetric production by TB3585 was 1.3-fold higher at 48th h and 1.7-fold higher at 96th h of growth as compared to the control TBV strain in TSB medium (Fig. 2B). This results in a specific production 2.2-times higher at 72 h and 3.1-times higher at 96 h fermentation, supporting a positive effect of additional copies of the ask gene on cephamycin C yields (Fig. 2C).
Figure 2.
Growth (A), volumetric (B) and specific (C) cephamycin C production of S. clavuligerus NRRL 3585 (open circles), TB3585 (open squares), TBV (open triangles), AK39 (closed circles), BA39 (closed squares), BAV (closed triangles) grown in TSB medium. The experiment was performed in duplicate flasks and triplicate samples were assayed from each flask.
The expression on a multicopy plasmid of the homologous ask gene in the hom-disrupted BA39 strain was expected to lead to a further increase in cephamycin C levels due to: (1) the funnelling of the aspartate flow directly to L-lysine rather than being shared by the two branches (Fig. 1) and (2) a reduced/null concerted feedback inhibition of the aspartokinase due to lack of L-threonine biosynthesis. However, antibiotic production by the hom minus transformants carrying the amplified ask gene was delayed and the stimulatory effect on cephamycin production exerted by the ask overexpression in the wild type strain was not observed in the hom minus background (Fig. 2, right).
Figure 1.
The aspartate pathway in S. clavuligerus.
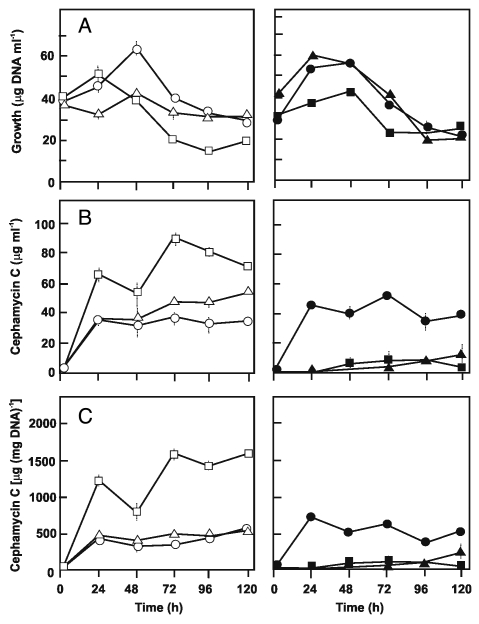
It was of interest to test if these results also occur in cells grown in defined medium. In our previous study, the growth of the AK39 strain was rather poor in chemically-defined medium, CDM, in spite of a two-fold increased specific level of cephamycin C.17 Some inhibitory effects of glycerol on cephamycin C production18,19 as well as improvement of growth and cephamycin C yields by L-asparagine20,21 have been previously reported. In the present study, the composition of CDM was submitted to several modifications to improve the growth of this strain and their transformants. The highest growth and antibiotic biosynthesis were obtained by replacing glycerol with sucrose, a carbon source not utilized by S. clavuligerus, but producing an homogeneous and disperse growth, and by increasing two-fold the L-asparagine concentration, since this amino acid is used as carbon and nitrogen source by S. clavuligerus. In this modified medium (mCDM), growth (Fig. 3A) and cephamycin C titers of the strains were lower than those in complex TSB medium, i.e., maximum cephamycin specific production by TB3585 was 1,890 µg mg−1 in TSB while it was 1,581 µg mg−1 in mCDM. Nevertheless, the antibiotic level of TB3585 relative to its control TBV was higher in mCDM with 3.3- and 1.9-fold increase of specific and volumetric production at 72 h, respectively (Fig. 3B and C). As expected, the beneficial effect of hom-disruption in the AK39 strain was more pronounced in mCDM than in complex TSB (1.8-fold versus 1.3-fold increase in specific production). However the antibiotic levels of the BAV and BA39 strains in mCDM medium were very low and of almost equal level, suggesting an adverse effect of the plasmid itself as well as the multiple copies of ask. It was shown earlier that metabolic changes which occur when cloned genes are expressed in auıxotrophic yeast strains may well arise from peculiarities of such hosts rather than the activity of the cloned proteins, hence avoidance of such auxotrophic host systems was strongly recommended.22 Pleiotropic depression of antibiotic titers in auxotrophic mutants of streptomyces were also mentioned.23 Our results supported these warnings.
