Summary
Identifying cis-regulatory elements is important to understand how human pancreatic islets modulate gene expression in physiologic or pathophysiologic (e.g., diabetic) conditions. We conducted genome-wide analysis of DNase I hypersensitive sites, histone H3 lysine methylation modifications (K4me1, K4me3, K79me2), and CCCTC factor (CTCF) binding in human islets. This identified ~18,000 putative promoters (several hundred unannotated and islet-active). Surprisingly, active promoter modifications were absent at genes encoding islet-specific hormones, suggesting a distinct regulatory mechanism. Of 34,039 distal (non-promoter) regulatory elements, 47% are islet-unique and 22% are CTCF-bound. In the 18 type 2 diabetes (T2D)-associated loci, we identified 118 putative regulatory elements and confirmed enhancer activity for 12/33 tested. Among 6 regulatory elements harboring T2D-associated variants, 2 exhibit significant allele-specific differences in activity. These findings present a global snapshot of the human islet epigenome and should provide functional context for non-coding variants emerging from genetic studies of T2D and other islet disorders.
Introduction
Type 2 diabetes (T2D) is a complex metabolic disorder that accounts for 85-95% of all cases of diabetes and afflicts hundreds of millions of people worldwide (http://www.diabetesatlas.org/content/diabetes). It is a leading cause of substantial morbidity and is characterized by defects in insulin sensitivity and secretion resulting from the progressive dysfunction and loss of beta cells in the pancreatic islets of Langerhans (Butler et al., 2007; Muoio and Newgard, 2008). Both genetic predisposition and environmental factors contribute to these islet defects. Islets constitute 1-2% of human pancreatic mass (Joslin and Kahn, 2005) and are composed of five endocrine cell types that secrete different hormones: alpha cells (glucagon), beta cells (insulin), delta cells (somatostatin), PP cells (pancreatic polypeptide Y), and epsilon cells (ghrelin). These cells sense changes in blood glucose concentration and respond by modulating the activity of multiple pathways, including insulin and glucagon secretion, to maintain glucose homeostasis (Joslin and Kahn, 2005). Several key transcription factors (TFs) that regulate these responses are known (Oliver-Krasinski and Stoffers, 2008). However, efforts to identify cis-regulatory elements upon which these and other factors act have been restricted primarily to promoter regions at specific loci (e.g., INS, PDX1) (Brink, 2003; Ohneda et al., 2000).
Results from genome-wide association studies (GWAS) of type 1 diabetes (Barrett et al., 2009), T2D (reviewed in (Prokopenko et al., 2008)) and related metabolic traits (Dupuis et al., 2010; Ingelsson et al., 2010; Prokopenko et al., 2009) suggest that genetic variation in cis-regulatory elements may play an important role in beta cell (dys)function and diabetes susceptibility (DeSilva and Frayling, 2010). Of the 18 most strongly associated single nucleotide polymorphisms (SNPs) in each of the T2D-associated loci, only 3 are missense variants; the remaining are non-coding (Prokopenko et al., 2008). Furthermore, there is evidence for allele-specific effects of two T2D-associated SNPs on the islet-expression level of nearby genes (TCF7L2, Lyssenko et al., 2007; MTNR1B, Lyssenko et al., 2009). However, the dearth of annotation of functional regulatory elements has limited the capacity to investigate the role of regulatory variation in complex diseases such as T2D.
Recent characterization of histone modifications and DNase hypersensitivity in cultured cells has identified chromatin signatures predictive of regulatory elements and actively transcribed regions (Boyle et al., 2008; Guenther et al., 2007; Heintzman et al., 2007). The data generated so far suggest that regulatory element location and usage vary substantially among cell types (Heintzman et al., 2009; Xi et al., 2007). Also, extensive chromatin profiling has been conducted in very few human primary tissues to date (Bhandare et al., 2010). In this study, we describe a comprehensive genome-wide epigenomic map of unstimulated human pancreatic islets. Using DNase- and ChIP-seq approaches, we identified DNase I hypersensitive sites that mark regions of open chromatin, loci enriched for active histone H3 lysine methylation modifications (H3K4me1, H3K4me3, and H3K79me2), and binding sites for the insulator CCCTC binding factor (CTCF). These profiles provide a detailed chromatin snapshot of regulatory elements and actively transcribed units in the islet. Moreover, they identify regulatory elements harboring T2D-associated variants in 6/18 loci. These data provide a valuable resource to understand and investigate cis-regulation in the human islet and to discover regulatory elements that may play an important role in diabetes susceptibility.
Results
Genome-wide characterization of open chromatin in the human pancreatic islet
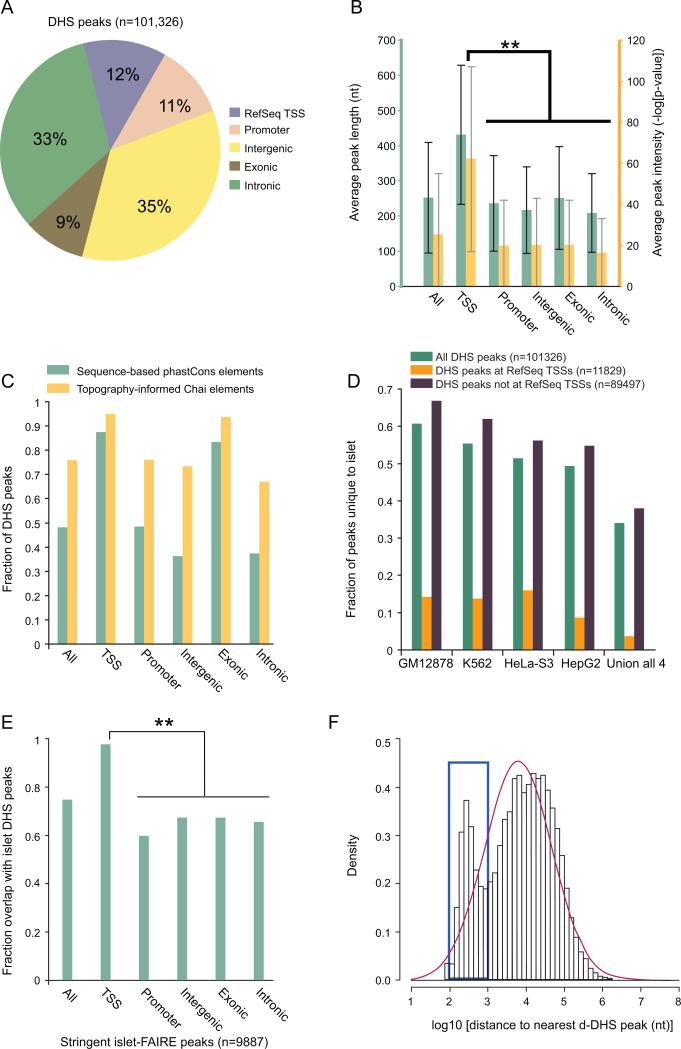
Active regulatory elements reside in open chromatin regions hypersensitive to DNase I digestion (ENCODE, 2007; Boyle et al., 2008; Crawford et al., 2004; Hesselberth et al., 2009; Sabo et al., 2004). To identify all DNase hypersensitive sites (DHS) in the human pancreatic islet, we performed DNase-seq (Boyle et al., 2008) and identified regions of the genome with significant enrichment of sequence reads using the MACS algorithm (Zhang et al., 2008; Methods). This approach identified 101,326 human islet DHS peaks (Table S1) covering ~27 million bases (~1% of the human genome). Consistent with observations in CD4+ T cells (Boyle et al., 2008), a substantive fraction of islet DHS peaks (23%, n=23,408) span annotated RefSeq transcription start sites (TSS) or are within regions 5kb upstream (Promoter), but the majority reside within currently un-annotated genomic regions that may harbor functional distal regulatory elements (Figure 1A). Peaks at TSSs are significantly longer and more intense than those at all other loci (Figure 1B). This observation supports the view that regions around TSSs are generally more susceptible to DNase I digestion than putative non-TSS regulatory elements (Boyle et al., 2008).
