Abstract
Purpose
Age-related macular degeneration (AMD) is the leading cause of blindness among older adults, in which oxidative damage may play a pivotal role. Paraoxonase 1 (PON1) protects against oxidative damage and has been evaluated for its involvement in aging diseases including AMD. This study investigated whether PON1 gene polymorphisms associate with AMD.
Design
Case-control association study.
Methods
We studied 1037 individuals with AMD subcategorized using AREDS criteria and 370 control subjects without retinal disease. Participants were primarily Caucasian of European descent. All exons of PON1 were evaluated by single strand conformation polymorphism and direct sequence analysis.
Results
Six missense changes (Leu55Met, Met127Arg, His155Arg, Gln192Arg, Gln192Glu, Ala252Gly) were identified in PON1. We observed a weak association of Leu55Met with an increased risk of wet AMD (P = 0.02), but not with dry AMD or when combining all patient categories. A significantly higher allele frequency for Gln192Arg was detected in controls than in the combined AMD patient population (P < 0.0001), and when category 2, 3 and 4 patients were separately considered (P = 0.004, P = 0.002 and P < 0.0001, respectively). For category 4 AMD, the Arg192 allele was significantly less prevalent in the wet form (P < 0.0001) but not in the dry form (P = 0.377).
Conclusion
We report a weak association of PON1 Leu55Met with an increased risk of wet AMD, replicating previous reports. Our findings indicate a protective role for Gln192Arg, particularly for patients with the wet form. Gln192Glu warrants consideration, as this variant alters the same amino acid as Gln192Arg and was identified only in Category 4 AMD patients. We believe that Met127Arg, His155Arg and Ala252Gly play minor roles in AMD susceptibility due to their limited frequency and/or location within the PON1 gene. The functional and biological mechanism by which Gln192Arg is acting to decrease AMD susceptibility remains to be determined.
Keywords: age-related macular degeneration, paraoxonase, oxidative damage, genetics, mutation
Introduction
Age-related macular degeneration (AMD) is the leading cause of irreversible visual impairment and blindness among older adults in the USA and throughout the developed world.1 The prevalence of AMD will increase even further as the number of people aged 65 years or older will increase by approximately 80% in the next 25 years.2–4 AMD is a progressive disorder of the photoreceptor - retinal pigment epithelium (RPE) - Bruch’s membrane - choriocapillaris complex. AMD typically starts with the formation of a few macular drusen, the lipoproteinaceous extracellular deposits localized between the RPE and Bruch’s membrane. Drusen are considered to be a major risk factor for developing end-stage AMD associated with irreversible vision loss, due to geographic atrophy (GA; also known as dry, atrophic or non-exudative AMD) or choroidal neovascularization (CNV; also known as wet or exudative AMD).
Although the clinical and histopathological characteristics of AMD have been described, little is known about the etiology and pathogenesis of this disorder. It is clear, however, that AMD is a complex disease caused by actions and interactions between multiple genes and environmental factors. Familial aggregation studies, twin studies, association studies and whole genome linkage scans provided clear evidence that AMD has a strong genetic component.5,6 The discoveries of AMD-associated single nucleotide polymorphisms (SNPs) in several genes involved in the regulation of the immune-mediated complement pathway, including complement factor H (CFH), complement factor B (BF), complement component 2 (C2), and complement component 3 (C3) provide an important opportunity to advance our understanding of AMD pathogenesis.7–12 The specific role of these variants in causing AMD remains, however, unknown.
In addition to inflammation, it has been postulated that oxidative damage plays a pivotal role in AMD progression. Epidemiological studies show that cigarette smoking significantly increases the risk of AMD2,13 and that dietary supplementation with zinc and antioxidants can slow AMD progression.14 During the past several years, an enzyme which protects against oxidative damage, Paraoxonase 1 (PON1), has been studied extensively for its involvement in several aging diseases including coronary artery disease, arthrosclerosis, Parkinson’s disease, Alzheimer’s disease, diabetes, cancer and AMD.15,16
PON1 belongs to the paraoxonase (PON) family of genes, which also includes PON2 and PON3. All PON genes are highly conserved in mammalian evolution.17 Based on their respective cDNA structures and deduced amino acid sequences, there is over 80% identity between human, mouse and rabbit PON1 proteins, and at least 60% identity between PON1, PON2, and PON3 genes of these species.18 The human PON genes lay in a tandem array on chromosome 7q21.3. Human PON1 is a high-density lipoprotein (HDL)-associated serum enzyme that is synthesized in the liver and protects lipoproteins from oxidative modifications via antioxidant activities and by hydrolyzing oxidized substrates including phospholipids, hydroperoxides, lactones and carboxylic acid esters.15
In vivo, there is a wide individual variation in serum PON1 concentration and enzymatic activity.17 This variability is attributed to genetic polymorphisms in the PON1 gene and also by environmental factors such as smoking, alcohol consumption and diet.15 The two most prevalent PON1 polymorphisms, Leu55Met and Gln192Arg, involve residues that are thought to influence enzymatic activity and concentration.19–22 Four additional SNPs in the promoter region of the PON1 gene (−107C→T, −162A→G, −824G→A,−907G→C) have also been reported to affect the expression and thus the serum concentration of the enzyme.23–25 Among them, the −107C→T polymorphism has been indicated as the most important genetic determinant of PON1 levels. Leu55Met is in linkage with the promoter region SNPs, including −107C→T, and consequently it is associated with variations of the enzyme concentration.15,26 Specifically, Leu55 is associated with higher concentrations of the enzyme, irrespective of the amino acid present at position 192.21,26 The Gln192Arg polymorphism is responsible for a striking difference in the substrate specificity and activity of the enzyme.19,20,27 Interestingly, several studies indicate that individuals with a specific genetic background, such as those carrying the Arg192 allele and having a high PON1 enzymatic activity, appear to have a survival advantage and seem to age more successfully.15 These individuals have been defined as centenarians that have avoided or delayed major age-related diseases due to a superior ability to counteract oxidative stress.15
Two PON1 SNPs (Leu55Met and Gln192Arg) have been reported to be associated with wet AMD in a study of 72 individuals of Japanese descent.28 In that study, significant differences in the distributions of both SNPs were found between cases and controls, suggesting that these PON1 gene polymorphisms represent a genetic risk factor for AMD. When the same two SNPs were evaluated in separate cohorts of 62 or 94 Caucasian patients primarily from Northern Ireland and Melbourne, Australia with United Kingdom ancestry with end-stage AMD (dry or wet), no significant association with AMD was found.29,30 Given the conflicting PON1 genetic data, we reasoned that investigating a much larger number of patients will help to clarify whether PON1 is associated in AMD pathogenesis. Therefore, we evaluated 1037 patients with AMD for Leu55Met and Gln192Arg, as well as other novel sequence variations.