Figure 3.
Growth (A), volumetric (B) and specific (C) cephamycin C production of S. clavuligerus NRRL 3585 (open circles), TB3585 (open squares), TBV (open triangles), AK39 (closed circles), BA39 (closed squares), BAV (closed triangles) grown in defined mCDM medium. The experiment was performed in duplicate flasks and triplicate samples were analyzed from each flask.
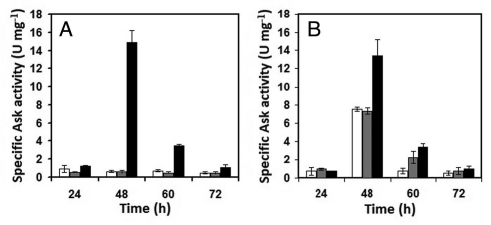
Expression of the multicopy ask gene in both strains was verified in aspartokinase assays on crude cell extracts of each strain grown in mCDM. Althought no noticeable differences were found at 24 h of fermentation, aspartokinase specific activities of the TB3585 and BA39 strains were 25- and 1.8-fold higher than TBV and BAV, respectively, at 48 h (Fig. 4).
Figure 4.
Specific Ask activities of (A) S. clavuligerus NRRL 3585 (white), TBV (grey), TB3585 (black) and (B) AK39 (white), BAV (grey), BA39 (black) grown in defined mCDM medium. The experiment was performed in duplicate flasks and triplicate samples were analyzed from each flask.
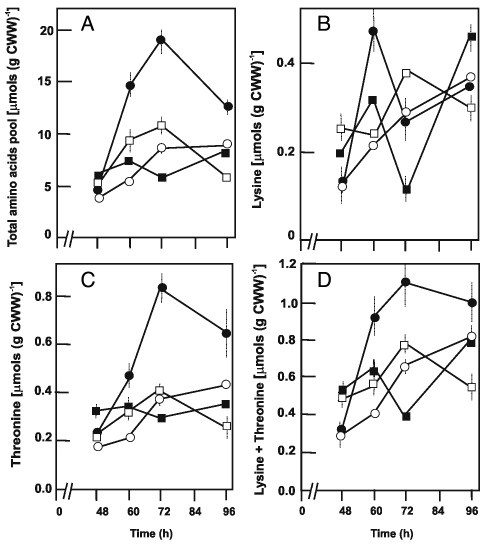
The intracellular free amino acid pools in the wild type strain and in the recombinants (TB3585, AK39 and BA39) were next determined in cultures grown in mCDM in order to relate aspartokinase overexpression with amino acid biosynthesis. Using asparagine as the sole carbon and nitrogen source, glycine was the predominating amino acid in the wild type strain followed by asparagine, glutamic acid and serine (data not shown). There were strong differences in the total pool content as well as in individual amino acids between strains. The glutamic acid and glycine contents of AK39 were 6 to 10 times higher compared to those of the wild type and TB3585 strains, and it always contained much less asparagine than the other strains. The TB3585 strain contained 2- to 4-fold more asparagine than the other strains. The highest intracellular amino acid content (2.2-fold) was found in AK39 (Fig. 5A). This strain synthesized appreciable amounts of lysine till 60 h (Fig. 5B) and extensively accumulated externally added threonine (Fig. 5C). Thus its intracellular lysine plus threonine contents were almost always much higher than that of the other strains (Fig. 5D).
Figure 5.