Figure 1. Analysis of DNase I hypersensitive sites in the islet genome.
(A) Distribution of DNase I hypersensitive (DHS) peaks across five genomic annotation sets. “Promoter” denotes proximal regions 5kb upstream of RefSeq transcription start sites (TSSs) that do not overlap the TSS. “Exonic” represents regions that overlap at least 1 base with an exon.
(B) Average length (teal) and intensity (yellow) of DHS peaks across five genomic annotation sets. Peaks at RefSeq transcription start sites (TSSs) are significantly longer and more intense than those elsewhere (** two-tailed paired Student's t-test p-value < 10-100). Error bars represent s.d. (s.d. measurements were often greater than the sample average due to highly skewed distributions, but error bars were cut off at zero for visualization).
(C) Sequence and structure constraint at DHS. DHS peaks at RefSeq TSSs are under substantially greater sequence constraint (assessed by phastCons vertebrate conservation scores) than intronic and intergenic DHS peaks. A large majority of DHS peaks within all genomic annotation sets are under strong structural constraint (assessed by the Chai algorithm (Parker et al., 2009)).
(D) Comparison of islet DHS peaks with peaks from 4 different human cell lines. Each data point represents the fraction of total peaks (n=101,326) unique to the human islet relative to each of the other 4 human cell types or all of them combined (Union of all 4). Roughly 35% are unique to the islet and 99% of these are not located at RefSeq TSSs. Varying levels of similarity across cell types may be at least partially explained by differences in the stage of cellular differentiation and/or sequencing depth.
(E) Overlap between DHS peaks and Formaldehyde-Assisted-Isolation-of-Regulatory-Elements (FAIRE) peaks. The overlap is significantly greater at RefSeq TSSs than elsewhere (**Fisher's exact test < 10-100).
(F) Logarithm-based distribution of the distance to the nearest distal DHS (d-DHS) peak among all d-DHS peaks. The blue box indicates an increased representation of peaks in the ~100-1000 bp range (clustered) relative to Gaussian expectation (red curve). This range is significantly enriched for islet-unique peaks (Fisher's exact test p=2.7 × 10-9). Comparison of d-DHS, FAIRE, and GLITR locations is found in Figure S1.
Approximately 48% (n=48,777) of all DHS peaks overlap phastCons vertebrate conserved elements (Siepel et al., 2005) (Figure 1C). Notably, ~87% (10,348/11,829) of peaks at TSSs overlap phastCons elements, compared to ~43% (38,429/89,497) at non-TSS loci (Figure 1C). This difference remains even after accounting for the longer peaks at TSSs (data not shown), supporting the model that TSS-proximal regions evolve under stronger sequence constraint than distal regulatory elements (Boyle et al., 2008). A recent study developed an algorithm (Chai) for topography-informed conservation analysis, which identified ~2-fold more bases in the human genome under evolutionary constraint compared to sequence-based methods (Parker et al., 2009). Accordingly, ~1.5 times as many (~76%) islet DHS peaks overlap these structurally constrained regions (Figure 1C).
To determine the extent of cell-type specificity of our islet DHS peaks, we obtained DNase-seq data generated for four different human cell lines: GM12878, K562, HeLa-S3 and HepG2 (Duke DNase, ENCODE Consortium (2007)). We identified DHS peaks for these cell lines (Methods) and found that roughly half of the islet peaks are shared with each individual non-islet cell type. Notably, ~35% (n=34,273) are completely unique to the islet (Figure 1D). Almost all (~99%) of these islet-unique peaks do not overlap RefSeq TSSs, which is consistent with the model that tissue-specific gene expression patterns are governed largely by distal cis-regulatory elements (Heintzman et al., 2009).
An independent method to map open chromatin is Formaldehyde Assisted Isolation of Regulatory Elements (FAIRE) (Giresi et al., 2007). Recently, this approach was used for human islets to identify three sets of candidate peaks, including “stringent” (n=9,887) and “liberal” (n=100,715) (Gaulton et al., 2010). Approximately 75% of the “stringent” islet FAIRE peaks overlap DHS peaks. However, this corresponds to only 7,360 peaks, which is far fewer than the predicted number of functional regulatory elements genome-wide (ENCODE Consortium, 2007). The overlap is significantly greater at TSSs compared to non-TSSs [97% vs. 65%] (Figure 1E). Comparing DHS peaks to the set of “liberal” islet FAIRE peaks, the overlap drops to ~29%. Therefore, the two approaches seem to identify distinct sets of non-TSS regulatory elements. Because it is difficult to assess the extent to which the dissimilarity between DHS and FAIRE data is explained by differences in islet sample purity, preparation methods, false positive signals, or population diversity (McDaniell et al., 2010), more controlled comparisons of these techniques will be necessary to elucidate inherent preferences of each for specific classes of open chromatin.
Though many of the mechanistic details are not clear, it is widely accepted that distal and promoter regulatory elements can exert coordinated control of gene transcription via physical interactions (Dekker, 2003; Miele and Dekker, 2008). Therefore, it has been hypothesized that distal cis-regulatory elements may cluster together to form functional modules (Blanchette et al., 2006). To assess the clustering of putative islet-active, distal cis-regulatory elements, we filtered from the islet DHS peaks (n=101,326) the regions that may represent promoters to identify a set of high confidence distal peaks (d-DHS, n=34,039; Table S2; Figure S1; Methods). For each d-DHS peak, we computed the distance to the nearest d-DHS peak and observed an increased representation in the ~100-1000 bp range (n=7,652) relative to the expectation from a normal distribution (Figure 1F). Furthermore, this set is significantly enriched for islet-unique peaks (p=2.7 × 10-9).
