Abstract
That commensal bacteria play an important role in human health is beyond doubt, and it is now widely accepted that humans function as super organisms, whose collective metabolic potential exceeds the sum of our individual eukaryotic and prokaryotic components. However, while it is has been established that the prokaryotic component of the human superorganism is amenable to manipulation by chemotherapeutic, dietary or microbial interventions, the significance of such alterations in terms of human health or well being is less well established. Prebiotics (non- digestible food ingredients that stimulate the growth and/or activity of bacteria in the digestive system) and probiotics (live microorganisms that when administered in adequate amounts, confer a health benefit on the host) are often bracketed among ‘alternative’ approaches to influencing human health, such as homeopathy, naturopathy, acupuncture and hypnotherapy. Others believe that prebiotics and probiotics have proven their effectiveness in properly conducted, clinically controlled human trials and therefore can be considered as evidence-based alternatives or adjuncts to conventional medicines. My journey from a position of total skepticism to ‘reluctant convert’ is the basis of this article, which should not be considered in any sense as a review of the literature but simply a personal account of this transition. While I am not bent on converting other doubters, I will recount some of the thought processes and evidence that has helped to form my current opinion.
Key words: probiotics, commensals, pharmabiotic, infection, bacteriocins, microbiota
Man will occasionally stumble over the truth, but usually manages to pick himself up, walk over or around it, and carry on—Winston S. Churchill.
Introduction
In 1988 I finished my PhD in University College Cork (UCC) in Ireland, having worked on the genetics of lactic acid bacteria (LAB) under the excellent supervision of Charlie Daly and Gerald Fitzgerald. The LAB, and in particular Lactococcus lactis, were becoming more genetically accessible through the pioneering work by Larry McKay and Todd Klaenhammer in the US, Gerard Venema and Willem de Vos in The Netherlands and Mike Gasson in the UK, among many other outstanding researchers. While my interest, similar to that of many contemporaries, was in designing and implementing genetic tools for the manipulation of our favorite genera, Lactococcus and Lactobacillus, we did this on the basis that their importance rested on their industrial applications. To my recollection, the fact that Lactobacillus spp. could also be found in the gut was not considered important, and we were solely focused on their beneficial roles in cheese ripening, yogurt production and their undesirable roles in the spoilage of beer and other commodities. A lack of awareness of a possible role for lactobacilli in the gut condemns my younger self for a failure to properly read the literature (a weakness which I unfortunately have never properly rectified) combined with a somewhat lactococcal-centered world view which prevailed at that time among LAB researchers. The first instance that I can recall mention of the potential probiotic qualities of lactobacilli was in 1987 when the late Khem Shahani of the University of Nebraska-Lincoln gave a presentation to the Second International Symposium on LAB in The Netherlands (which I attended as a member of the Venema lab, having been fortunate to spend a year in Groningen as part of my PhD). While the lecture introduced many of us to the concept of probiotics, from memory it was regarded by those interested in molecular microbiology as a quaint but super ficial topic, certainly not a serious research area likely to generate exciting data or to yield funding opportunities. At that time, I (and at least some others) regarded probiotic research as peripheral and largely based on uncontrolled experiments and apocryphal stories. We prided ourselves on the knowledge that we were rigorously elucidating the genetic and mechanistic basis of truly important industrially relevant phenotypes, many of which had the added bonus of being plasmid-encoded. It would also be fair to say that health effects were not something that most LAB researchers were equipped to measure, and the age of the health conscious consumer (the worried well?) was yet to dawn.