Total amounts of the intracellular free amino acids (A) and intracellular pool of lysine (B), threonine (C) and lysine plus threonine (D) in S. clavuligerus NRRL 3585 (open circles), TB3585 (open squares), AK39 (closed circles), BA39 (closed squares) grown in defined mCDM medium. The experiment was performed in duplicate flasks and triplicate samples were analyzed from each flask.
The intracellular pool levels of aspartate pathway amino acids accorded well with the Ask activity levels in TB3585 and AK39 in that the drastic increment of Ask specific activity at 48 h was followed by a sharp increase in the intracellular pool levels of lysine and threonine till 72 h during which Ask activity gradually decreased. Cephamycin C levels also increased in this interval, particularly in TB3585. Thus, the initial production of antibiotic within first 24 h of cultivation in both wild-type and recombinant strains seemed to be independent of Ask expression level and intracellular pools of amino acids.
Metabolic engineering has proven to be a rational alternative to classical strain improvement and today the β-lactam industry combines the two approaches. However, there are few studies aimed at enhancing cephamycin C yields by the manipulation of the aspartate pathway in S. clavuligerus. Malmberg et al. applied a metabolic engineering strategy which involved the introduction of an additional copy of the L-lysine ε-aminotransferase (lat) gene into the chromosome of S. clavuligerus.24 Its product (LAT) constitutes the first step in cephamycin C biosynthesis in which L-lysine coming from the aspartate pathway is eventually converted to α-AAA. This manipulation resulted in a 2- to 4-fold increase in specific cephamycin C production. Pérez-Llarena et al. demonstrated that amplification of the ccaR gene on a multicopy plasmid resulted in a two to three fold increase in cephamycin and clavulanic acid yields.25 ccaR gene is located within the cephamycin gene cluster of S. clavuligerus and its product has a regulatory function in cephamycin C and clavulanic acid biosynthesis.25 This gene carries upstream of the promoter a 26 bp ARE sequence (AREccaR) for binding of a butyrolactone receptor protein.26 When grown in starch-asparagine (SA) medium a mutant, S. clavuligerus ΔareB, unable to produce the butyrolactone receptor protein, produced 1.4- to 3-fold more clavulanic acid, but the cephamycin C yield remained similar to that of the wild-type strain.
In the present study, we demonstrated that the amplification of the ask gene in S. clavuligerus TB3585 results in a significant increase in the volumetric and specific production of cephamycin C, after 24 h growth in complex TSB medium. Although the growth and antibiotic formation were lower in defined medium, the pattern followed by the different strains was similar to that of cultures grown in complex medium.
The lack of the effect of ask amplification on cephamycin C production in a hom minus background might be due to multiple reasons. The pool of amino acids in this background, including those of lysine and threonine, was much higher than in other strains. This might affect the aspartokinase activity through a concerted feedback inhibition. Lysine + threonine pool increased 3.3-fold between 48 h and 72 h in mCDM (Fig. 5D) in this strain while its Ask activity decreased by 14-fold (Fig. 4B). Mendelovitz and Aharonowitz reported that threonine at 10 mM concentration inhibited cephamycin production by 41% and different amino acids, as methionine or 2,6-diaminopimelate, affected the antibiotic production by inhibition of different enzymes of the pathway.16 Therefore, since the amino acid pool in the hom minus strain is higher, the aspartokinase, as well as other enzymes of the aspartate pathway leading to α-aminoadipic acid formation might be subjected to different types of regulation in this strain. Besides α-aminoadipic acid formation, aspartate pathway provides the methyl group (via S-adenosyl-L-methionine) and sulfur atom (transsulfuration) to cephamycin C as contributed by methionine, involves cysteine as an intermediate plus shares common enzymes with the valine biosynthetic pathway; cysteine and valine being the part of the antibiotic.27–30 If the aspartokinase regulation were the main responsible for a lack of further stimulation of cephamycin C production, the cloning of a feedback deregulated aspartokinase gene from a S-(2-aminoethyl)-L-cysteine resistant mutant might solve the problem.
