Abstract
Distinct from most alginate-assimilating bacteria that secrete polysaccharide lyases extracellularly, a gram-negative bacterium, Sphingomonas sp. A1 (strain A1), can directly incorporate alginate into its cytoplasm, without degradation, through a “superchannel” consisting of a mouth-like pit on the cell surface, periplasmic binding proteins, and a cytoplasmic membrane-bound ATP-binding cassette transporter. Flagellin homologues function as cell surface alginate receptors essential for expressing the superchannel. Cytoplasmic alginate lyases with different substrate specificities and action modes degrade the polysaccharide to its constituent monosaccharides. The resultant monosaccharides, α-keto acids, are converted to a reduced form by NADPH-dependent reductase, and are finally metabolized in the TCA cycle. Transplantation of the strain A1 superchannel to xenobiotic-degrading sphingomonads enhances bioremediation through the propagation of bacteria with an elevated transport activity. Furthermore, strain A1 cells transformed with Zymomonas mobilis genes for pyruvate decarboxylase and alcohol dehydrogenase II produce considerable amounts of biofuel ethanol from alginate when grown statically.
Key words: sphingomonas, alginate, pit, flagellin, ABC transporter, polysaccharide lyase, bioremediation, biofuel
Introduction
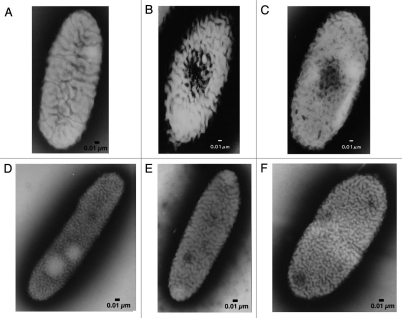
Bacteria belonging to the genus Sphingomonas are gram- negative, yellow-pigmented, aerobic rods.1 Unlike other gram-negative bacteria, sphingomonads contain glycosphingolipids (GSL) with long-chain base dihydrosphingosine in the outer membrane, but have no lipopolysaccharides (LPS).2 GSL confer the property of high hydrophobicity on the cell surface of sphingomonads. In view of these unusual cell surface characteristics, the genus Sphingomonas was reclassified from pseudomonades in 1990 as a new member of the α-4 subgroup of the Proteobacteria.3 More recently, the genus Sphingomonas has been further regrouped into five related genera, namely, Sphingomonas sensu strictu, Sphingobium, Novosphingobium, Sphingopyxis and Sphingosinicella.4,5 Some sphingomonads are peculiar due to (1) the presence of many large pleat-like structures on the cell surface (Fig. 1A and D–F);6,7 (2) the extraordinary metabolic ability to degrade various refractory environmental pollutants, notably xenobiotics such as dioxin, biphenyl and bisphenol;8–11 and (3) the production of useful biopolymers such as gellan and related polysaccharides.12,13 Sphingomonas sp. A1 (strain A1) was isolated from soil as an alginate-assimilating bacterium expressing intracellular alginate lyases.6 Strain A1 also contains GSL in the outer membrane and pleat structures on the cell surface (Fig. 1A).
Figure 1.
Electron micrographs of sphingomonads. (A) Strain A1 cell grown in the absence of alginate. (B) Strain A1 cell grown on alginate. (C) p6 gene-disruptant cell grown on alginate. (D) Sphingomonas mali cell. (E) Sphingomonas paucimobilis cell. (F) Sphingomonas parapaucimobilis cell.