Genome-wide characterization of TSSs in the islet genome via H3K4me3 ChIP-seq
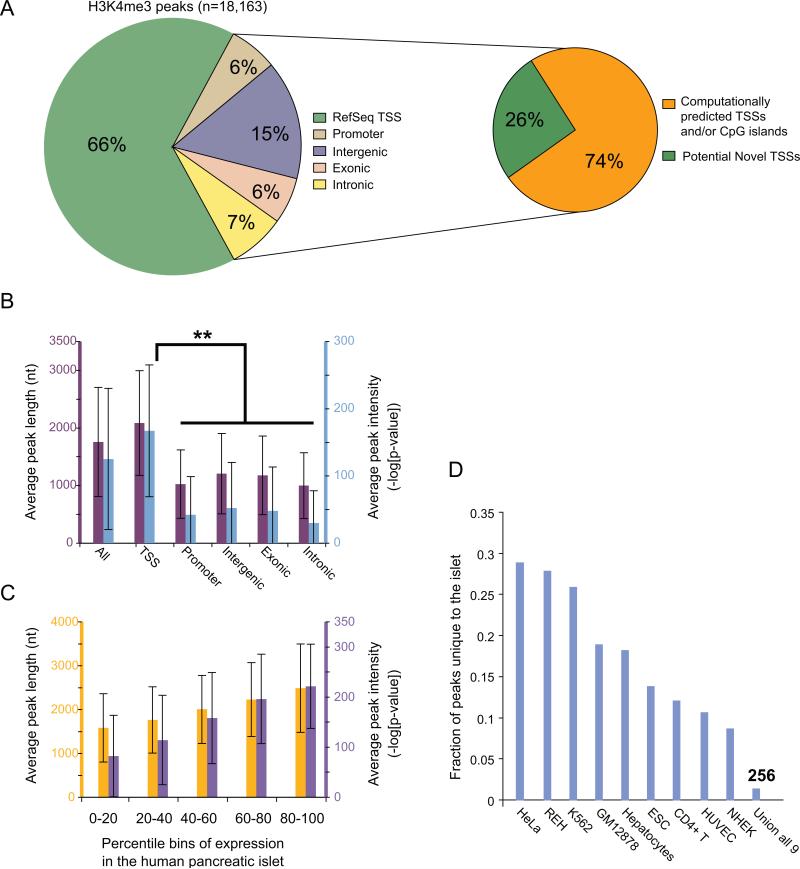
To characterize human islet TSSs, we conducted ChIP-seq analysis of histone 3 lysine 4 trimethylation (H3K4me3) in four different human islet samples. H3K4me3 is enriched at CpG islands (Bernstein et al., 2007), TSSs (Li et al., 2007), and sites of active transcription (Kouzarides, 2007). Enriched regions present in all four islet samples, but absent from three mock-IP (anti-GFP) experiments, were designated as “H3K4me3 peaks.” This method identified 18,163 human islet H3K4me3 peaks (Table S3) covering ~1% of the genome.
As expected, approximately 2/3 (n=11,973) of H3K4me3 peaks overlap RefSeq TSSs (Figure 2A). Greater than 70% of the remaining, unannotated peaks (n=6,190) overlap computationally predicted TSSs and/or CpG islands. However, the significantly lower average length and intensity of unannotated H3K4me3 peaks compared to those at RefSeq TSSs (Figure 2B) suggests that at least some of these peaks may indicate weakly active TSSs, inactive but poised TSSs (Barski et al., 2007; Guenther et al., 2007; Mikkelsen et al., 2007), remnants of transcriptional activity from the developmental past or prior environmental stimulation (Barski et al., 2009), or chromatin looping with distal regulatory regions. While a subset of peaks could be false positive signals, this is unlikely as it would require a technical artifact that is consistent across all four islet samples.
Figure 2. Analysis of histone 3 Lysine 4 tri-methylation (H3K4me3) loci in the islet genome.
(A) Distribution of H3K4me3 peaks across five genomic annotation sets as described in Figure 1A. 2/3 of the peaks span RefSeq transcription start sites (TSSs: left pie chart). Non-RefSeq H3K4me3 peaks are enriched for computationally predicted TSS and/or CpG islands (right pie chart). Additional information is provided in Figure S3.
(B) Average length (purple) and intensity (blue) of H3K4me3 peaks across five genomic annotation sets as described in Figure 1B. The average length and intensity of peaks is significantly higher at TSSs (** two-tailed paired Student's t-test p-value < 10-100). Error bars represent s.d.
(C) Relationship between average H3K4me3 peak length (yellow)/intensity (purple) and average gene expression level. Error bars represent s.d.
(D) Comparison of islet H3K4me3 peaks with peaks from 9 different human cell types. Each data point represents the fraction of total peaks (n=18,163) unique to the human islet relative to each of the other 9 human cell types or all of them combined (Union all 9). ~1.5% of the peaks are unique to the islet. Varying levels of similarity across cell types may be at least partially explained by differences in the stage of cellular differentiation and/or sequencing depth.
Previous genome-wide profiling studies have reported a positive correlation between the intensity of H3K4me3 signal and gene expression level (Barski et al., 2007; Guenther et al., 2007). To test this observation in islets, we downloaded human islet gene expression data from http://T1Dbase.org (Kutlu et al., 2009), partitioned gene expression into quintiles and computed the average H3K4me3 signal length and intensity at the TSSs of genes within each bin. Although the average H3K4me3 peak length and intensity monotonically increases with gene expression, there is great variability within each expression bin (Figure 2C). Surprisingly, of the 245 most highly islet-expressed genes in this data set, 18% (n=45) have either no or extremely low associated H3K4me3 signal. Notably, 71% (32/45) also lacked a DHS peak (data not shown). Gene Ontology (GO) analysis revealed that these 45 genes are most significantly enriched for the molecular function of hormone activity (p = 0.029 after Bonferroni correction for multiple testing; Methods). These genes include insulin (INS), glucagon (GCG), islet amyloid polypeptide (IAPP), pancreatic polypeptide preprotein (PPY), somatostatin (SST) and transthyretin (TTR). We confirmed by RT-qPCR that INS, GCG, and SST are robustly expressed (Figure S2), so it is unlikely that low H3K4me3 at these TSSs is due to technical artifacts or adverse effects of the islet shipment or handling process. Because these genes are <10 kb in length, we considered the possibility that weak H3K4me3 signal is simply associated with short genes. However, the proportion of short genes (<10 kb in length) within the set of “most highly expressed with no/low H3K4me3 signal” (66.7%, 30/45) is not statistically different from the proportion of short genes within the entire set of most highly expressed (69.8%, 171/245). This result suggests that the transcriptional regulation of islet hormones and other related, highly islet-expressed genes occurs through a distinct mechanism as compared to most other genes.
H3K4me3 ChIP-chip (human embryonic stem cells, hepatocytes, REH cells (Guenther et al., 2007)) or ChIP-seq (human CD4+ T cells (Barski et al, 2007); GM12878, HUVEC, NHEK, K562 and HeLa cell lines (Broad Institute ChIP-seq, Bernstein lab, ENCODE, 2007)) data are available for nine different human cell types. Comparisons between islet and each other cell type indicated that, on average, 10–30% of the islet peaks are unique (Figure 2D). Not surprisingly, this value drops to ~1.5% (n=256) when compared with all nine cell types together. Only 34 of the 256 islet-unique peaks correspond to TSSs of annotated RefSeq genes, and these are enriched for known pancreatic beta cell functions such as secretion (p=9.3 × 10-3) and Ca2+ dependent exocytosis (p=6.6 × 10-3) (Table 1). Furthermore, several of the genes (SLC30A8, GCK) harbor genetic variants that confer significant risk for T2D and elevated plasma fasting glucose levels (Dupuis et al., 2010; Ingelsson et al., 2010; Prokopenko et al., 2009; Prokopenko et al., 2008). The remaining 222 islet-unique peaks may represent alternative TSSs of genes with function in developing and/or mature islets, or TSSs of unannotated coding or non-coding transcription units.
Table 1. Examples of islet-unique H3K4me3 peaks.