Materials and Methods
Ascertainment of patients
Blood samples were collected at the Cole Eye Institute (CEI), Cleveland Clinic and the VA Eye Clinic. We collected a total of 1037 index cases with AMD, each diagnosed through ophthalmologic examination by a clinician in the Clinical Study Group and subcategorized into one of four experimental groups using the criteria defined by the Age-Related Eye Disease Study.14 AMD disease progression was categorized based on fundus examination. Briefly, AMD Category 1 subjects have five or less small (<63 μm) drusen/eye and visual acuity of 20/30 or better in both eyes. AMD Category 2 patients exhibited early-stage disease with multiple small drusen, single or nonextensive intermediate (63–124 μm) drusen, RPE pigmentary abnormalities, or any combination of these, in one or both eyes and visual acuity of 20/30 or better in both eyes. AMD Category 3 patients exhibited mid-stage disease with at least one eye having visual acuity 20/30 or better, one or more large (125 μm) drusen, and extensive intermediate drusen and/or geographic atrophy that did not involve the macula. Category 3 patients lacked advanced AMD in either eye. AMD Category 4 patients exhibited advanced AMD with substantial CNV or GA involving the macula in one or both eyes. Patients with a diagnosis of exudative or neovascular AMD in at least one eye were classified as wet. Control donors lacked macular drusen and exhibited no clinical evidence of any retinal disorder. 661 patients were obtained from CEI and 376 patients were obtained from the VA. Of the 1037 patients recruited, 132 were defined as Category 1 AMD, 191 were Category 2, 127 were Category 3 and 587 were Category 4. Of the 587 Category 4 patients, 76 had dry or atrophic AMD, 493 had wet or exudative AMD and we were unable to categorize 18 unequivocally as dry or wet AMD. Approximately 95% of our patient population were Caucasian of European descent while ~5% were African American and <1% were Asian. The age range was 53 to 105 years, with a mean age of 77.0 years. A total of 370 unrelated individuals without retinal disease or a family history of retinal disease was used as normal control subjects; 84 were obtained from CEI and 286 were obtained from the VA. Approximately 97% of normal controls were Caucasian of European decent, ~ 3 % were African American and <1% were Asian. The age range was 54 to 94 years, with a mean age of 76.3 years. Study population characteristics including mean age, gender and health history are summarized for each AMD Category in Table 1. Information was obtained by questionnaire administered at the time of consent. Leukocyte nuclei were prepared from the blood samples followed by DNA purification using standard protocols.
Table 1.
Characteristics and Health History of AMD Patient Population.
| Cat 1 | Cat 2 | Cat 3 | Cat 4 | All | |
|---|---|---|---|---|---|
| N = 132 | N = 191 | N = 127 | N = 587 | N = 1037 | |
| Females | 19/132 = 14.4% | 38/191 = 19.9% | 54/127 = 42.5% | 303/587 = 51.6% | 414/1037 = 39.9% |
| Males | 113/132 = 85.6% | 153/191 = 80.1% | 73/127 = 57.5% | 284/587 = 48.4% | 623/1037 = 60.1% |
| Mean Age | 72.6 | 75.3 | 77.0 | 79.0 | 77.0 |
| Smoker | 91/111 = 82.0% | 139/178 = 78.1% | 55/88 = 62.5% | 258/477 = 54.1% | 543/854 = 63.6% |
| Diabetic | 23/36 = 63.9% | 35/121 = 28.9% | 10/71 = 14.1% | 56/442 = 12.7% | 124/670 = 18.5% |
| Hypertensive | 62/67 = 92.5% | 100/146 = 68.5% | 51/77 = 66.2% | 267/462 = 57.8% | 480/752 = 63.8% |
| Hyperlipidemic | 44/52 = 84.6% | 50/128 = 39.1% | 35/80 = 43.7% | 170/447 = 38.0% | 299/707 = 42.3% |
| Cardiovascular Disease | 86/89 = 96.6% | 90/150 = 60.0% | 61/84 = 72.6% | 176/465 = 37.8% | 413/788 = 52.4% |
| Neurologic Disease | 2/16 = 12.5% | 10/106 = 9.4% | 6/69 = 8.7% | 16/433 = 3.7% | 34/624 = 5.4% |
AMD: age-related macular degeneration
All numbers are presented as % of total reported for each category.
Mutation screening
For mutation detection, PCR products corresponding to the PON1 coding sequence (GenBank RefSeq NM_000446) were amplified from genomic DNA and analyzed by the single-strand conformation polymorphism (SSCP) technique. Twelve primer pairs were designed to cover the nine coding exons as well as the immediately flanking intron sequences and are listed in Table 2 along with the PCR conditions. Two primer pairs were designed for exons 4, 6 and 9 in order to split the exons into two fragments to yield smaller sized amplicons for SSCP. The buffer pH, Mg++ concentration, annealing temperature, and presence or absence of 10% dimethyl sulfoxide were tailored to each primer pair to yield optimal amplification.
Table 2.