Alginate is a linear polysaccharide produced by brown seaweeds and certain bacteria. The polymer consists of two monosaccharides, β-d-mannuronate (M) and its C5 epimer α-l-guluronate (G). These two monosaccharides are arranged in three different configurations (Fig. 2): polyM, polyG, and heteropolymeric random sequences (polyMG).14 Brown seaweeds, particularly gulfweed (genus Sargassum), produce alginate as a major component of their cell walls. Gulfweed is cultivable and alginate is easily purified from this seaweed. Consequently, alginate is expected to become a resource for biofuel production due to its abundance in marine biomass.15 Since brown seaweed alginate chelates metal ions and thereby forms a highly viscous solution, this polysaccharide is widely used in the food, chemical and pharmaceutical industries as a reagent for stabilization, thickening and emulsification.16 In the case of the bacterial alginate, an opportunistic pathogen, Pseudomonas aeruginosa, produces alginate-containing extracellular biofilms that are involved in the expression of virulence factors during lung infections in cystic fibrosis patients.17 Since these biofilms often protect P. aeruginosa cells from antibiotics and phagocytic cells, bacterial biofilm-dependent infections are particularly difficult to treat.18
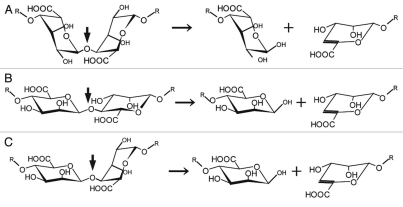
Figure 2.
Structure of alginate molecules and scheme of the alginate lyase reaction. (A) Disaccharide in polyG. (B) Disaccharide in polyM. (C) Disaccharide in polyMG. Left, disaccharide in each polymer; right, products of the alginate lyase reactions. Thick and thin arrows indicate the sites cleaved by alginate lyases and progression of the enzyme reactions, respectively.
Alginate lyases are therefore promising as degrading enzymes for the processing of edible seaweed alginate and the removal of bacterial biofilm alginate. These enzymes are produced by alginate- assimilating microbes isolated from the soil, sea or waste water, and also from some marine algae and mollusks.19 Polysaccharide lyases, including alginate lyases, commonly recognize uronic acid residues in polysaccharides, catalyze the β-elimination reaction, and produce unsaturated saccharides with C=C double bonds at C4 and C5 sites in nonreducing terminal uronic acid residues. Since alginate is a heteropolysaccharide consisting of M and G, alginate lyases are often categorized from the viewpoint of their reaction mode, either endo or exo, and substrate specificity, i.e., a preference for polyM, polyG and/or polyMG (Fig. 2).
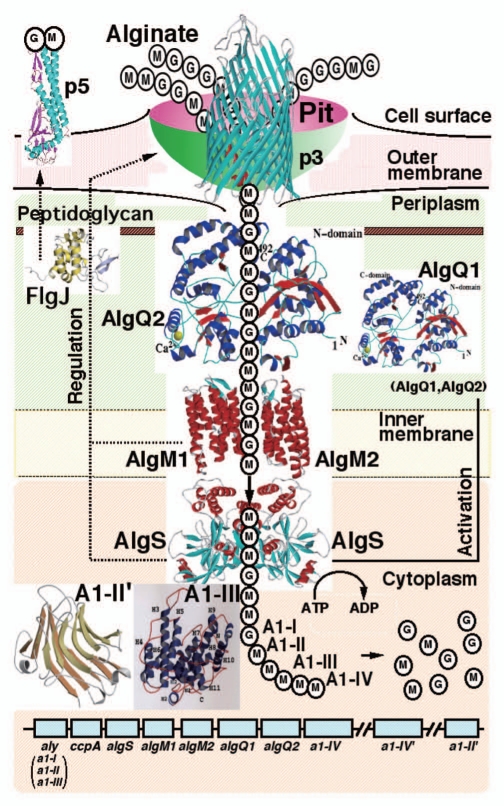
Most alginate-assimilating bacteria secrete degrading enzymes, i.e., alginate lyases, into the extracellular fraction or periplasm, and incorporate the resultant alginate oligosaccharides across the cytoplasmic membrane. In contrast, strain A1 has an unexplored specific and elegant system, the “superchannel,” on the cell surface for alginate import without degradation (Fig. 3).20 Subsequent to alginate import by the superchannel, four cytoplasmic alginate lyases with different substrate specificities and action modes degrade alginate to its constituent monosaccharides. These lyases therefore constitute a degradation system in the cytoplasm for alginate heteropolysaccharide metabolism.