9 examples among the 34 islet-unique peaks that are at RefSeq transcription start sites (TSSs). The corresponding genes have known pancreatic islet function (such as insulin secretion) and some harbor genetic variants that confer significant risk for type 2 diabetes (SLC30A8 and GCK).
| Gene symbol |
Relevance to islet biology |
|---|---|
| GCK | • Involved in glucose metabolism |
| • T2D GWAS locus (Zeggini et al., 2008) | |
| |
• Harbors an islet-specific promoter (Magnuson, 1990) |
| SLC30A8 | • Involved in cation (Zn+) transport important for insulin secretion (Chimienti et al., 2004) |
| • T2D GWAS locus (Zeggini et al., 2008) | |
| |
• Exhibits islet-specific expression (Chimienti et al., 2004) |
|
REG1A
|
• Derived from regenerating islets (Terazono et al., 1988) |
| FFAR1 | • Exhibits islet-specific expression (Bartoov-Shifman et al., 2007) |
| |
• Regulates insulin secretion (Itoh et al., 2003) |
|
SYT4
|
• Involved in Ca2+ dependent trafficking and exocytosis of secretory vesicles (Tsuboi and Rutter, 2003) |
|
KCNK16
|
• Exhibits pancreas specific expression (Girard et al., 2001) |
|
ELAVL4
|
• Regulates cell proliferation (Joseph et al., 1998) |
|
UCN3
|
• Regulates glucose-stimulated insulin secretion (Li et al., 2007) |
| PRSS1 | • Harbors mutations that underlie hereditary pancreatitis and pancreatic cancer (Teich et al., 1998) |
Identification of unannotated islet-active TSSs
H3K4me3 peaks in unannotated genomic space (n=6190) are TSS candidates. Because H3K4me3 may also be enriched at inactive TSSs (Guenther et al., 2007), we adopted a two-step approach to identify the subset of these 6,190 peaks that are likely to be active in the human islet (Figure S3A). First, we developed an algorithm that uses DHS peaks to assign directionality to H3K4me3 peaks (Methods). DHS peaks tend to be sharply focused around the TSS, while H3K4me3 peaks are broader and extend well into the body of the transcription unit. We hypothesized that the location of the DHS peak relative to the H3K4me3 peak could predict the directionality of the underlying gene. Using the strongest DHS peak within an H3K4me3 peak, this simple algorithm performed at ~90% accuracy on annotated RefSeq genes known to be expressed in the human islet (Methods). Interestingly, the majority (~80%) of the incorrectly assigned TSSs (based on current annotation) harbored multiple DHS peaks, positioned on either end of the H3K4me3 peak. These H3K4me3 peaks are slightly (~200 nt) longer than those for which the orientation was correctly assigned, increasing the likelihood of overlapping non-TSS-related DHS peaks, which can confound the prediction algorithm. Many of these non-TSS DHS peaks may correspond to CTCF binding sites that are located on the opposite side of the DHS with respect to the TSS (Boyle et al., 2008) and RNA polymerase (Pol) III bound loci found in chromatin domains occupied by Pol II and associated with enhancer-binding factors (Oler et al., 2010). We observe examples of each case in our dataset (Figure S4).
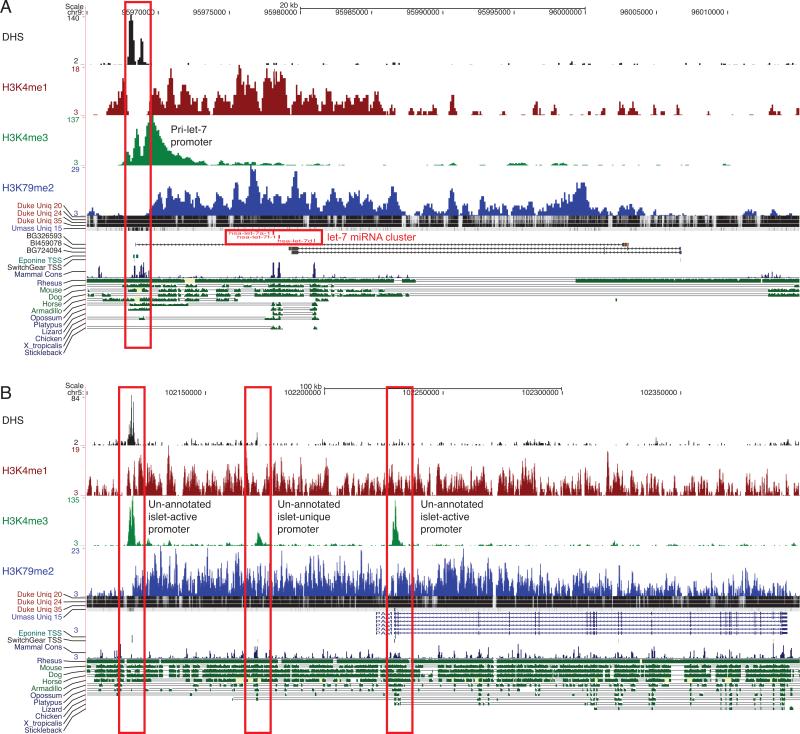
Second, we performed ChIP-seq to profile genome-wide histone 3 lysine 79 dimethylation (H3K79me2), which is enriched in actively transcribed regions (Guenther et al., 2007). If the relative density of H3K79me2 reads on either side of an H3K4me3 peak was consistent with its predicted directionality (as determined from the pattern of the DHS and H3K4me3 signal), then the underlying TSS was classified as islet-active. Intragenic TSSs are difficult to assess using this method because the H3K79me2 signal may be due to transcription from an upstream TSS. Restricting the analysis to intergenic space, we identified 263 candidates for unannotated, islet-active TSSs (Table S4), of which 75% (n=196) overlap CpG islands and/or computationally predicted TSSs (Figure S3A). These candidates include islet-active TSSs for non-coding RNAs such as the let-7a-1 cluster of microRNAs (Figure 3A) and the miR-1179/miR-7-2 cluster (Figure S3B). We also identified putative alternative TSSs for genes with important islet function such as pancreatic peptidylglycine alpha-amidating monooxygenase (PAM), which encodes for an islet secretory granule membrane protein (Figure 3B). Finally, we identified an active promoter locus that is contained within a recently reported type 1 diabetes (T1D) associated region on chromosome 12 (index SNP rs1701704). This promoter could underlie an unannotated transcript or could be an alternative promoter for the downstream gene Ikaros family zinc finger 4 (IKZF4) (Figure S3C), which is considered a strong functional candidate for T1D (Hakonarson et al., 2008).
Figure 3. Identifying unannotated islet-active transcription start sites (TSSs).
(A) Candidate islet-active TSS for the primary transcript of the ubiquitous let-7a-1/7d/7f-1 microRNA cluster. The TSS (red box; DHS+, H3K4me3+, H3K4me1-) is ~10kb upstream of the 5’-most microRNA in the cluster, and the full-length primary transcript (H3K79me2+) of ~35kb matches a known EST (BSG326593). This EST likely represents a non-coding RNA primary transcript from which the let-7 cluster of miRNAs are processed (Marson et al., 2008). The strategy for predicting TSSs is shown in Figure S3A.
(B) Two candidate islet-active alternative TSSs (red boxes) for the gene PAM, which encodes an islet secretory granule membrane protein. One of the candidate TSSs is also islet-unique and occurs between the annotated TSS and an un-annotated islet-active TSS. Examples of confounding factors for predicting islet-active TSSs are shown in Figure S4.