Polymerase Chain Reaction Conditions and Primers Used to Amplify PON1 in AMD Patient and Control Populations.
| Exon | Sense Primer | Antisense Primer | Anneal Temp. °C | Buffer pH | MgCl2 mM | % DMSO | Fragment (bp) |
|---|---|---|---|---|---|---|---|
| 1 | tcctagcccgtcggtgtctgcgcc | ggctcgtggagctggcagggagtg | 64 | 8.4 | 1.5 | 10 | 190 |
| 2 | aacttttgtggactgcctaccttt | ggcacattaatcacacaaagagca | 52 | 8.6 | 1.5 | 0 | 228 |
| 3 | ggatccacatcctgcaataatatg | aaagacttaaactgccagtcctag | 50 | 8.6 | 1.5 | 0 | 180 |
| 4.1 | agcgcctctattagcatttgaagt | tcaaatttacttccagtgatcccc | 52 | 8.6 | 1.5 | 0 | 187 |
| 4.2 | gaagaagatccaacagtgttggaa | agagggtaagtttaaaacccagag | 50 | 8.6 | 1.5 | 0 | 242 |
| 5 | gtgagagcttagttaatgtttcat | actaaaagtggattaactatccgc | 46 | 8.4 | 1.5 | 10 | 238 |
| 6.1 | cctgtaatgttcaataccttcacc | ctatagtagacaacatacgaccac | 54 | 8.6 | 1.5 | 0 | 185 |
| 6.2 | gatcactattttcttgacccctac | ctcctgagaatctgagtaaatcca | 52 | 8.4 | 1.5 | 0 | 200 |
| 7 | cttaatctctcaagttgtgttact | attaagcagtccttttttcttcac | 50 | 8.4 | 1.5 | 0 | 233 |
| 8 | ctctgatatattttgctccatcac | aacagtagctgggaataaagtcac | 54 | 8.4 | 1.5 | 0 | 254 |
| 9.1 | gaccctattatgcataaggttgtt | actgtgccattttctgcataaacc | 52 | 8.6 | 1.5 | 0 | 142 |
| 9.2 | tccacaggtgcttcgaatccagaa | ctaagaacactgggtcctcggaat | 56 | 8.4 | 1.5 | 10 | 248 |
PON1: Paraoxonase 1
AMD: age-related macular degeneration
SSCP was performed as previously described.33 Variant bands detected by SSCP were analyzed by sequencing the corresponding PCR-amplified DNA segments using the Quick Start sequencing kit (Beckman-Coulter) following the manufacturer’s protocol. A CEQ-2000 automated sequencer (Beckman-Coulter) was used to resolve sequences.
Statistical Analysis
The χ2 test was used for comparisons of genotype frequencies and to analyze Hardy-Weinberg equilibrium. A two-tailed Fisher’s exact test was used for comparisons of allele frequencies. Logistic regression analysis was used to estimate odds ratios and 95% confidence intervals. P values <0.05 were defined as nominally significant and were further subjected to Bonferroni correction to account for multiple comparisons testing. Following correction for the six SNPs examined, the significance level for single SNP tests was 0.008 (0.05/6).
Computational assessment of missense mutations
Two sequence homology based programs were used to predict the functional impact of missense changes identified in this study: PolyPhen (Polymorphism Phenotyping, http://genetics.bwh.harvard.edu/pph/) and PMut (http://mmb2.pcb.ub.es:8080/PMut/). PolyPhen analyzes the impact of an amino acid polymorphism on protein structure, and predicts whether that amino acid change is likely to be deleterious to protein function.34 The prediction is based on the position specific independent counts (PSIC) score derived from multiple sequence alignments of observations. PolyPhen scores of >2.0 indicate the polymorphism is probably damaging to protein function, scores of 1.5 – 2.0 are possibly damaging, and scores of <1.5 are likely benign. PMut allows the accurate pathological prediction of single amino acid mutations based on the use of neural networks.35 Following the input of a reference sequence and the amino acid substitution of interest, the algorithm provides an answer and a reliability index. An output value >0.5 is predicted to be a pathological mutation and a value <0.5 is neutral. The reliability is considered good with a score of six and greater and is highly reliable at the maximum score of nine.
Results
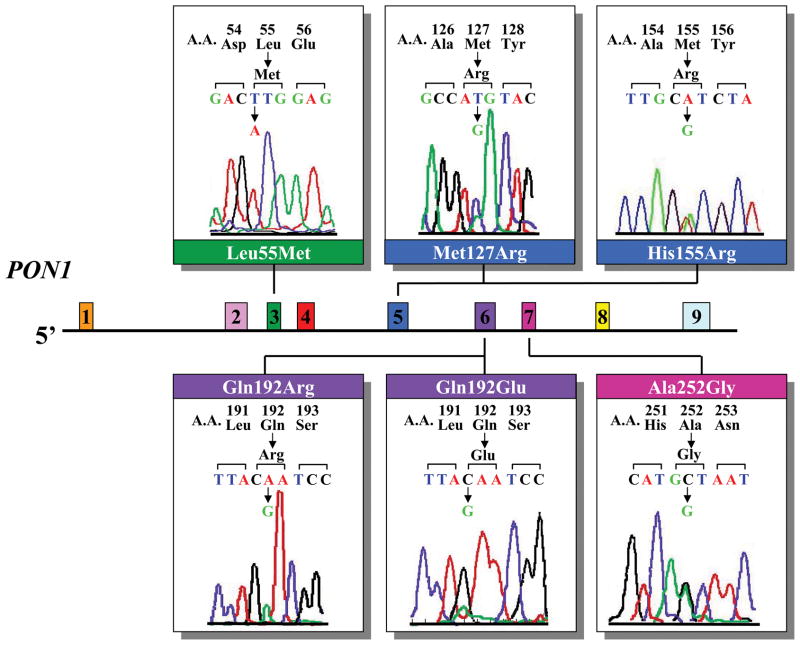
We initially evaluated a cohort of 377 unrelated patients with AMD. In these patients, all nine PON1 coding exons were screened by SSCP analysis. Six missense changes (Leu55Met, Met127Arg, His155Arg, Gln192Arg, Gln192Glu, Ala252Gly) were identified (Table 3). An example of each DNA sequence change and its location on a schematic representation of PON1 are shown in Figure 1. The Leu55Met and Gln192Arg sequence changes have been previously reported and influence the concentration and/or activity of PON1. The other four missense changes (Met127Arg, His155Arg, Gln192Glu, Ala252Gly) are novel.
Table 3.