Figure 3.
Strain A1 superchannel for alginate import and degradation. G, L-guluronate; M, D-mannuronate; aly, gene for alginate lyases (A1-I, A1-II and A1-III); ccpA, catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II', alginate lyase A1-II' gene; a1-IV', alginate lyase A1-IV' gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Details of each protein function are described in the text.
This article deals with the structural and functional aspects of the strain A1 supersystem for alginate import and metabolism. Application of the bacterial supersystem in bioremediation and biofuel production is also discussed.
Pit-Forming Bacterium
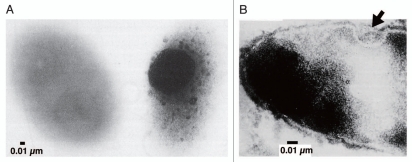
As is often observed in sphingomonads, strain A1 has pleat structures on the cell surface (Fig. 1A).6 However, distinct from other sphingomonads, strain A1 cells when grown on alginate form a mouth-like pit on the cell surface through the reorganization and/or fluidity of the pleat structures. The size of the pit can reach approximately 0.1 µm in diameter (Fig. 1B).7 Conversely, no pit is formed by strain A1 cells grown in alginate-free medium. Thus, pit formation is dependent on the presence of alginate in the external milieu. The size and regulation of pit formation are similar to those of certain eukaryotic organelle. To the best of our knowledge, this is the first discovery of a pit-forming bacterium in the history of microbiology. Thin sections of strain A1 cells grown on alginate show specific sites where the cell membrane sinks into the cytoplasm (Fig. 4A). Electron microscopy of strain A1 cells treated with alginate-staining dye has revealed that alginate is concentrated in these pits, suggesting that the pit functions as a funnel or concentrator of alginate (Fig. 4B). Strain A1 cells produce alginate lyases exclusively in the cytoplasm, indicating that a direct import system for macromolecule alginate must be installed in the cell envelope, spanning the outer and inner membranes and the periplasm. In addition to alginate, strain A1 cells can assimilate pectin through the action of cytoplasmic pectin-degrading enzymes. Pectin is a polyuronate consisting of galacturonic acid, which is similar to alginate. This suggests that strain A1 cells have become somewhat specialized to incorporate polysaccharides such as alginate and pectin during their growth and have developed a direct import system to transport these molecules.
Figure 4.
Pit function. (A) Alginate detection on the cell surface by mucopolysaccharide staining (left, strain A1 cell grown in the absence of alginate; right, strain A1 cell grown on alginate). (B) Thin section of strain A1 cell grown on alginate (arrow indicates the position corresponding to the pit).
Genomics, Transcriptomics and Proteomics
Strain A1 cells contain a chromosome as a circular double-stranded DNA consisting of 4,622,788 base pairs.21 The genome encodes approximately 4,800 genes and the GC content is relatively high (62%) among gram-negative bacteria. Thirty percent of the total genes of strain A1 show a significant identity with genes encoded in the genome of P. aeruginosa,22 suggesting that strain A1 has evolved from pseudomonads. More than half of the total genes (58%) are categorized in the class of function-unknown genes. Molecular characterization of function-unknown genes will provide useful information on the molecules constituting the supersystem responsible for alginate import and metabolism. A large number of genes for polyuronate (alginate and pectin)-degrading enzymes are encoded in the genome of strain A1, and certain ATP-binding cassette (ABC) transporter genes are found in the vicinity of these enzyme genes. This indicates that strain A1 cells have a gene cluster for polyuronate import and degradation.
Independent of the chromosome, strain A1 cells also harbor the pA1 plasmid.23 The pA1 plasmid consists of 46,557 base pairs coding for 49 genes with a GC content of 65%. Homology analysis indicates that pA1 is significantly similar to the self-transmissible promiscuous incompatibility (Inc) group P-1β plasmid. However, pA1 is peculiar among IncP-1 plasmids in that it contains no inserted mobile genetic elements.