Identification of distal cis-regulatory elements
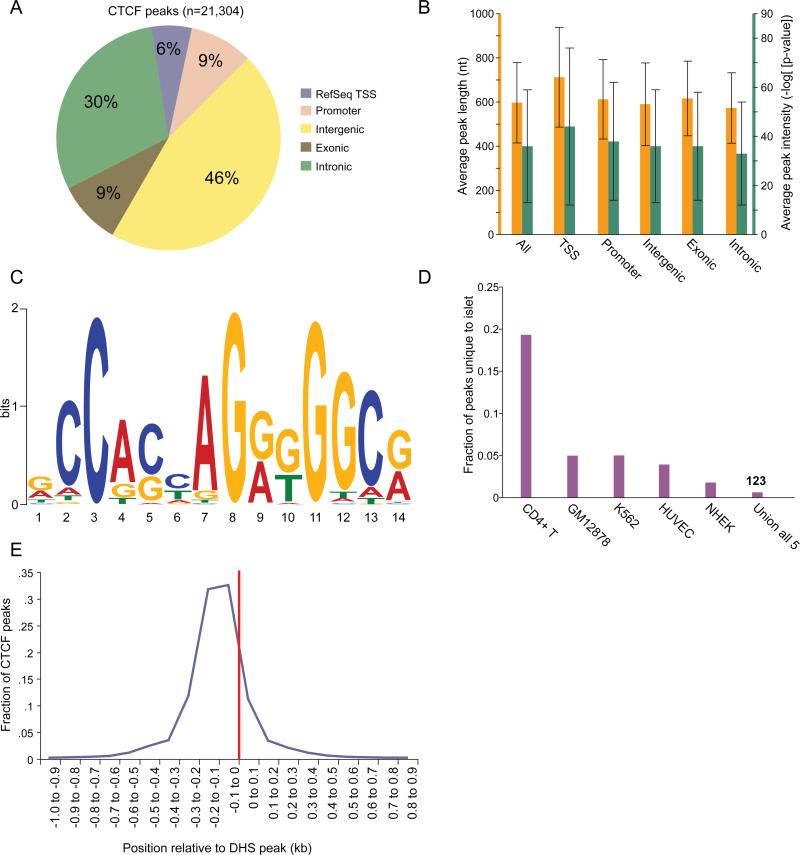
Sites bound by the CCCTC binding factor (CTCF) are an important class of cis-regulatory elements that can mediate insulator or other regulatory activities (Phillips and Corces, 2009). To generate a genome-wide CTCF binding site profile in the human islet, we performed ChIP-seq and designated enriched regions as “CTCF peaks” (n=21,304, Table S5; Methods). We assessed the genomic distribution of peaks (Figure 4A), computed the average peak intensity/length across various genomic categories (Figure 4B), and identified the most significantly over-represented motif within the peaks using MEME (Figure 4C; Supplemental Methods). The results corroborate those from previously described studies in other cell types (Kim et al., 2007; Jothi et al., 2008; Cuddapah et al., 2009). Further, only 0.6% (n=123) of CTCF peaks were islet-unique (Figure 4D). Finally, we observed that among the 77% of CTCF peaks that overlap 22% of DHS peaks, the CTCF peaks are positioned near the center of the DHS peak with a slight 5’-shift (Figure 4E).
Figure 4. Profiling of binding sites for the CCCTC-binding factor (CTCF).
(A) Distribution of CTCF peaks across five genomic annotation sets as described in Figure 1A.
(B) Average length (orange) and intensity (green) of CTCF peaks across five genomic annotation sets is fairly uniform. Error bars represent s.d.
(C) Motif determined by MEME (Bailey and Elkan, 1994) using the top 10% of CTCF peaks.
(D) Comparison of islet CTCF peaks with peaks from 5 different cell types. Each data point represents the fraction of total peaks (n=21,304) unique to the human islet relative to each of the other 5 human cell types or all of them combined (Union of all 5). Less than 1% of the peaks are unique to the islet (n=123). Varying levels of similarity across cell types may be at least partially explained by differences in the stage of cellular differentiation and/or sequencing depth.
(E) Positioning of CTCF peaks relative to the center of overlapping DHS peaks (red line). Almost all CTCF peaks that overlap DHS peaks are within 200 bp of the DHS peak center.
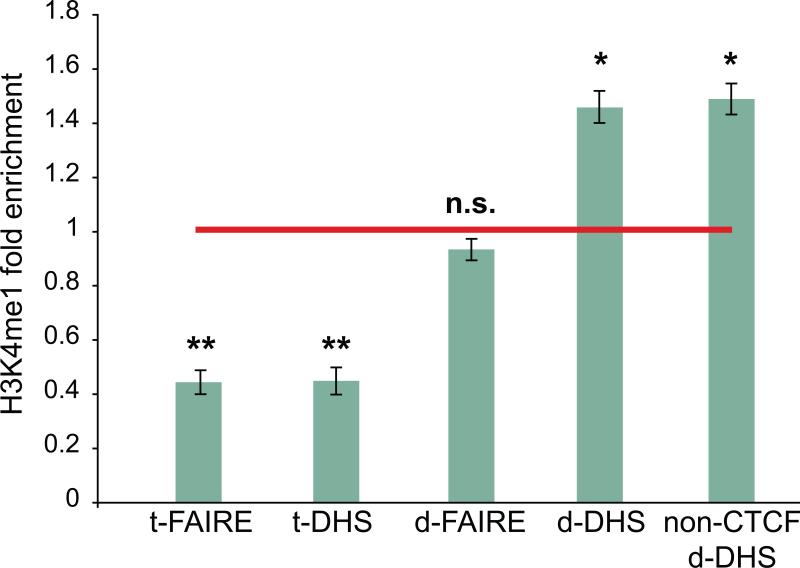
Previous studies have observed depletion of mono-methylated histone 3 lysine 4 (H3K4me1) at TSSs and enrichment at putative enhancers such as distal STAT1 and EP300 sites (ENCODE, 2007; Heintzman et al., 2009; Heintzman et al., 2007; Robertson et al., 2008) and non-promoter DHS (Barski et al., 2007; Robertson et al., 2008; Wang et al., 2008). To profile H3K4me1 across the human islet genome, we repeated the ChIP-seq strategy described above for three islet samples. We computed the average ratio of the density of extended H3K4me1 sequence reads in DHS peaks at RefSeq TSSs (t-DHS, n=11,829) and d-DHS peaks (n=34,039; Methods) to the density in flanking control regions that do not harbor DHS signal (Methods). t-DHS peaks are significantly depleted for H3K4me1, whereas d-DHS peaks are significantly enriched (Figure 5). Further, there was no significant difference in H3K4me1 enrichment between CTCF positive and CTCF negative d-DHS. Although we detected depletion of H3K4me1 at t-FAIRE peaks, there was no enrichment at d-FAIRE peaks (Figure 5).
Figure 5. Representation analysis of histone H3 lysine 4 monomethylation in candidate regulatory regions.
DNase I hypersensitive site (DHS) and Formaldehyde Assisted Isolation of Regulatory Element (FAIRE) peaks at RefSeq TSSs (t-DHS and t-FAIRE, respective) are significantly depleted for H3K4me1 signal (** two-tailed paired Student's t-test p-value < 0.005) and DHS peaks at distal, candidate regulatory elements (d-DHS) are enriched for H3K4me1 signal (* two-tailed paired Student's t-test p-value < 0.01). Error bars represent s.d. among three islet samples. FAIRE data was obtained from Gaulton et al. (2010). Representation analysis of additional histone modifications is shown in Figure S5.
We did not detect dramatically different H3K4me1 enrichment levels between intergenic and intragenic d-DHS peaks (Figure S5). Interestingly, although the average H3K4me3 read density in d-DHS peaks was ~3-fold less than that of H3K4me1, d-DHS peaks were still enriched for H3K4me3 signal relative to flanking control regions (Figure S5). These observations are consistent with the previous finding that although H3K4me1 often marks distal regulatory regions, a substantial portion is also associated with H3K4me3 signal (Robertson et al., 2008). Overall, the enrichment of active histone modifications suggests that islet d-DHS peaks are strong candidates for putative regulatory elements. Fifty published index SNPs (http://www.genome.gov/gwastudies/) and their linkage disequilibrium partners (r2 > 0.6) for diabetes (T1D, T2D) and related quantitative traits (fasting glucose, fasting insulin) are found within 500 bp of non-promoter d-DHS peaks (Table S9; Methods), suggesting that these SNPs may contribute to diabetes or altered islet physiology by modulating regulatory element activity.