Sequence Alterations Identified in PON1 in AMD Patient and Control Populations.
| Exon | Sequence Change | Protein Change | Allele Frequency AMD Patients | Allele Frequency Normals |
|---|---|---|---|---|
| 3 | TTG→ATG | Leu55Met | 693/1838 = 37.70% | 250/736 = 33.97% |
| 5 | ATG → AGG | Met127Arg | 3/1774 = 0.17% | 1/714 = 0.14% |
| 5 | CAT → CGT | His155Arg | 1/1774 = 0.06% | 0/714 = 0.0% |
| 6 | CAA → CGA | Gln192Arg | 565/1918 = 29.45% | 284/740 = 38.38% |
| 6 | CAA → GAA | Gln192Glu | 7/1932 = 0.36% | 0/740 = 0.0% |
| 7 | GCT → GGT | Ala252Gly | 1/1894 = 0.05% | 0/730 = 0.0% |
PON1: Paraoxonase 1
AMD: age-related macular degeneration
Figure 1.
Schematic representation of the PON1 gene showing the six sequence changes identified in AMD patients. Each of the nine exons is shown as a separate colored box. A representative sequence change is aligned to the corresponding exon.
We next expanded our evaluation of these six variations by screening an additional 660 patients with AMD. Table 3 summarizes the frequency for each of these variants in our AMD and age-matched control cohorts. All six variants were identified in Caucasians.
Leu55Met Polymorphism
The Leu55Met polymorphism is a common variant present in the NCBI SNP database (rs854560). In our samples, this SNP had an allele frequency of 37.7% in patients (N = 919) and 34.0% in controls (N = 368) (P = 0.078). Allele and genotype data for the Leu55Met polymorphism are given in Table 4. There was no significant difference between the observed genotype frequencies for both the patient and control populations with those expected under Hardy-Weinberg equilibrium (χ2 = 0.20, P = 0.91 in patients; χ2 = 0.01, P = 0.99 in controls). Compared with controls, the frequency of the A allele was not significantly higher in all AMD cases (Categories 1 – 4) or in phenotypic subgroups defined by AREDS as Categories 1, 2, 3 and 4. However, if Category 1 patients are not included in the AMD cohort, this allele was significantly higher in patients than controls (P = 0.030). This association does not remain significant following a conservative Bonferroni correction of P < 0.008. The frequency of the AA genotype was also not significantly higher in all AMD cases when combining Categories 1 – 4 or Categories 2 – 4 or in the four phenotypic subgroups as compared with controls. Table 5 shows that when the Category 4 wet and dry AMD patients were separately compared to controls, the minor allele frequency of this variant was significantly more prevalent for wet AMD patients (N = 436; P = 0.020) but not for dry AMD patients (N = 70; P = 0.328). Similarly, the genotypic frequency of the AA genotype was significantly more prevalent than controls for wet AMD patients (P = 0.040) but not for dry AMD patients (P = 0.341). However, neither of these associations remains significant following a conservative Bonferroni correction of P < 0.008.
Table 4.
Allele and Genotype Frequencies for the Leu55Met PON1 Polymorphism Identified in AMD Patient and Control Populations.
| Controls | Cat 1 | Cat 2 | Cat 3 | Cat 4 | All AMD (Cat 2 – 4) | All AMD (Cat 1 – 4) | |
|---|---|---|---|---|---|---|---|
| N = 368 | N = 117 | N = 175 | N = 105 | N = 522 | N = 802 | N = 919 | |
| Alleles | |||||||
| T | 486 (66.0) | 161 (68.8) | 212 (60.6) | 125 (59.5) | 647 (62.0) | 984 (61.3) | 1145 (62.3) |
| A | 250 (34.0) | 73 (31.2) | 138 (39.4) | 85 (40.5) | 397 (38.0) | 620 (38.7) | 693 (37.7) |
| Significance* | P = 0.474 | P = 0.090 | P = 0.086 | P = 0.080 | P = 0.030 | P = 0.078 | |
| Genotypes | |||||||
| TT | 161 (43.7) | 56 (47.8) | 64 (36.6) | 34 (32.4) | 198 (37.9) | 296 (36.9) | 352 (38.3) |
| TA | 164 (44.6) | 49 (41.9) | 84 (48.0) | 57 (54.3) | 251 (48.1) | 392 (48.9) | 441 (48.0) |
| AA | 43 (11.7) | 12 (10.3) | 27 (15.4) | 14 (13.3) | 73 (14.0) | 114 (14.2) | 126 (13.7) |
| Significance* | P = 0.726 | P = 0.215 | P = 0.111 | P = 0.196 | P = 0.074 | P = 0.179 | |
Comparison with control frequencies, Fisher’s Exact (alleles) and χ2 test (genotypes).
Data expressed as the number of subjects (% of entire group).
PON1: Paraoxonase 1
AMD: age-related macular degeneration
Table 5.
Allele and Genotype Frequencies for the PON1 Leu55Met Polymorphism in End-Stage AMD Patients.
| Controls | Cat 4 - Dry | Cat 4 - Wet | |
|---|---|---|---|
| N = 368 | N = 70 | N = 436 | |
| Alleles | |||
| T | 486 (66.0) | 99 (70.7) | 526 (60.3) |
| A | 250 (34.0) | 41 (29.3) | 346 (39.7) |
| Significance* | P = 0.328 | P = 0.020 | |
| Genotypes | |||
| TT | 161 (43.7) | 37 (52.9) | 153 (35.1) |
| TA | 164 (44.6) | 25 (35.7) | 220 (50.5) |
| AA | 43 (11.7) | 8 (11.4) | 63 (14.4) |
| Significance* | P = 0.341 | P = 0.040 | |
Comparison with control frequencies, Fisher’s Exact (alleles) and χ2 test (genotypes).
Data expressed as the number of subjects (% of entire group).