Sweet Carolina
I promptly forgot about probiotics and went back to work on lactococcal genetics, this time as a post-doctoral scientist at North Carolina State University, working with Todd Klaenhammer. To a modern reader, Todd is synonymous with probiotic research, having done more than anyone to promote the type of rigorous research I hope to come back to later in this article, but at that time he was more renowned for his work on lactococcal phage resistance and on bacteriocins. However, he was also the direct successor at NC State to Dr. Marvin Speck, a pioneer of probiotic research1 and the developer of the Sweet Acidophilus™ milk that was available in local supermarkets. I had the good fortune to meet this most gentle of men when he visited the laboratory on a number of occasions before his death in 2003, but even those encounters did not send me back to the literature. While I remained sceptical of the benefits of probiotics, I should admit to buying Sweet Acidophilus™ milk for our 18 month old son when he was suffering from a gastrointestinal upset, thus confirming that in extremis most of us are willing to set aside science in favor of hope. The NC State Food Science Department was understandably proud of Dr. Speck's contributions, and Todd took up the baton by continuing to work on Lactobacillus acidophilus and other lactobacilli. In fact, I narrowly missed working on a project devoted to generating genetic tools for lactobacilli (a project that went to John Luchansky who had just joined the lab) and was assigned to another project on the genetic basis of bacteriophage resistance in lactococci. Another scientist who crossed my path at this time was Gerald Tannock, a pioneer of rigorous experimentation on the behavior of lactobacilli in the gut, who spent some time on sabbatical in Todd's lab, working with John Luchansky on the Lactobacillus genetic tools project.2 So, while Todd was obviously alive to the possibility of probiotic interventions and a role for these important bacteria in human health, and others in the lab were working hard on developing tools to allow the subsequent dissection of these phenomena, I managed to leave the group wholly uncontaminated by such convictions and aware of little outside of my immediate project. I should not miss this opportunity to acknowledge that Todd had a huge influence on my development as a scientist and his elevation to the ranks of the US National Academy of Sciences speaks for his contributions across a number of fields, including probiotic research. With a very neat symmetry Todd's inaugural article for the Academy presented the genome sequence of Lactobacillus acidophilus, the same organism used by Speck some decades earlier in the development of Sweet Acidophilus™ milk.3
Passing Up on Probiotics
I returned to Ireland to take up a position as senior research officer with Teagasc in Moorepark, continuing the work on lactococcal genetics. We were also interested in another phenomenon common among members of the LAB, the production of antimicrobial peptides, or bacteriocins (of which more later). During the two years I spent in Moorepark (1990–1992), I was contacted by a European consortium to determine whether I might have an interest in collaborating on probiotic research. Due largely to my continuing skepticism about the scientific basis of probiotics, I was unable to generate the enthusiasm necessary to convince myself or others of the value of this type of research. Thus, I missed the chance to get in on the ground floor of probiotic research in Europe. As it turns out, while I missed out on probiotics at this stage, Moorepark did not and through the efforts of Paul Ross and Catherine Stanton have made Moorepark a center of excellence in probiotic research in the intervening years. Indeed, my future colleague at UCC, the immunologist Kevin Collins, took full advantage of the opportunity to get involved in probiotic research and he started a research programe at UCC that blossomed and has culminated in the founding of the Alimentary Pharmabiotic Centre (APC) which now has over 100 scientists working on the human microbiome (apc.ucc.ie).
In 1992 I took up a position on the faculty of the Microbiology Department at my alma mater, UCC. Following the advice of the departmental Chair, Seamus Condon, I elected to initiate a program on microbial food safety. While Seamus and I both thought I would leave LAB research far behind, two important events conspired to draw me back into that familiar world. One was the fact that my successor at Moorepark was an old friend, Paul Ross, and we agreed to collaborate to finish off some of the work I had initiated on LAB bacteriocins, particularly focusing on their role in food safety. In the event, Paul and I have had a long and enjoyable collaboration over the intervening years, initially solely on bacteriocins, but one that has come to include many other areas in recent times. Secondly, I was determined to develop a program in food safety research and settled on Listeria monocytogenes as an ideal organism for this purpose. Listeria lagged behind the state of the art in the LAB in terms of genetic accessibility, but it seemed to me that many of the LAB genetic tools would transfer readily to this new target, again bringing me back into contact with my old colleagues in LAB research. I was also greatly helped at that time by hiring an outstanding scientist as a post-doctoral fellow, Cormac Gahan. Cormac, now a colleague and close collaborator on the faculty at UCC and in the APC, was familiar with animal infection models and had the necessary immunology background that meant for the first time the laboratory could venture into matters of health and disease, beginning with murine models of Listeria infection and eventually extending to a number of other gastrointestinal and respiratory pathogens and additional hosts such as pigs and cattle.