Materials and Methods
Bacterial strains, plasmids, media and culture conditions.
Bacterial strains and plasmids used are described in Table 1. E. coli cultures were grown in either Luria broth (LB; Q-Biogene, CA) or on Luria agar at 37°C. S. clavuligerus was maintained on sporulation agar31 supplemented with CoCl2.6H2O (20 µg ml−1). For isolation of plasmid DNA and protoplast preparation, seed culture media containing trypticase soy broth (TSB, Oxoid, UK) supplemented with 0.5% (w/v) maltose (Merck, Germany) was inoculated with spore stocks of S. clavuligerus and incubated at 28°C on a rotary shaker (220 rpm) in baffled flasks for 48–60 h. Five ml of this seed culture were inoculated into 50 ml of 2:3 (v/v) mixture of TSB and yeast extract-malt extract medium (YEME,31) supplemented with 0.3% (v/v) glycine (Merck) and 3 mM MgCl2 and incubated at 28°C for 24 h. In the case of plasmid containing cultures, ampicillin (100 µg ml−1 for E. coli, Sigma), or thiostrepton (50 µg ml−1 for S. clavuligerus, Sigma) was added to the medium. For antibiotic selection, Streptomyces colonies were plated on trypticase soy agar (TSA) supplemented with 8 µg ml−1 thiostrepton. For cephamycin C assay, the cultures were grown in TSB or a modified chemically defined medium (mCDM) which was prepared as in Malmberg et al.24 with some modifications; the amount of L-asparagine (Sigma) was increased from 2 g l−1 to 4 g l−1 and 10 g l−1 glycerol was replaced with 25 g l−1 sucrose (Merck). mCDM was supplemented with L-methionine (Sigma) and L-threonine (50 mg l−1 each; Sigma) for growth of S. clavuligerus AK39, S. clavuligerus BA39 and S. clavuligerus BAV. Seed cultures were grown in 50 ml of TSB until mid-log phase and centrifuged at 3,200 rpm for 10 min at 4°C. The pellets were washed with TSB or mCDM and resuspended in 50 ml of fresh media. The cultures were grown in duplicate flasks at 28°C at 220 rpm for 120 h.
DNA manipulations.
Plasmid DNA was isolated using Qiagen Plasmid Midi kits (Qiagen, Germany). DNA fragments were isolated from agarose gels using Qiagen Quickgel extraction kits. Restriction enzyme digestion of DNA was carried out according to suppliers' recommendations. PCR products which were used in construction of recombinant plasmids were cloned in vector pGEM-T (Promega, WI) prior to transfer into the desired plasmid. Transformation of E. coli strains was performed by standard procedures.33 E. coli plasmids that were used in the construction of recombinant S. clavuligerus plasmids were propagated in E. coli ET12567 to avoid restriction barriers.
Cloning of S. clavuligerus ask gene into pIJ486.
Two oligonucleotide primers, 5′-TTC TAG AGT TCG TCC GGC TGC CGG T-3′ and 5′-GGA TAT CCT ACC GCC CAC TTC CCG C-3′, were used to generate a 1,506 bp DNA product comprising ask from the S. clavuligerus ask-asd operon.11 The PCR amplification was as follows: 95°C (10 min) and 30 cycles of 95°C (1 min), 63°C (1 min) and 72°C (2 min). The amplified DNA was cloned into pGEM-T, released using XbaI and EcoRV and inserted into XbaI-EcoRV digested pBluescript II KS (Stratagene, Germany) to generate pTBKS. pTBKS was then digested with XbaI and HindIII and the resulting ask gene was inserted into XbaI-HindIII digested pIJ486 to create pTB486.
Transformation of S. clavuligerus.
S. clavuligerus transformations were performed via PEG mediated protoplast transformation according to Kieser et al.31 with slight modifications. Transformants were regenerated on R2YE medium at 26°C for 48 h and then each plate was overlaid with thiostrepton containing 2.5 ml soft nutrient agar.