PON1: Paraoxonase 1
AMD: age-related macular degeneration
Gln192Arg Polymorphism
The Gln192Arg polymorphism is also present in the NCBI SNP database (rs662). Allele and genotype data for the Gln192Arg polymorphism are shown in Table 6. In our study population, this SNP had a significantly different (P < 0.0001) allele frequency of 38.4% in controls (N = 370) and 29.5% in patients (N = 959). For both the patient and control populations, there was no significant difference between the observed genotype frequencies with those expected under Hardy-Weinberg equilibrium (χ2 = 4.57, P = 0.10 in patients; χ2 = 5.21, P = 0.07 in controls). The frequency of the G allele was significantly higher in controls than in all AMD patients when combining Categories 1 – 4 or Categories 2 – 4 (P < 0.0001, P < 0.0001; respectively). This difference is significant for AMD Categories 2, 3 and 4 as compared to controls (P = 0.004, P = 0.002, P < 0.0001, respectively) but not for AMD Category 1 patients (P = 0.197). The frequency of the GG genotype was also significantly higher in controls as compared with all AMD cases, both Categories 1 – 4 and Categories 2 – 4, and in the three more severe AMD subgroups individually. Similarly, Category 1 did not appear different than controls. Table 7 shows that when only the Category 4 wet AMD patients were considered (N = 457), the minor allele frequency of this variant is significantly higher in controls as compared to patients (P < 0.0001); whereas controls did not show a statistical difference when compared to Category 4 dry AMD patients (N = 65) (P = 0.377). The associations observed with Gln192Arg remain significant after Bonferroni correction. These results indicate that the Gln192Arg SNP in PON1 is associated with a decreased susceptibility to AMD.
Table 6.
Allele and Genotype Frequencies for the Gln192Arg PON1 Polymorphism in AMD Patient and Control Populations.
| Controls | Cat 1 | Cat 2 | Cat 3 | Cat 4 | All AMD (Cat 2 – 4) | All AMD (Cat 1 – 4) | |
|---|---|---|---|---|---|---|---|
| N = 370 | N = 123 | N = 179 | N = 118 | N = 539 | N = 836 | N = 959 | |
| Alleles | |||||||
| A | 456 (61.6) | 163 (66.3) | 253 (70.7) | 171 (72.5) | 766 (71.1) | 1190 (71.2) | 1353 (70.5) |
| G | 284 (38.4) | 83 (33.7) | 105 (29.3) | 65 (27.5) | 312 (28.9) | 482 (28.8) | 565 (29.5) |
| Significance* | P = 0.197 | P = 0.004 | P = 0.002 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |
| Genotypes | |||||||
| AA | 155 (41.9) | 56 (45.5) | 88 (49.2) | 58 (49.2) | 256 (47.5) | 402 (48.1) | 458 (47.7) |
| AG | 146 (39.5) | 51 (41.5) | 77 (43.0) | 55 (46.6) | 254 (47.1) | 386 (46.2) | 437 (45.6) |
| GG | 69 (18.6) | 16 (13.0) | 14 (7.8) | 5 (4.2) | 29 (5.4) | 48 (5.7) | 64 (6.7) |
| Significance* | P = 0.353 | P = 0.006 | P = 0.0007 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |
Comparison with control frequencies, Fisher’s Exact (alleles) and χ2 test (genotypes).
Data expressed as the number of subjects (% of entire group).
PON1: Paraoxonase 1
AMD: age-related macular degeneration
Table 7.
Allele and Genotype Frequencies for the PON1 Gln192Arg Polymorphism in End-Stage AMD Patients.
| Controls | Cat 4 - Dry | Cat 4 - Wet | |
|---|---|---|---|
| N = 370 | N = 65 | N = 457 | |
| Alleles | |||
| A | 456 (61.6) | 86 (66.3) | 657 (70.7) |
| G | 284 (38.4) | 44 (33.7) | 257 (29.3) |
| Significance* | P = 0.377 | P < 0.0001 | |
| Genotypes | |||
| AA | 155 (41.9) | 27 (41.6) | 222 (48.6) |
| AG | 146 (39.5) | 32 (49.2) | 213 (46.6) |
| GG | 69 (18.6) | 6 (9.2) | 22 (4.8) |
| Significance* | P = 0.126 | P < 0.0001 | |
Comparison with control frequencies, Fisher’s Exact (alleles) and χ2 test (genotypes).
Data expressed as the number of subjects (% of entire group).
PON1: Paraoxonase 1
AMD: age-related macular degeneration
The results of logistic regression analysis for the Gln192Arg SNP are given in Table 8. There was no significant difference in the AMD odds ratio between the AA (non-risk alleles) and AG genotypes, whether the comparison was based on all AMD patients taken together (Categories 1 – 4 or Categories 2 – 4) or each AMD category examined separately. In comparison, the odds ratio (OR) for the GG genotype was significantly different from the AA genotype when comparisons were made for combined Categories 1 – 4 AMD patients (OR = 0.31, CI 0.21 – 0.46, P < 0.0001), combined Categories 2 – 4 AMD patients (OR = 0.27, CI 0.18 – 0.41, P < 0.0001), AMD Categories 2 (OR = 0.36, CI 0.19 – 0.67, P = 0.001), 3 (OR = 0.20, CI 0.08 – 0.49, P < 0.0001) and 4 (OR = 0.25, CI 0.16 – 0.41, P < 0.0001), but not for AMD Category 1.
Table 8.
Odds Ratios for AMD Associated with the PON1 Gln192Arg Genotype.
| AMD Type | # Controls | # Cases | Odds of AMD (95% CI) Compared with the AA Genotype |
|
|---|---|---|---|---|
| AG | GG | |||
| Cat 1 | 370 | 123 | 0.967 (0.623–1.501) | 0.642 (0.346–1.191) |
| P = 0.911 | P = 0.180 | |||
| Cat 2 | 370 | 179 | 0.929 (0.636–1.358) | 0.357 (0.192–0.667) |
| P = 0.771 | P = 0.001 | |||
| Cat 3 | 370 | 118 | 1.007 (0.654–1.550) | 0.197 (0.077–0.490) |
| P = 1.000 | P < 0.0001 | |||
| Cat 4 | 370 | 539 | 1.053 (0.792–1.400) | 0.254 (0.158–0.409) |
| P = 0.771 | P < 0.0001 | |||
| All (Cat 2 – 4) | 370 | 836 | 1.019 (0.782–1.329) | 0.268 (0.178–0.405) |
| P = 0.892 | P < 0.0001 | |||
| All (Cat 1 – 4) | 370 | 959 | 1.013 (0.780–1.315) | 0.314 (0.214–0.461) |
| P = 0.947 | P < 0.0001 | |||
PON1: Paraoxonase 1
AMD: age-related macular degeneration
Novel Sequence Changes
Two novel missense changes were identified in exon 5, Met127Arg and His155Arg (Table 3). The Met127Arg change was detected heterozygously in three patients, representing a minor allele frequency of 0.17%. Each patient had a different level of AMD severity (one each in Categories 1, 2 and 4). This change was also found heterozygously in one normal control yielding a minor allele frequency of 0.14%, similar to that seen in AMD patients. In silico analysis using PolyPhen predicts this missense change to be possibly damaging to the protein. A large difference (1.950) was noted in PSIC scores between the allelic variants. PMut analysis also predicts Met127Arg to be likely pathological (output value of 0.8) with good reliability (score of 5).