The Case for Probiotics
The work on pathogens over the next few years led me to consolidate my very jaundiced view of probiotics and their associated claims. While infection microbiologists used Koch's postulates to rigorously identify pathogens, conducted experiments where the end-points were measurable degrees of morbidity and even mortality, identified virulence factors and elucidated pathogenic strategies, our probiotic colleagues seemed to lack these hard endpoints. ‘Well being’ is difficult to measure in a mouse, and it seemed to me that the immunological responses exhibited by animals or cell lines on exposure to probiotic strains (some cytokines went up while others went down!) could be rationalized in any number of ways, all of which seemed to support the probiotic hypothesis under investigation. In pathogen research one could not call something a virulence factor without demonstrating some key characteristics (not least of all a diminution in virulence when the factor was abolished), but it seemed that probiotic factors abounded without any such formal proof (adherence, bile resistance bacteriocin production, etc.). The term probiotic was, extended to include whole genera (Lactobacillus and Bifidobacterium), but it seemed impossible to identify a non-probiotic control for most experiments. From the lofty world of pathogen research, probiotics seemed to suffer from a lack of rigor, and yet…
Something that at first surprised and then intrigued me was the number of highly respected colleagues beginning to work in this field. Todd Klaenhammer, Paul Ross and Kevin Collins I have already mentioned, but the probiotics research effort at UCC now also included Fergus Shanahan and Eamonn Quigley, two gastroenterologists recently returned from the US with stellar research reputations. Charlie Daly and Gerald Fitzgerald were also now working on probiotics. These scientists were unlikely in my view to get involved in anything that lacked rigor, and so I began to pay a little bit more attention to this growing research field. At around this time, the UCC probiotics team started a company, Alimentary Health Limited (AH), to exploit the growing demand for probiotics with demonstrable health benefits. For purposes of clarification I should say I had no involvement or conflict of interest with this very successful spin-off company other than watching with a mixture of envy and delight as it grew under the guidance of an old friend Barry Kiely. While our infection work and our bacteriocin group flourished, I was aware through departmental seminars, casual conversations and peer reviewed publications that AH were screening for probiotics, and were even conducting human clinical trials with strains to demonstrate efficacy in irritable bowel syndrome, a growing problem in developed societies.4,5 I am not the right person to chronicle the AH story, but it is interesting to note that based on the positive outcome of those trials, a product (Align®) based on a Bifidobacterium infantis strain isolated UCC has been successfully launched in the US in a capsule format.
The Alimentary Pharmabiotic Centre (APC)—What's in a Name?
In 2002 The Irish government, through Science Foundation Ireland (SFI), initiated a funding scheme in which they called for large collaborative groups to set up centers of excellence in partnership with industry. The ad hoc grouping of scientists working on probiotics at UCC and Moorepark, together with AH as an industrial partner, seemed ideally placed to take advantage of this scheme and to put their informal research collaborations on a firm strategic basis. Fergus Shanahan undertook to lead this attempt, and many hours were spent on brainstorming, troubleshooting and identifying potential scientific themes and principal investigators to lead the individual programs. I was asked to participate and was delighted to agree to act as a principal investigator in two of the six themes, bacteriocins and pathogenicity. These two themes were particularly relevant in that they allowed us to continue our work on bacteriocins (together with Paul Ross) on the basis that bacteriocins are potential probiotic factors and could well play a role in determining the composition of the microbiota and in preventing infection. The pathogenicity theme was interesting since many of the ideas and techniques that had been developed to study pathogens could be immediately applicable to probiotics (in a continuing collaboration with Cormac Gahan). Probiotics and pathogens face many similar challenges in the gut, though with a different health outcome. One topic of considerable debate was the name of our proposed center. Fergus always insisted that the APC is not a probiotic research center, but that we are more concerned with exploiting the potential health benefits of gut commensals, the ‘hidden’ organ of our human superorganism.6 This was not simply semantics, since there is a very significant distinction between probiotic and commensal, but it did also stem from a feeling that the term ‘probiotic’ had become more of a marketing tool than a useful scientific descriptor. In the end, we settled on the term ‘pharmabiotic,’ which we defined as bacterial cells of human origin, or their products, with a proven pharmacological role in health or disease.7 After an exhausting peer-review process, SFI informed us that our application was successful and the APC got off the ground in 2003 and currently (after a renewal process in 2008) has over 100 scientists working on pharmabiotic research in UCC and Moorepark.