Gene expression analysis using DNA microarrays (unpublished data) has demonstrated that genes encoding the supersystem for alginate import and metabolism, i.e., cell surface alginate-binding proteins, outer membrane-bound transporters, periplasmic alginate-binding proteins, inner membrane-bound ABC transporter, and cytoplasmic alginate lyases and α-keto acid-metabolizing enzymes, are significantly upregulated (3- to 70-fold) in strain A1 cells grown on alginate in comparison with those grown on alginate-free medium. This is convincing evidence that the supersystem in strain A1 cells is specialized for alginate import and metabolism.
Two-dimensional polyacrylamide gel electrophoresis-dependent proteome analysis has indicated that eight proteins (p1–p8) are inducibly expressed in the outer membrane of strain A1 cells grown on alginate.24,25 Each of these proteins is described in detail below.
Superchannel for Alginate Import
Cell surface receptors.
Two flagellin homologues, p5 (40 kDa) and p6 (31 kDa), are inducibly expressed on the cell surface of strain A1.25 This is surprising given that strain A1 exhibits neither cell motility nor flagella formation. Bacterial flagellin is well characterized as a component of the helical filament of the bacterial flagellum. A p6 gene disruptant constructed by insertion of a kanamycin-resistance cassette in the p6 gene shows significant growth retardation in a medium containing alginate as the sole carbon source. The p5 and p6 flagellin homologues might be essential for cell viability because the simultaneous disruption of both genes results in growth failure. There are significant differences in cell surface structures, i.e., pit formation and pleat shape, between the p6 gene disruptant and the wild-type of strain A1. In the p6 gene disruptant, pit formation is incomplete and the cell surface changes from a pleated structure to a network structure (Fig. 1C). This clearly indicates that the flagellin homologues regulate cell surface structures in strain A1. In order to gain a better understanding of the structure and function of strain A1 flagellin homologues, we purified and characterized these proteins following their overexpression in E. coli cells.25
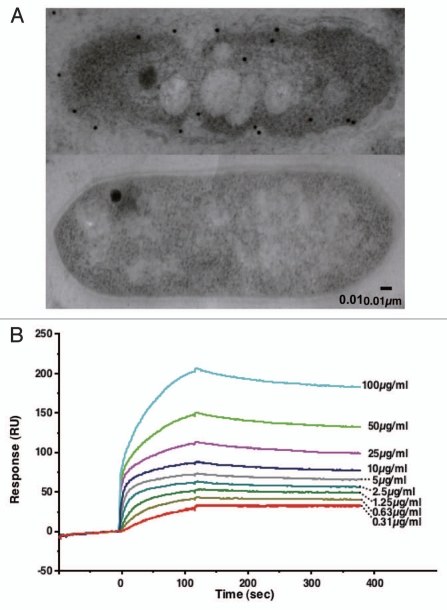
The flagellin homologue p5 was found to be inducibly expressed in alginate-grown strain A1 cells and exclusively localized in the cell envelope by immunoelectron microscopy using anti-p5 antibody (Fig. 5A upper). In strain A1 cells grown in alginate-free medium there is hardly any expression of flagellin homologues on the cell surface (Fig. 5A lower). To clarify the intrinsic function of flagellin homologues as cell surface proteins, the interaction between p5 and alginate was analyzed using a surface plasmon resonance (SPR) biosensor (Fig. 5B). Flagellin homologues exhibit a potent alginate-binding ability, and their affinity is specific for alginate. Two binding sites for alginate were found to be located in flagellin homologues based on the interaction profile. In the case of p5 binding to alginate, the dissociation constants (Kd) at the two sites were determined to be 1.3 × 10−7 M and 2.6 × 10−9 M. This high alginate-binding ability of p5 strongly suggests that strain A1 flagellin homologues function as cell surface receptors for alginate similar to the mammalian CD44 transmembrane receptor for hyaluronan.26
Figure 5.