The His155Arg sequence change was identified heterozygously in one Category 1 patient and was not found in normal controls, representing a minor allele frequency of 0.06%. PolyPhen predicts this missense change to be probably damaging. A large difference (2.934) was calculated in PSIC scores between the allelic variants. This difference indicates that the observed substitution is rarely or never observed in the PON protein family and is predictive of a hydrophobicity charge change at a buried site within the protein. PMut analysis also predicts His155Arg to be pathological (output value of 0.7) with good reliability (score of 4).
While both bioinformatic algorithms predict these two missense changes to be pathological, available structure-function analysis of PON1 indicates that neither amino acids 127 or 155 are involved in functional properties of the enzyme such as hydrolytic activity or substrate specificity. Although methionine to arginine alters a hydrophobic, non-polar amino acid to a positively charged residue, Met127 is not conserved across the three human PON genes or across PON1 in different species.16–18 His155Arg is also likely to be a rare variant since both amino acids have similar positively charged side chains and this residue is not conserved across the family of PON genes.16–18 The possibility that either of these low frequency variants is pathological will require further study of different and larger patient and control populations.
The Gln192Glu sequence variant involves the same amino acid altered by the Gln192Arg polymorphism. This change was found heterozygously in seven Category 4 patients, representing a minor allele frequency of 0.36% in patients and was not identified in any normal controls (Table 3). This difference was not statistically significant (P = 0.201). Gln192Arg changes an uncharged amino acid to a positively charged amino acid and is responsible for altering the substrate specificity and activity of PON1.19,20,27 Gln192Glu changes this residue to a negatively charged amino acid, possibly altering similar properties of the enzyme. Interestingly, in silico analysis using PolyPhen predicts Gln192Glu to be a benign alteration with only a small difference (0.095) noted in PSIC scores between the allelic variants. A similar benign result was predicted between Gln192Arg variants with a small PSIC score difference (1.475). PMut analysis predicts both Gln192Glu and Gln192Arg to be neutral changes not affecting the function of the protein. Output values were 0.04 with high reliability (score of 9) for Gln192Glu and 0.23 with good reliability (score of 5) for Gln192Arg. Whether Gln192Glu is a rare AMD-associated PON1 variant will require additional study.
Finally, the novel sequence variant, Ala252Gly, was found heterozygously in one Category 4 patient yielding a minor allele frequency of 0.05% (Table 3). It was not found in normal controls. Both computational assessments of this missense change predict this variant to be benign (PSIC score difference of 1.242) and neutral (output value of 0.3 with good reliability). This residue is not conserved across the PON gene family and is therefore considered to be a rare variant not associated with AMD.
All of the patients identified as having one of the four novel PON1 sequence change were Caucasian, eliminating the possibility that these rare polymorphisms were related to ethnicity or race.
Discussion
We report six sequence changes in PON1 in patients with AMD. Two of the variants are common polymorphisms reported to protect against oxidative stress in aging disorders (Leu55Met and Gln192Arg) and four are novel missense changes (Met127Arg, His155Arg, Gln192Glu and Ala252Gly). The two most common variants of PON1 have been studied extensively in cardiovascular disease, cancer, Parkinson’s disease, Alzheimer’s disease, and other aging disorders.15,16 While there is a large amount of data on the influence of these SNPs on age-related diseases, the results are conflicting with some reports suggesting positive associations while others indicating no associations.
Both Leu55Met and Gln192Arg were linked to the development of wet AMD in a previous study of 72 Japanese patients.28 In two subsequent studies, examining 62 or 94 Caucasian patients, neither polymorphism was associated with end-stage AMD.29,30 In the current study, we evaluated a substantially larger primarily Caucasian cohort and stratified our AMD cases into phenotypic categories defined by the AREDS study group. We believe that selection bias in our case-control association study was avoided since cases and controls derived from the same source populations, and no significant deviations from Hardy-Weinberg equilibrium were noted.
Although Leu55Met has been shown to be associated with a higher concentration and stability of PON1,21,26 our results do not support an association between this SNP and an increased risk of AMD, for our entire patient cohort (P = 0.078) or for groups defined by AREDS criteria. Nevertheless, when Category 4 or end-stage AMD patients were subdivided into wet versus dry, we observed a weak association of the PON1 Leu55Met SNP with an increased risk of wet AMD (P = 0.02) (Table 5). Overall, our results replicate other findings of a minor association of Met55 with AMD in Caucasian populations.