Proof of Concept
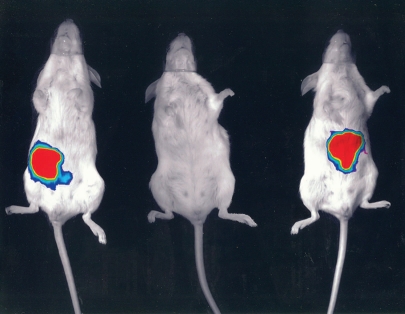
Along with our colleagues in the APC, we were determined to try to identify the precise molecular mechanism by which gut commensals could influence the health of the host, and particularly in our case to try to establish whether an introduced commensal could affect the outcome of a subsequent infection. We, like many others, were able to establish in a variety of models, ranging from human cell lines to mice to pigs and even cattle, that the introduction of a commensal organism like Lactobacillus or Bifidobacterium could affect disease outcome upon subsequent infection. Taking into account that we were measuring hard endpoints of morbidity and mortality, the evidence was irrefutable that the introduction of specific commensal organisms could reduce the impact of pathogenic species on the health of the host. Importantly, other related commensals, including members of the same genus and even species, had no such effect. So firsthand I was faced with hard evidence that lactobacilli and bifidobacteria could confer a significant health benefit, at least in the prevention of gastrointestinal infection in animal models. One example that has been crucial in convincing me of the benefits of bacterial interventions was conducted largely by Sinead Corr, a PhD student in the lab working with Cormac, myself and Paul O'Toole, another PI in the APC. Sinead established that feeding a mouse Lactobacillus salivarius UCC118 prior to infection with L. monocytogenes was crucial to the outcome of the infection.8 It appealed to the infection microbiologist in me that we had the hardest endpoint of all in these experiments—mice that did not receive strain UCC118 died of the Listeria infection (even if we fed them other lactobacilli or bifidobacteria), while the mice that received the commensal strain not only lived, they actually displayed no symptoms whatever. The outcome was both dramatic and strain specific. Serendipitously, strain UCC118 produces a bacteriocin that can kill Listeria, and so this was an obvious possible mechanistic basis for the effect. Without going through the actual experiments, Sinead (together with Yin Li and Pat Casey of the APC) were able to conclusively show that the protection afforded by UCC118 is entirely due to the production of the bacteriocin in the gut, and that a bacteriocin negative mutant no longer protected against infection (Fig. 1). In the face of this evidence I finally abandoned my last vestiges of my former scepticism and accepted that the introduction of a single well characterized commensal strain can have the most profound influence on the subsequent sensitivity of a host to infection and that the precise molecular mechanism can also be defined, at least in the model that I have described.
Figure 1.
A lux tagged Listeria monocytogenes emits light from the gastrointestinal tract 30 minutes after oral inoculation of a placebo fed mouse (left). No Listeria can be detected in a mouse that had received Lactobacillus salivarius UCC118 prior to infection (center), while a mouse fed a bacteriocin negative version of UCC118 is also infected (right).
Pharmabiotics
Determining the mechanistic basis by which commensal bacteria can influence health has been the objective of the APC since its inception, and our identification of bacteriocins as a potential tool in determining the outcome of infection led us to explore other possibilities of using bacteriocins as pharmabiotics in addition to using the producing cells. We were able to show that incorporating a powerful bacteriocin produced by strains of Lactococcus lactis into teat seals was effective in preventing mastitis in dairy cattle.9 Although this is obviously not a gut application (and Lactococcus is not normally considered to be a commensal organism), it did confirm that bacteriocins could work under physiological conditions and in real infections in large animals. We have also shown that introducing the live organism can also be used to treat ongoing natural mastitis infections in cattle, albeit probably by a mechanism involving activation of the immune system rather than solely by direct antagonism between bacteriocin and pathogen.10,11 Other work using commensal bacteria to prevent salmonellosis in a porcine model13 simply added to the growing amount of in-house data that commensal strains and their products can impact the outcome of infections. One significant concern is that the introduced bacterium or bacteriocin could have a detrimental effect on the existing host microbiota, much as is the case with standard antibiotic therapies. The advent of pyro sequencing as a tool to study the composition of the gut microbiota, particularly the fine work done in Jeff Gordon's laboratory in St. Louis among others,13 has allowed us to determine the impact of introducing bacteriocins and bacteriocin producers into model gut systems. While much of the work is unpublished, we are convinced that such interventions can have a significant and probably undesirable impact on the existing microbiota, and the targeting of pathogens with narrow spectrum bacteriocins may have significant advantages over standard therapies or broad host range bacteriocins.