Measurement of aspartokinase (Ask) activity.
Aspartokinase (Ask) activity was measured in triplicates according to the procedure of Follettie et al.34 by determining the rate of formation of aspartyl-hydroxamate. Background activity was measured in the absence of aspartate. Ask specific activity was measured as nanomoles of aspartyl-hydroxamate formed per milligram protein per min.
Growth determination.
Cultural growth was determined by DNA quantification with diphenylamine method adapted from Burton.35 2 ml of cell culture was centrifuged at 13,200 rpm for 10 min and the pellet was resuspended in 1 ml of 0.85% NaCl (Merck). 0.5 ml of 1 N perchloric acid (Merck) was added into 0.5 ml of sample and incubated at 70°C for 20 min. To the mixture was added 1 ml of diphenylamine (Sigma) reagent and incubated overnight at 30°C. The mixture was centrifuged at 13,200 rpm for 10 min and the absorption was measured at 600 nm. DNA concentrations were calculated from a regression equation obtained by assaying known concentrations of herring sperm DNA (Sigma). The values were multiplied by a factor of four and the DNA concentration was defined as µg of DNA per ml of culture.
Bioassay of β-lactam antibiotics.
β-lactam antibiotic bioassays were conducted by agar plate diffusion with E. coli ESS 3235 as indicator organism.18 Zones of inhibition were measured and cephamycin C concentrations in samples calculated using a standard curve constructed with cephalosporin C (Sigma). Cephamycin concentration is calculated as cephalosporin C equivalents.
Determination of intracellular free amino acid concentrations.
Cells were grown in mCDM, and samples were collected at intervals during cultivation. Intracellular free amino acids were extracted as described by Yılmaz et al.17 Following automated pre-column derivatization with o-phthaladehyde (OPA), amino acid analysis was performed by conventional reverse-phase HPLC (Shimadzu VP series) on a Novapac C18 column (Waters) followed by Xuorescence detection. Amino acid concentrations were expressed as micromoles of amino acid extracted from 1 g wet weight of bacteria.
Acknowledgements
The authors would like to thank Dr. Sedef Tunca for her kind interest during the early stages of this work. We also thank Ibrahim Sertdemir and Memet Deveci for their technical assistance.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/11244
References
- 1.Zhang J, Wolfe S, Demain AL. Biochemical studies on the activity of delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Streptomyces clavuligerus. Biochem J. 1992;283:691–698. doi: 10.1042/bj2830691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kern BA, Hendlin D, Inamine E. L-lysine-ε-aminotransferase involved in cephamycin C synthesis in Streptomyces lactamdurans. Antimicrob Agents Chemother. 1980;17:679–685. doi: 10.1128/aac.17.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malmberg LH, Hu WS, Sherman DH. Effects of enhanced lysine ε-aminotransferase activity on cephamycin biosynthesis in Streptomyces clavuligerus. Appl Microbiol Biotechnol. 1995;44:198–205. doi: 10.1007/BF00164502. [DOI] [PubMed] [Google Scholar]
- 4.Rius N, Maeda K, Demain AL. Induction of L-lysine-ε-aminotransferase by L-lysine in Streptomyces clavuligerus, producer of cephalosporins. FEMS Microbiol Lett. 1996;144:207–211. doi: 10.1111/j.1574-6968.1996.tb08532.x. [DOI] [PubMed] [Google Scholar]
- 5.Malumbres M, Martin JF. Molecular control mechanisms of lysine and threonine biosynthesis in amino acid-producing corynebacteria: redirecting carbon flow. FEMS Microbiol Lett. 1996;143:103–114. doi: 10.1111/j.1574-6968.1996.tb08468.x. [DOI] [PubMed] [Google Scholar]