Based on the limited structure-function data available for PON1, we speculate that, in comparison to Leu55, the Met55 allozyme confers lower plasma concentration and stability of PON1 and thereby reduces its oxidative protective properties. Oxidative stress, either from endogenous sources such as inflammation and cellular metabolism or exogenous sources including intense light exposure and environmental toxins, has long been proposed to contribute to the risk of AMD.36,37 This is supported by indirect evidence that smoking significantly increases the risk of AMD13 and that antioxidant vitamins and zinc can slow the progression of the disease for select individuals.14 Since PON1 functions to protect against oxidative stress,15 a decrease in PON1 activity would likely increase oxidative stress and the risk of AMD. This hypothesis is supported by studies showing that serum PON1 activity was significantly lower in AMD patients than in controls.31,32
The Gln192Arg polymorphism impacts PON1 substrate specificities and activity rates.19, 20,27 In fact, a meta-analysis of 43 studies examining multiple SNPs for PON1 in their relation to cardiovascular disease found the most promising variant to be Gln192Arg.38 The two allozymes possess considerably different affinity for and catalytic activity with a number of substrates, with the Gln192 variant demonstrating an overall lesser activity than Arg192.15,16,39 In addition, individuals with the Arg192 variant had higher levels of PON1 activity, and lower levels of systemic oxidative stress, measured by plasma levels of multiple oxidized fatty acids indicating that this variant strongly influences quantitative measures of oxidative damage.39 While Ikeda et al.28 found a higher frequency of the Arg192 variant in their Japanese cohort of wet AMD, two subsequent studies found no association between this SNP and AMD in Caucasian patients.29,30 Our results from primarily Caucasian patients indicate a statistically higher allele frequency for the Arg192 allele in normal controls than in the combined AMD patient population or the AREDS categories considered separately (Table 6). Interestingly, when Category 4 AMD was further stratified into wet versus dry, the Arg192 allele was statistically lower in the wet form but not the dry form of AMD (Table 7). While the number of patients with wet AMD in our cohort (N = 457) is substantially larger than the number of patients with dry AMD (N = 65), our results suggest that the Arg192 allele may have a protective effect or a decreased susceptibility to AMD, particularly for end-stage patients with the exudative form.
While the mechanism(s) for PON1-mediated protection against AMD remains to be determined, our current findings support a role for the Gln192Arg SNP in decreasing susceptibility to AMD. Although it is not entirely clear how this SNP may function to protect against AMD, a recent study found that individuals with the Arg192 alloenzyme exhibited higher levels of PON1 activity and lower levels of systemic oxidative stress.39
We also identified four additional novel sequence changes, three of which we believe are not associated with an increased or decreased susceptibility to AMD due to their limited frequency and/or location within the PON1 gene (Met127Arg, His155Arg and Ala252Gly). The fourth new variant, Gln192Glu, changes an uncharged amino acid to a negatively charged residue whereas Gln192Arg changes the uncharged residue at this position to a positively charged amino acid. As noted above, structure function studies indicate that this position contributes to the substrate specificity and activity of PON1. Given that this variant was identified only in Category 4 AMD patients, albeit at low frequency, it is possible that this SNP also alters similar properties of the PON1 enzyme as Gln192Arg. Functional studies are needed to support and confirm this possibility.
One of the main issues in association studies is how to evaluate the significance of multiple comparisons of SNPs. The Bonferroni correction is commonly applied; however, it has been criticized as being too conservative. An alternative approach would be to reproduce findings in a second replication cohort. This approach is less stringent and more powerful; however, we did not have access to a second dataset and so we applied the Bonferroni correction. Significant associations for this study therefore require an adjusted P < 0.008. Following this correction, we eliminated the significant finding of an association between the PON1 Leu55Met SNP with an increased risk of wet AMD. We note that this correction may be a limitation but without verification through a second dataset, this approach is the most efficacious.
The strengths of this study include the large number of patients with AMD and the availability of information on the functional consequences of the common PON1 SNPs, Leu55Met and Gln192Arg. The mechanism by which PON1 Gln192Arg might protect from AMD is not clear. We predict that the Arg192 alloenzyme will confer a higher level of enzymatic activity and thereby may counteract oxidative damage more effectively. Seeking an association between AMD and PON1 genotypes which are more protective against oxidative modification or damage is not a definitive test. The real test of this hypothesis would be to determine whether the concentration and/or activity of the enzyme is increased or decreased in individuals with AMD. The extrapolation from genotype to phenotype is complex in this type of situation where interindividual variation in enzyme levels and activity is influenced by multiple genetic and environmental factors (smoking, alcohol consumption, pharmacological agents, diet, and vitamins15). A future study evaluating the relationship between AMD and PON1 genotype and phenotype will be important for fully understanding the role of PON1 in AMD pathogenesis.40 If a positive relation is noted, strategies to increase PON1 activity levels may be a therapeutic option for AMD.
In sum, we have referred to the association of the Gln192Arg variant with AMD as protective solely because the minor allele of this polymorphism is more frequent in controls than in cases. However, the functional and biological mechanism by which this mutation is acting to influence the risk of protection of AMD remains to be determined. Clearly, statistical analyses can only indicate potential genetic associations, and additional work will be required to determine whether Gln192Arg decreases AMD susceptibility and to identify the mechanism(s) by which this influence might be realized.
Acknowledgments
The Clinical Genomic and Proteomic AMD Study Group was composed of the following clinicians who performed ophthalmic patient examinations: Peter K Kaiser MD1, Gregory S Kosmorsky DO1, Roger HS Langston, MD1, Hilel Lewis MD1, Andrew P Schachat MD1, Jonathan E Sears MD1, Arun Singh MD1, Scott D Smith MD1, Elias I Traboulsi MD1 and Stacia S Yaniglos OD2,4. The Study Group also included the following individuals who obtained written informed consent and blood specimens from patients: Elisa Bala MD2, Sue Crowe BS1, and Ellen Simpson RN1. The authors thank all patients and control individuals for participation in this study.
This work was supported in part by US National Institute of Health grants EY015638, EY016072, BRTT 05-29 from the State of Ohio, a Foundation Fighting Blindness Center Grant, a Research to Prevent Blindness (RPB) Center Grant, a RPB Sybil B. Harrington Special Scholar Award to SAH and the VA Medical Research Service. The authors indicate no financial conflict of interest. Contributions of Authors: Involved in conception and design of study (GJTP, GMS, NSP, SAH); analysis and interpretation of data (GJTP, GMS, NSP, SAH); writing the article (SAH, NSP); critical revision of the article (GJTP, GMS); final approval of the article (GJTP, GMS, NSP, SAH); data collection (EB, SC, ES); provision of patients (PKK, GSK, RHSL, HL, APS, JES, AS, SDS, EIT, SSY); and obtaining funding (SAH, NSP). This study conformed to the tenets of the Declaration of Helsinki and was approved by the Internal Review Boards of the Cleveland Clinic and the Louis Stokes Cleveland Department of Veterans Affairs Medical Center. Written informed consent for research was obtained from each participant.