Bioengineered Bugs
The mounting data that orally introducing commensals or their products can impact on host health begs some obvious questions; what is the best form of delivery and will we be able to improve the benefits through genetic engineering? For food manufacturers, the answer is probably easy at this time. Since food cannot be used to treat disease under existing regulations it is likely that only natural food grade isolates will be used in the foreseeable future in this sector. Having said that, Roy Sleator (a former post-doctoral scientist in our lab, now a PI in the APC and the editor of this journal) has shown that we can improve the robustness, gastric survival and colonization of commensal strains by introducing genetic determinants from pathogens that help commensals to overcome stresses encountered during processing and in the gut14,15—a strategy we have termed patho-biotechnology.16 For example, improving bile resistance of bifidobacteria leads to higher colonization levels in murine models.17 If specific health benefits can be associated with such bioengineering they may well outweigh any perceived reservations among consumers due to public resistance to genetically modified organisms in the food supply. But what of using bioengineered bugs in pharmaceutical applications? Commensal organisms offer significant possibilities as delivery platforms for vaccines, therapeutic agents and in the treatment of disease.16 An excellent example is that in which Lothar Steidler (a former PI in the APC) engineered strains of Lactococcus lactis to produce the anti-inflammatory cytokine IL10 in biologically significant amounts.18 Lothar has overseen trials using his particular bioengineered bug (genetically disabled to prevent growth outside of the body), and can be regarded as a pioneer in the therapeutic use of genetically modified LAB. Will we even see bioengineered pathogens elevated to ‘probiotic’ status if they can be attenuated and used as delivery vehicles for immune stimulation?19
Conclusions
The journey from skeptic to convert is rarely as a result of a ‘Road to Damascus’ conversion, but is more likely to be due to a dawning realization that previously held opinions are ill-founded. Such a conversion is most likely to result from paying attention to the opinions of people you hold in high regard, eliminating pre-conceived notions, and looking anew at the evidence. That has certainly been true in my case. I characterized myself earlier as a ‘reluctant’ convert to the concept of probiotics, and I hold to that view. It has always seemed to me that it is a disservice to the excellent research that has been conducted on individual strains to simply label whole genera as probiotics, and as a result I would prefer the term pharmabiotic to be reserved for those strains with proven health effects. We should also bear in mind that any agent that has a pharmacological effect that is beneficial for one particular target group may well be contra-indicated for some individuals with a different underlying pathophysiology. I have already mentioned that there may well be collateral damage on the existing microbiota following the introduction of antimicrobial peptides, as another example of potential adverse effects. In this regard comprehensive safety studies in large populations will be important for any strain which is to be marketed to the general population. Whatever the case for including commensal strains in our general diet, I propose that sufficient evidence has emerged to convince even the most jaded sceptic that our commensal passengers represent an arsenal of potential health benefits; whether in the form of living cells, cell components or bacterial products. In that respect I am a zealot rather than a reluctant convert, and I am convinced that many of the next generation of therapies for chronic and acute conditions will come from within our human superorganism.