Application of chromatin profiles to T2D susceptibility loci
To identify regulatory elements and transcripts that may underlie molecular mechanisms of T2D, we analyzed the chromatin profiles in the 18 GWAS-derived genomic loci conferring risk for T2D (Prokopenko et al., 2008). The genomic boundaries of each association signal (Table S6) were defined by the Spotter algorithm (Methods). The chromatin profiles do not predict any alternative promoters or unannotated/non-coding transcripts in these regions. However, they do identify 118 d-DHS peaks, which represent putative distal regulatory elements (Table S7; Methods). About one quarter of these elements (n=28) are bound by CTCF in the islet. Six of the 118 elements contain one or more T2D-associated SNPs (index SNP or SNP with r2 > 0.6; Table S8). These six include a previously identified element containing the index SNP rs7903146 in the TCF7L2 locus (Gaulton et al., 2010). The remaining five map to the IGF2BP2, KCNQ1, WFS1, FTO, and CDC123/CAMK1D loci. Only the CDC123/CAMK1D element is bound by CTCF in the islet.
Validation of putative islet regulatory elements in T2D loci
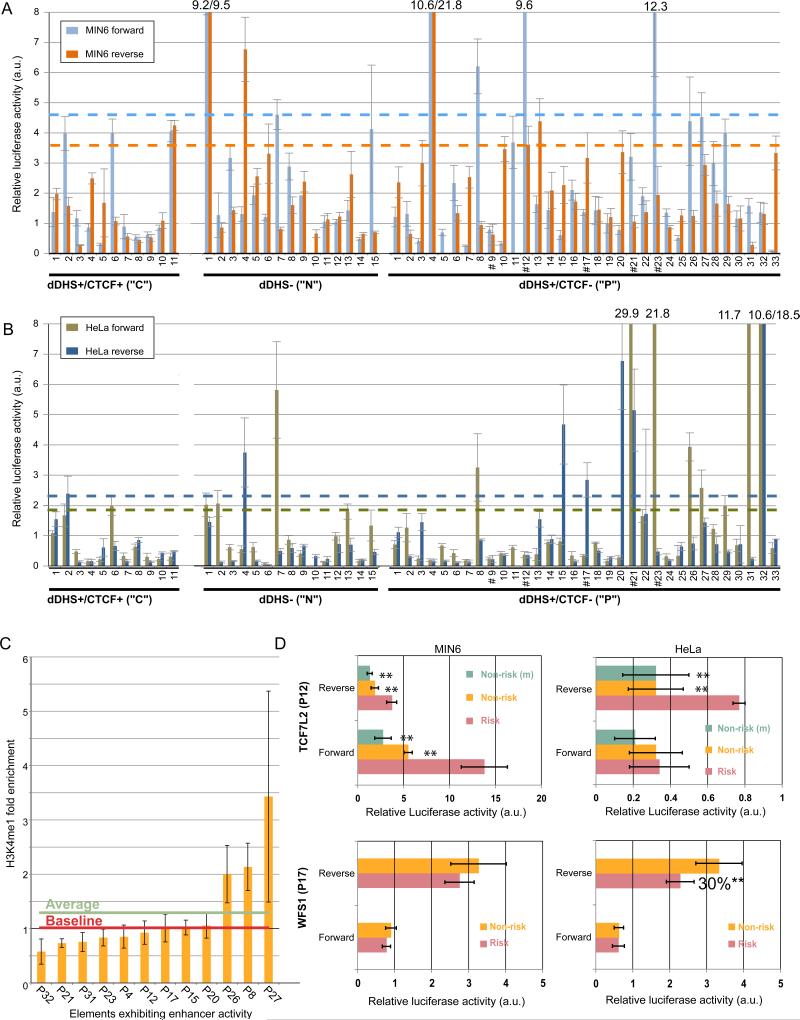
To determine whether predicted regulatory elements in the islet can function as enhancers, we cloned two classes of elements containing d-DHS peaks into luciferase reporter vectors (Figure 6): those bound by CTCF (“C”, n=11) and those that are not (“P”, n=33). We also cloned a number of non-DHS, non-CTCF controls (“N”, n=15). Because human islet cell lines are not available, we tested these elements for enhancer activity in murine pancreatic MIN6 (Figure 6A) and HeLa (Figure 6B) cell lines. Only ~15% (4/26) of the negative controls exhibited enhancer activity in any orientation or cell type (Figures 6A and 6B; ~9% (1/11) of “C” elements and 20% (3/15) of “N” elements). In contrast, ~2.5 fold more “P” elements demonstrated enhancer activity (12/33). This positive rate (36.4%) is comparable to that of predicted HeLa enhancers (Heintzman et al., 2009) that exhibited increased luciferase activity in our HeLa reporter assays (38.5%, 5/13).
Figure 6. Luciferase reporter activity validates putative enhancer elements.
(A) Relative luciferase activity of constructs in 3 element classes tested in MIN6 cells. Genomic locations of elements are found in Table S13. Blue and orange dashed lines indicate 2.33 standard deviations (p=0.01) (Heintzman et al., 2009) above the median activity of tested CTCF-bound regions for elements cloned in the forward or reverse orientations, respectively. Data represent the mean ± s.d. of 3 replicates each for 2 separate clones (6 total measurements). C=d-DHS+/CTCF+ element. N=d-DHS-/CTCF-. P=d-DHS+/CTCF- element. # marks elements containing T2D-associated SNPs. Numbers above the bars indicate the luciferase activity for elements beyond the scale of the y axis; a.u. denotes arbitrary units.
(B) Relative luciferase activity of constructs in 3 element classes tested in HeLa cells. Data are analyzed and annotated as in (A); a.u.=arbitrary units.
(C) H3K4me1 representation in the 12 elements exhibiting enhancer activity. Though the overall average enrichment of H3K4me1 is ~1.3 fold (green line), only 3/12 elements are above baseline (red line). Error bars represent s.d. among three islet samples.
(D) Relative luciferase activity of TCF7L2 (P12) and WFS1 (P17) elements in MIN6 (left panels) or HeLa (right panels) cells containing the risk or non-risk alleles of T2D-associated SNPs. For TCF7L2, (m) denotes a mutation generated by site-directed mutagenesis from the risk to non-risk allele. Data represent the mean ± s.d. of 3 replicates each from at least 2 independent clones. **= 2-tailed unpaired Student's t-test p<0.01 a.u.=arbitrary units. Additional allelic analysis is shown in Figure S6.
Four of 12 “P” elements exhibiting enhancer activity (P4, KCNJ11/ABCC8; P12, TCF7L2; P17, WFS1; P20, HHEX/IDE) are unique to the islet; 1 of these (P17, WFS1) is also undetected by at least three other methods for the prediction of regulatory element potential: PReMod (Ferretti et al., 2007), phastCons (Siepel et al., 2005) and islet-FAIRE (Gaulton et al., 2010). The average H3K4me1 enrichment among the 12 d-DHS peaks in the elements exhibiting enhancer activity was similar to that computed for all d-DHS (~1.3 fold; Figure 6C). However, there was large variation in H3K4me1 enrichment among individual elements (0.6-3.4 fold), with only 3/12 enriched above baseline (1.0; Figure 6C).