References
- 1.Stone EM. Macular degeneration. Annu Rev Med. 2007;58:477–90. doi: 10.1146/annurev.med.58.111405.133335. [DOI] [PubMed] [Google Scholar]
- 2.Evans JR. Risk factors for age-related macular degeneration. Prog Retin Eye Res. 2001;20:227–253. doi: 10.1016/s1350-9462(00)00023-9. [DOI] [PubMed] [Google Scholar]
- 3.Hollman FW, Mulder TJ, Kallan JE. Popul Proj Branch Popul Div. US Census Bureau; Wash, D.C.: 2003. Methodology and assumptions for the population projects of the United States: 1999–2100; pp. 1–34. [Google Scholar]
- 4.Friedman DS, O’Colmain BJ, Muñoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 5.Swaroop A, Branham KE, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits. Hum Mol Genet. 2007;16:174–182. doi: 10.1093/hmg/ddm212. [DOI] [PubMed] [Google Scholar]
- 6.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51:316–363. doi: 10.1016/j.survophthal.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 9.Edwards AO, Ritter R, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 10.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7053–7054. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yates JR, Sepp T, Matharu BK, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 13.Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA. 1996;276:1141–1146. [PubMed] [Google Scholar]
- 14.Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age- related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchegiani F, Marra M, Olivieri F, et al. Paraoxonase 1: genetics and activities during aging. Rejuvenation Res. 2008;11:113–127. doi: 10.1089/rej.2007.0582. [DOI] [PubMed] [Google Scholar]
- 16.Draganov DI, La Du BN. Pharmacogenetics of paraoxonases: a brief review. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:78–88. doi: 10.1007/s00210-003-0833-1. [DOI] [PubMed] [Google Scholar]
- 17.Furlong CE, Li W-F, Shih DM, Lusis AJ, Richter RJ, Costa LG. Genetic factors in susceptibility: Serum PON1 variation between individuals and species. Hum Ecol Risk Assess. 2002;8:31–43. [Google Scholar]
- 18.Primo-Parmo SL, Sorenson RC, Teiber J, La Du BN. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics. 1996;33:498–507. doi: 10.1006/geno.1996.0225. [DOI] [PubMed] [Google Scholar]
- 19.Adkins S, Gan KN, Mody M, La Du BN. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am J Hum Genet. 1993;52:598–608. [PMC free article] [PubMed] [Google Scholar]
- 20.Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–76. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- 21.Garin MC, James RW, Dussoix P, et al. Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J Clin Invest. 1997;99:62–66. doi: 10.1172/JCI119134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iselius L, Evans DA, Playfer JR. Genetics of plasma paroxonase activity. J Med Genet. 1982;19:424–426. doi: 10.1136/jmg.19.6.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leviev I, James RW. Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arterioscler Thromb Vasc Biol. 2000;20:516–521. doi: 10.1161/01.atv.20.2.516. [DOI] [PubMed] [Google Scholar]
- 24.Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Jarvik GP, Furlong CE. Effects of 5_ regulatory- region polymorphisms on paraoxonasegene (PON1) expression. Am J Hum Genet. 2001;68:1428–1436. doi: 10.1086/320600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deakin S, Leviev I, Brulhart-Meynet MC, James RW. Paraoxonase-1 promoter haplotypes and serum paraoxonase: a predominant role for polymorphic position-107, implicating the Sp1 transcription factor. Biochem J. 2003;372:643–649. doi: 10.1042/BJ20021670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leviev I, Deakin S, James RW. Decreased stability of the M54 isoform of paraoxonase as a contributary factor to variations in human serum paraoxonase concentrations. J Lipid Res. 2001;42:528–535. [PubMed] [Google Scholar]
- 27.Davies HG, Richter RJ, Keifer M, Broomfield CA, Sowalla J, Furlong CE. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda T, Obayashi H, Hasegawa G, et al. Paraoxonase gene polymorphisms and plasma oxidaized low-density lipoprotein level as possible risk factors for exudative age-related macular degeneration. Am J Ophthalmol. 2001;132:191–195. doi: 10.1016/s0002-9394(01)00975-8. [DOI] [PubMed] [Google Scholar]
- 29.Baird PN, Chu D, Guida E, Vu HT, Guymer R. Association of the M55L and Q192R Paraoxonase Gene Polymorphisms With Age-related Macular Degeneration. Am J Ophthalmol. 2004;138:665–666. doi: 10.1016/j.ajo.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 30.Esfandiary H, Chakravarthy U, Patterson C, Young I, Hughes AE. Association study of detoxification genes in age related macular degeneration. Br J Ophthalmol. 2005;89:470–474. doi: 10.1136/bjo.2004.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baskol G, Karakucuk S, Oner AO, et al. Serum paraoxonase 1 activity and lipid peroxidation levels in patients with age-related macular degeneration. Ophthalmologica. 2006;220:12–16. doi: 10.1159/000089269. [DOI] [PubMed] [Google Scholar]
- 32.Ates O, Azizi S, Alp HH, et al. Decreased serum paraoxonase 1 activity and increased serum homocysteine and malondialdehyde levels in age-related macular degeneration. Tohoku J Exp Med. 2009;217:17–22. doi: 10.1620/tjem.217.17. [DOI] [PubMed] [Google Scholar]
- 33.Xi Q, Pauer GJ, Traboulsi EI, Hagstrom SA. Mutation screen of the TUB gene in patients with retinitis pigmentosa and Leber congenital amaurosis. Exp Eye Res. 2006;83:569–573. doi: 10.1016/j.exer.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrer-Costa C, Gelpí JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 36.Beatty S, Koh H, Phil M, Henson D, Boulton M. Surv Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 37.Winkler BS, Boulton ME, Gottsch JD, Sternberg P. Mol Vis. 1999;5:32. [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler JG, Keavney BD, Watkins H, Collins R, Danesh J. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls: meta-analysis of 43 studies. Lancet. 2004;363:689–695. doi: 10.1016/S0140-6736(04)15642-0. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharyya T, Nicholls SJ, Topol EJ, et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008;299:1265–1276. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter RJ, Furlong CE. Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics. 1999;9:745–753. [PubMed] [Google Scholar]