Acknowledgements
I would like to acknowledge the many scientists, both named and unnamed in this article, who have influenced my thinking on this topic; the colleagues and friends with whom I have collaborated over the years; my colleagues at the APC and our funding agency Science Foundation Ireland. Finally, my thanks go to Dymphna, Cian, Grainne, Daragh and Ultan for their support.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/10796
References
- 1.Speck ML. Interactions among lactobacilli and man. J Dairy Sci. 1976;59:338–343. doi: 10.3168/jds.S0022-0302(76)84207-5. [DOI] [PubMed] [Google Scholar]
- 2.Tannock GW, Luchansky JB, Miller L, Connell H, Thode-Andersen S, Mercer AA, et al. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid. 1994;31:60–71. doi: 10.1006/plas.1994.1007. [DOI] [PubMed] [Google Scholar]
- 3.Hitt E. Biography of Todd R. Klaenhammer. Proc Natl Acad Sci USA. 2005;102:3903–3905. doi: 10.1073/pnas.0500351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, et al. Lactobacillus and Bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 5.Whorwell PJ, Altringer L, Morel J, Bond Y, Charbonneau D, O'Mahony L, et al. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol. 2006;101:1581–1590. doi: 10.1111/j.1572-0241.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 6.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shanahan F, Stanton C, Ross P, Hill C. Pharmabiotics, Bioactives from mining host-microbe-dietary interactions. Funct Food Rev. 2009;1:20–25. [Google Scholar]
- 8.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CGM. From the cover: Bacteriocin production as a mechanism for the anti-infective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crispie F, Twomey D, Flynn J, Hill C, Ross RP, Meaney W. The lantibiotic lacticin 3147 produced in a milk-based medium improves the efficacy of a bismuth-based teat seal in cattle deliberately infected with Staphylococcus aureus. J Dairy Res. 2005;72:159–167. doi: 10.1017/s0022029905000816. [DOI] [PubMed] [Google Scholar]
- 10.Klosterman K, Crispie F, Flynn J, Ross RP, Hill C, Meaney W. Intramammary infusion of a live culture of Lactococcus lactis for treatment of bovine mastitis: comparison to antibiotic treatment in field trials. J Dairy Res. 2008;75:365–373. doi: 10.1017/S0022029908003373. [DOI] [PubMed] [Google Scholar]
- 11.Crispie F, Alonso-Gomez M, O'Loughlin C, Klostermann K, Flynn J, Arkins S, et al. Intramammary infusion of a live culture for treatment of bovine mastitis: effect of live lactococci on the mammary immune response. J Dairy Res. 2008;75:374–384. doi: 10.1017/S0022029908003385. [DOI] [PubMed] [Google Scholar]
- 12.Casey PG, Gardiner GE, Casey G, Bradshaw B, Lawlor PG, Lynch PB, et al. A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella typhimurium. Appl Environ Microbiol. 2007;73:1858–1863. doi: 10.1128/AEM.01840-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic Analysis of the Human Distal Gut Microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheehan V, Sleator RD, Fitzgerald GF, Hill C. Heterologous expression of BetL, a betaine uptake system, enhances the stress tolerance of Lactobacillus salivarius. Appl Environ Microbiol. 2006;72:2170–2177. doi: 10.1128/AEM.72.3.2170-2177.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheehan VM, Sleator RD, Hill C, Fitzgerald GF. Improving gastric transit, gastrointestinal persistence and therapeutic efficacy of the probiotic strain Bifidobacterium breve UCC2003. Microbiology. 2007;153:3563–3571. doi: 10.1099/mic.0.2007/006510-0. [DOI] [PubMed] [Google Scholar]
- 16.Sleator RD, Hill C. Pathobiotechnology. Landes Bioscience. 2008 [Google Scholar]
- 17.Watson D, Sleator RD, Gahan CGM, Hill C. Enhancing bile tolerance improves survival and persistence of Bifidobacterium and Lactococcus in the murine gastrointestinal tract. BMC Microbiology. 2008;8:176. doi: 10.1186/1471-2180-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, et al. Treatment of Murine Colitis by Lactococcus lactis Secreting Interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 19.Jia Q, Lee BY, Clemens DL, Bowen RA, Horwitz MA. Recombinant attenuated Listeria monocytogenes vaccine expressing Francisella tularensis IglC induces protection in mice against aerosolized Type A F. tularensis. Vaccine. 2009;27:1216–1229. doi: 10.1016/j.vaccine.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]