Allele-specific analysis of 5 regulatory elements containing T2D-associated SNPs
Five “P” elements tested contain T2D-associated SNPs (Figures 6A,B: P9 (IGF2BP2), P12 (TCF7L2), P17 (WFS1), P21 (KCNQ1), P23 (FTO)). Notably, four out of the five elements (all except P9) exhibited enhancer activity in at least one orientation and cell type tested. To assess allele- or haplotype-specific effect(s) of T2D-associated variants on enhancer activity, we cloned these four regions from the genomic DNA of individuals with risk and non-risk genotypes/haplotypes and compared luciferase reporter activity (Figure 6D and Figure S6A). We confirmed significantly stronger enhancer activity for the TCF7L2 element (P12) containing the rs7903146 risk allele relative to the non-risk allele (~3-fold, Figure 6D) (Gaulton et al., 2010). TCF7L2 allelic enhancer effects were specific to the MIN6 cell line (Figure 6D, compare MIN6 and HeLa). Sequencing of the TCF7L2 inserts from each haplotype revealed two variant bases, a novel variant (C/G at Chr10:114,747,977; hg18) and rs7903146; only rs7903146 mediated allele-specific effects on enhancer activity (Figure 6D, compare Risk to Non-risk and Non-risk(m); Figure S6B). We also identified a haplotypic effect on enhancer activity for the WFS1 element (P17), which contains four SNPs (rs4689397, rs6823148, rs881796, and rs4234731). The risk haplotype exhibited ~30% lower activity than non-risk in HeLa cells (Figure 6D).
Discussion
In this study, we describe the most comprehensive characterization to date of the epigenomic profile of unstimulated human pancreatic islets. Using DNase- and ChIP-seq techniques, we profiled open chromatin, CTCF binding sites, H3K4me3, H3K4me1, and H3K79me2 across the entire genome in human islets. Integrated analysis of these large-scale datasets identified ~18,000 putative TSSs, ~30% of which were previously unannotated by RefSeq. Further computational genomic analyses revealed that at least several hundred of these are islet-active TSSs, including those for major islet miRNAs previously implicated in the control of glucose homeostasis (Lynn, 2009). Interestingly, active chromatin marks (H3K4me3, DHS, H3K79me2) were absent from a subset of highly islet-expressed genes, including those encoding islet-specific hormones (INS, GCG, SST, IAPP, PPY, and TTR). This observation suggests that some genes critical for islet function have an unconventional promoter chromatin signature indicative of a unique transcriptional control mechanism. Mutskov and Felsenfeld (2009) have proposed such a model based on detailed analysis of the INS locus in human islets.
We also identified ~34,000 candidate distal regulatory elements in human islets. A substantial number of these putative elements were clustered (<1000 bp from each other). Comparisons with other cell types indicated that these clustered elements are significantly enriched for islet-unique sites and thus may represent islet-specific regulatory modules worthy of more extensive future investigation. Based on CTCF binding profiles, ~22% of the ~34,000 candidate distal regulatory elements are predicted insulator sites. Previous studies have reported that the H3K4me1 signal is enriched in distal regulatory elements (Heintzman and Ren, 2007, 2009). Though our analyses confirm this finding in aggregate, we show that H3K4me1 enrichment may not be a reliable predictor of regulatory activity for individual elements.
Fifty SNPs associated with islet-related diseases and traits map to within 500 bp of a candidate non-promoter regulatory element. Focusing on T2D, 4 of 12 elements that function as enhancers in vitro (FTO, KCNQ1, TCF7L2, and WFS1 loci) harbor T2D-associated SNPs, including 2 (TCF7L2 and WFS1 loci) that exhibit significant allele-specific differences in activity. These results suggest that altered enhancer activity plays a role in the molecular mechanism underlying at least a subset of T2D genetic association signals.
These datasets should provide functional context for non-coding variants identified through additional association, targeted resequencing, or whole genome sequencing studies. Further analysis of the repertoire of regulatory elements in the human islet will enhance the understanding of gene regulation in the islet and should offer additional insight into the molecular mechanisms that underlie diabetes susceptibility.
Accession Numbers
The Accession number for the sequencing data is GSE23784. This is an umbrella accession number that links to all of the individual ChIP-seq and DNase-seq data sets.
Experimental Procedures
Human Islets
Fresh human pancreatic islets were obtained from the ICR Basic Science Islet Distribution Program and National Disease Research Interchange (NDRI). Islet viability and purity were assessed by the distribution centers and are shown along with phenotypic/clinical information of each donor in Table S10. Islets were warmed to 37 °C and washed with calcium and magnesium free Dulbecco's Phosphate Buffered Saline (Invitrogen) prior to crosslinking. For ChIP studies, cells were crosslinked for 20 minutes in 1% formaldehyde at room temperature, frozen in liquid nitrogen, and stored at -80°C.
DNase-seq and DHS Peak Identification
For DNase-seq experiments, fresh pancreatic islets were disaggregated to achieve single cell suspension. Islets were washed with pre-warmed 1X PBS once and resuspended with dissociation solution (1 ml of 1X PBS, 50 μl of Dispase I stock: 0.05 U/μl, Roche). Islet suspension was transferred to a 6-well culture dish, incubated at 37 °C for 30 mins, dissociated with a 2 ml sterile pipette, and incubated for another 30 minutes. This incubation-agitation cycle was repeated 4-5 times until > 90% of islets were disaggregated into single cells. Cells were washed with pre-warmed 1X PBS once, and prepared for DNase-seq experiments as previously described (Song and Crawford, 2010). Libraries from three primary human islet samples (Table S10) were sequenced using the Illumina GA2 platform. Peaks were identified using MACS (Supplemental Methods; (Zhang et al., 2008)).
Chromatin Immunoprecipitation and Illumina GAII Sequencing (ChIP-seq)
Chromatin immunoprecipitation (ChIP) assays were carried out as previously described (Scacheri et al., 2006) with the following modifications. Intact nuclei were isolated and chromatin was sheared on ice using a Branson 450 Sonifier (constant duty cycle, output 4; 12-16 cycles of 20 second sonication with 1 minute rest between cycles) to a size of 200-1000bp. Antibodies used for ChIP were anti-H3K4me3 (ab8580, Abcam), anti-H3K4me1 (ab8895, Abcam), anti-H3K79me2 (ab3594 Abcam,), anti-CTCF (ab70303, Abcam; 07-729, Millipore), and anti-GFP (sc-8334, Santa Cruz).
Islet ChIP-seq libraries were prepared and sequenced using the Illumina GA2 protocol and platform. The number of sequencing lanes, clusters, aligned reads, repeat-filtered reads (no satellite reads), and unique starts is shown for each islet and ChIP experiment in Table S12. MACS (Zhang et al., 2008) was used to call H3K4me3 and CTCF peaks (Supplemental Methods).
Genome-wide Analysis of Chromatin Marks
Perl and R scripts were written to perform the genomic characterization and comparative analysis of DHS, H3K4me3, and CTCF peaks. Unless otherwise noted, functional annotation datasets (including RefSeq and UCSC known genes, predicted TSSs and bidirectional promoters, phastCons elements, CpG islands, ChIP-seq datasets) were downloaded from the UCSC Table Browser on 11/01/2009 (http://genome.ucsc.edu/cgi-bin/hgTables).