- 6.Kalinowski J, Bachmann B, Thierbach G, Puhler A. Aspartokinase genes lysCα and lysCβ overlap and are adjacent to the aspartate β-semialdehyde dehydrogenase gene asd in Corynebacterium glutamicum. Mol Gen Genet. 1990;224:317–324. doi: 10.1007/BF00262424. [DOI] [PubMed] [Google Scholar]
- 7.Chen NY, Jiang SQ, Klein DA, Paulus H. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem. 1993;268:9448–9465. [PubMed] [Google Scholar]
- 8.Cirillo JD, Weisbrod TR, Pascopella L, Bloom BR, Jacobs WR., Jr Isolation and characterization of the aspartokinase and aspartate semialdehyde dehydrogenase operon from mycobacteria. Mol Microbiol. 1994;11:629–639. doi: 10.1111/j.1365-2958.1994.tb00342.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Jiang W, Zhao G, Yang Y, Chiao J. Sequence analysis and expression of the aspartokinase and aspartate semialdehyde dehydrogenase operon from rifamycin SV-producing Amycolatopsis mediterranei. Gene. 1999;237:413–419. doi: 10.1016/s0378-1119(99)00307-8. [DOI] [PubMed] [Google Scholar]
- 10.Hernando-Rico V, Martin JF, Santamarta I, Liras P. Structure of the ask-asd operon and formation of aspartokinase subunits in the cephamycin producer “Amycolatopsis lactamdurans”. Microbiology. 2001;147:1547–1555. doi: 10.1099/00221287-147-6-1547. [DOI] [PubMed] [Google Scholar]
- 11.Tunca S, Yilmaz EI, Piret J, Liras P, Ozcengiz G. Cloning, characterization and heterologous expression of the aspartokinase and aspartate semialdehyde dehydrogenase genes of cephamycin C-producer Streptomyces clavuligerus. Res Microbiol. 2004;155:525–534. doi: 10.1016/j.resmic.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Le Y, He J, Vining LC. Streptomyces akiyoshiensis differs from other gram-positive bacteria in the organization of a core biosynthetic pathway gene for aspartate family amino acids. Microbiology. 1996;142:791–798. doi: 10.1099/00221287-142-4-791. [DOI] [PubMed] [Google Scholar]
- 13.Theze J, Margarita D, Cohen GN, Borne F, Patte JC. Mapping of the structural genes of three aspartokinases and of the two homoserine dehydrogenases of Escherichia coli K12. J Bacteriol. 1974;117:133–143. doi: 10.1128/jb.117.1.133-143.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JJ, Hu FM, Chen NY, Paulus H. Comparison of the three aspartokinase isoenzymes in Bacillus subtilis Marburg and 168. J Bacteriol. 1990;172:701–708. doi: 10.1128/jb.172.2.701-708.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang WW, Jiang WH, Zhao GP, Yang YL, Chiao JS. Expression in Escherichia coli, purification and kinetic analysis of the aspartokinase and aspartate semialdehyde dehydrogenase from the rifamycin SV-producing Amycolatopsis mediterranei U32. Appl Microbiol Biotechnol. 2000;54:52–58. doi: 10.1007/s002530000345. [DOI] [PubMed] [Google Scholar]
- 16.Mendelovitz S, Aharonowitz Y. Regulation of cephamycin C synthesis, aspartokinase, dihydrodipicolinic acid synthetase and homoserine dehydrogenase by aspartic acid family amino acids in Streptomyces clavuligerus. Antimicrob Agents Chemother. 1982;21:74–84. doi: 10.1128/aac.21.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yılmaz EI, Çaydasi AK, Özcengiz G. Targeted disruption of homoserine dehydrogenase gene and its effect on cephamycin C production in Streptomyces clavuligerus. J Ind Microbiol Biotechnol. 2008;35:1–7. doi: 10.1007/s10295-007-0259-8. [DOI] [PubMed] [Google Scholar]