For “computationally predicted TSSs”, both the Eponine and the Switchgear datasets from the UCSC Table Browser were utilized. Human pancreatic islet gene expression data was downloaded from T1DBase (http://T1Dbase.org) and expression data for other tissues was downloaded from BioGPS Human U133A/GNF1H Gene Atlas (http://biogps.gnf.org/downloads/). Islet-selective gene expression was defined as at least 3-fold greater expression in the islet relative to any other tissue represented. Genome-wide results of the Chai algorithm were determined according to the parameters in Parker et al. (2009) and islet-FAIRE datasets were obtained from Gaulton et al. (2010). Gene Ontology analyses were performed using the web-based tool NIH David 6.7 (http://david.abcc.ncifcrf.gov/). For the DHS peak clustering analysis (Figure 1F) and the histone modification enrichment/depletion analysis (Figure 5, Figure S5), we stringently defined distal DHS peaks (d-DHS) as those that are not within H3K4me3 peaks and ≥ 5kb away from RefSeq TSSs, UCSC Known Gene TSSs, Eponine or Switchgear computationally predicted TSSs and CpG islands, yielding 34,039 d-DHS. To select regulatory elements to test for enhancer activity (Figure 6), the definition of d-DHS was slightly loosened (≥ 5kb upstream and ≥ 1kb downstream from known and predicted TSSs and CpG islands). P-values for statistical comparisons were computed using either the two-tailed paired Student's t-test or the Fisher's exact test. Details of the remaining computational analyses are described in Supplemental Methods.
Molecular Cloning
Putative regulatory elements were amplified from human genomic DNA with primers designed using Primer Tile (http://research.nhgri.nih.gov/tools/). Element boundaries were determined by manual H3K4me1 profile inspection. Coordinates of amplified elements and primer sequences for amplification are found in Table S13. Putative regulatory elements were cloned using the Gateway system (Invitrogen). Generation of Gateway-compatible vectors is described in Supplemental Methods. Variants of interest were introduced using Quikchange Lightning (Stratagene). Mutagenesis primer sequences are available upon request. Mutagenesis was confirmed by direct sequencing.
Transfection and Dual Luciferase Assays
Cells were seeded in 96 well plates (40,000 cells/well HeLa, 60,000 cells/well MIN6) and co-transfected with 0.072 pmol Gateway-modified firefly (pGL 4.23, Promega) and 2 ng renilla (pRL-TK, Promega) vectors using Lipofectamine 2000 (Invitrogen). Two vector preparations per insert orientation were tested. Transfections were performed in triplicate.
Cells were lysed in 1x passive lysis buffer (Promega) 36-48 hours post-transfection and dual luciferase assays were run on a Centro/Centro XS3 Microplate Luminometer LB 960 (Berthold). Firefly values were normalized to Renilla to control for differences in cell number or transfection efficiency. Luciferase assays were performed in triplicate. For each element tested, at least two independent vector preparations were used. Activity was defined as 2.33 standard deviations (p=0.01) above the median activity of negative controls (Heintzman et al., 2009), defined as CTCF-bound elements in this study.
Supplementary Material
Acknowledgements
Human pancreatic islets used in this study were obtained through the ICR Basic Science Islet Distribution Program (University of Minnesota, University of Alabama-Birmingham, University of Illinois, University of Miami, Northwestern University) and the National Disease Research Interchange (NDRI). We thank Fangfei Ye and Lisa Bukovnik at the Duke IGSP Sequencing Core Facility for sequencing DNase libraries, the DIAGRAM Consortium for helpful discussion regarding variants in the KCNQ1 locus, and members of the Collins and Boehnke labs for insightful discussions during the study and critical comments on the manuscript. Special thanks to Cristen Willer and Greg Keele for help with statistical analyses of ChIP/GWAS data. This study was supported by the NIH Division of Intramural Research/NHGRI project number Z01-HG000024 (F.S.C.), by NIH grant DK062370 (M.B.), and by an NIH/NHGRI ENCODE Consortium grant U54HG004563 (G.E.C. and T.S.F.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- ~18,000 promoters and ~34,000 distal regulatory elements predicted in human islets
- 6 non-promoter regulatory elements harbor type 2 diabetes-associated variants
- Reporter assays verify 40% of putative enhancers and detect allele-specific effects
- Islet hormone genes lack typical active histone modifications of expressed genes
References
- ENCODE Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009 doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bhandare R, Schug J, Le Lay J, Fox A, Smirnova O, Liu C, Naji A, Kaestner KH. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 2010;20:428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, Deblois G, Giguere V, Ferretti V, Bergeron D, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Egana V, Rosero S, Molano RD, Pileggi A, Ricordi C, Dominguez-Bendala J, Pastori RL. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem Biophys Res Commun. 2008;366:922–926. doi: 10.1016/j.bbrc.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink C. Promoter elements in endocrine pancreas development and hormone regulation. Cell Mol Life Sci. 2003;60:1033–1048. doi: 10.1007/s00018-003-2247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler PC, Meier JJ, Butler AE, Bhushan A. The replication of beta cells in normal physiology, in disease and for therapy. Nat Clin Pract Endocrinol Metab. 2007;3:758–768. doi: 10.1038/ncpendmet0647. [DOI] [PubMed] [Google Scholar]
- Correa-Medina M, Bravo-Egana V, Rosero S, Ricordi C, Edlund H, Diez J, Pastori RL. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr Patterns. 2009;9:193–199. doi: 10.1016/j.gep.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc Natl Acad Sci U S A. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva NM, Frayling TM. Novel biological insights emerging from genetic studies of type 2 diabetes and related metabolic traits. Curr Opin Lipidol. 2010;21:44–50. doi: 10.1097/MOL.0b013e328334fdb6. [DOI] [PubMed] [Google Scholar]
- Dekker J. A closer look at long-range chromosomal interactions. Trends Biochem Sci. 2003;28:277–280. doi: 10.1016/S0968-0004(03)00089-6. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti V, Poitras C, Bergeron D, Coulombe B, Robert F, Blanchette M. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–126. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonarson H, Qu HQ, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–1146. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelsson E, Langenberg C, Hivert MF, Prokopenko I, Lyssenko V, Dupuis J, Magi R, Sharp S, Jackson AU, Assimes TL, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59:1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslin EP, Kahn CR. Joslin's diabetes mellitus. 14th edn Lippincott Williams & Willkins; Philadelphia, Pa: 2005. [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–475. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, Eizirik DL, Welsh N, Goodman N, Hood L. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lynn FC. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452–459. doi: 10.1016/j.tem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, Sjogren M, Ling C, Eriksson KF, Lethagen AL, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P, Bugliani M, Saxena R, Fex M, Pulizzi N, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- Mutskov V, Felsenfeld G. The human insulin gene is part of a large open chromatin domain specific for human islets. Proc Natl Acad Sci U S A. 2009;106:17419–17424. doi: 10.1073/pnas.0909288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohneda K, Ee H, German M. Regulation of insulin gene transcription. Semin Cell Dev Biol. 2000;11:227–233. doi: 10.1006/scdb.2000.0171. [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1801. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SCJ, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA Topography Correlates with Functional Noncoding Regions of the Human Genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends in Genetics. 2008;24:613–621. doi: 10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–1917. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Hawrylycz M, Wallace JC, Humbert R, Yu M, Shafer A, Kawamoto J, Hall R, Mack J, Dorschner MO, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc Natl Acad Sci U S A. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE. DNase-Seq: A High-Resolution Technique for Mapping Active Gene Regulatory Elements across the Genome from Mammalian Cells. Cold Spring Harb Protoc 2010. 2010 doi: 10.1101/pdb.prot5384. pdb.prot5384- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.