- 18.Aharonowitz Y, Demain AL. Carbon catabolite regulation of cephalosporin production in Streptomyces clavuligerus. Antimicrob Agents Chemother. 1978;14:159–164. doi: 10.1128/aac.14.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saudagar PS, Singhal RS. Optimization of nutritional requirements and feeding strategies for clavulanic acid production by Streptomyces clavuligerus. Bioresource Technol. 2007;98:2010–2017. doi: 10.1016/j.biortech.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Chmiel A, Brzeszczynska A, Kabza B. Biosynthesis of cephamycin by resting cells of Streptomyces lactamdurans L 2/6. Acta Microbiol Pol. 1986;35:251–257. [PubMed] [Google Scholar]
- 21.Khaoua S, Lebrihi A, Germain P, Lefebvre G. Cephamycin C biosynthesis in Streptomyces cattleya: nitrogen source regulation. Appl Microbiol Biotechnol. 1991;35:253–257. doi: 10.1007/BF00184697. [DOI] [PubMed] [Google Scholar]
- 22.Çakar ZP, Sauer U, Bailey JE. Metabolic engineering of yeast: the perils of auxotrophic hosts. Biotechnol Lett. 1999;21:611–616. [Google Scholar]
- 23.Hopwood DA, Merrick MJ. Genetics of antibiotic production. Bacteriol Rev. 1977;41:595–635. doi: 10.1128/br.41.3.595-635.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmberg LH, Hu WS, Sherman DH. Precursor flux control through targeted chromosomal insertion of the lysine epsilon-aminotransferase (lat) gene in cephamycin C biosynthesis. J Bacteriol. 1993;175:6916–6924. doi: 10.1128/jb.175.21.6916-6924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Llarena FJ, Liras P, Rodriguez-Garcia A, Martin JF. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J Bacteriol. 1997;179:2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santamarta I, López-García MT, Pérez-Redondo R, Koekman B, Martín JF, Liras P. Connecting primary and secondary metabolism: AreB, an IclR-like protein, binds the ARE(ccaR) sequence of S. clavuligerus and modulates leucine biosynthesis and cephamycin C and clavulanic acid production. Mol Microbiol. 2007;66:511–524. doi: 10.1111/j.1365-2958.2007.05937.x. [DOI] [PubMed] [Google Scholar]
- 27.Aharonowitz Y, Mendelovitz S, Kirenberg F, Kuper V. Regulatory mutants of Streptomyces clavuligerus affected in free diaminopimelic acid content and antibiotic biosynthesis. J Bacteriol. 1984;157:337–340. doi: 10.1128/jb.157.1.337-340.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nüesch J, Heim J, Treichler HJ. The biosynthesis of sulfur-containing β-lactam antibiotics. Ann Rev Microbiol. 1987;41:51–75. doi: 10.1146/annurev.mi.41.100187.000411. [DOI] [PubMed] [Google Scholar]
- 29.Martin JF. New aspects of genes and enzymes for β-lactam antibiotic biosynthesis. Appl Microbiol Biotechnol. 1998;50:1–15. doi: 10.1007/s002530051249. [DOI] [PubMed] [Google Scholar]
- 30.Öster LM, Lester DR, van Scheltinga AT, Svenda M, van Lun M, Généreux C, et al. Insights into cephamycin biosynthesis: the crystal structure of CmcI from Streptomyces clavuligerus. J Mol Biol. 2006;358:546–558. doi: 10.1016/j.jmb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 32.Higgins CE, Kastner RE. Streptomyces clavuligerus sp. nov., a β-lactam antibiotic producer. Int J Syst Bacteriol. 1971;21:326–331. [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Follettie MT, Peoples OP, Agoropoulou C, Sinskey AJ. Gene structure and expression of the Corynebacterium flavum N13 ask-asd operon. J Bacteriol. 1993;175:4096–4103. doi: 10.1128/jb.175.13.4096-4103.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burton K. Determination of DNA concentration with diphenylamine. In: Grossman L, Moldave K, editors. Methods in Enzymology. 12B. New York: Academic Press; 1968. pp. 163–166. [Google Scholar]