Flagellin homologue as a cell surface alginate receptor. (A) Localization of flagellin homologue p5 in strain A1 cells by immunogold electron microscopy (upper, strain A1 cell grown on alginate; lower, strain A1 cell grown in the absence of alginate). (B) Interaction of p5 and alginate using an SPR biosensor. p5 purified from E. coli cells was immobilized on a sensor chip. The binding of p5 to alginate is dependent on alginate concentration (0.31–100 µg/ml).
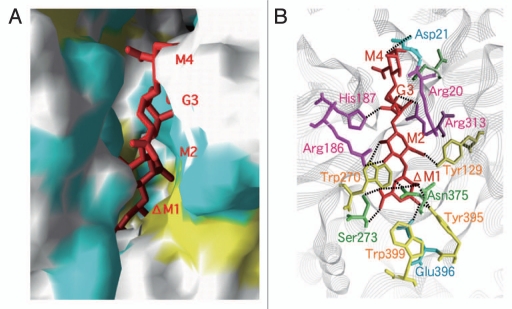
The crystal structure of a truncated p5 (p5ΔN53C45) was determined at 2.0 Å resolution to clarify its peculiar characteristics (alginate binding and cell surface localization) (Fig. 3).27 p5ΔN53C45 denotes a p5 mutant lacking the N-terminal 53 residues and the C-terminal 45 residues. Bacterial flagellin molecules are divided to three regions based on the sequence alignment. Two regions (flagellin_N and flagellin_C motifs) at the N- and C-termini, respectively, are well conserved in bacterial flagellins, whereas there is some variation in the central domain (flagellin_IN motif). Thus, bacterial flagellins are often grouped into two categories, i.e., flagellin_IN motif-free and flagellin_IN motif-including flagellins. p5ΔN53C45 was the first flagellin_IN motif-including flagellin to be structurally determined, although the crystal structure of the flagellin_IN motif-free flagellin from Salmonella typhimurium had previously been determined.28 p5ΔN53C45 consists of two structural elements, an α-domain rich in α-helices and a β-domain rich in β-strands. The α-domain includes the N- and C-terminal regions, whereas the β-domain consists of the central region. The α-domain resembles the D1 domain of the S. typhimurium flagellin, whereas the β-domain is similar to the finger domain of the bacteriophage T4 baseplate protein.29 Through deletion mutant analysis, the N-terminal residues 20–40 and C-terminal residues 353–363 were found to be responsible for alginate binding, suggesting that the edge of the α-domain constitutes the alginate-binding site (Fig. 3).
Although strain A1 cells form no flagellum, flagellum-related genes, such as flg and fli clusters, responsible for the formation of flagellar basal body and hook,30 occur in the genome of strain A1. This suggests that strain A1 flagellin homologues are transported to the cell surface by the flagellar basal body-dependent type III secretion system. Indeed, a flgJ-disruptant appears to inhibit the expression of flagellin homologues on the cell surface. FlgJ is involved in the rod assembly of the flagellar basal body and hydrolysis of peptidoglycan.31 In order to obtain clues regarding the peculiar localization of flagellin homologues, the three-dimensional structure of FlgJ-C, the peptidoglycan hydrolase domain, was determined at a 1.74 Å resolution by X-ray crystallography (Fig. 3).32 The enzyme was found to consist of two lobes, α and β. A deep cleft located between the two lobes can accommodate polymer molecules, suggesting that the active site is located in the cleft. The involvement of FlgJ in the secretion of flagellin homologues is now being structurally characterized.
Cell surface alginate-binding proteins.
The p7 cell surface protein (28 kDa) is inducibly expressed in strain A1 cells grown on alginate.25 To clarify the function of p7, the protein was expressed in E. coli cells, purified, and characterized.33 SPR biosensor analysis revealed that p7 binds to alginate most efficiently at neutral pH with a Kd of 3.6 × 10−8 M. p7 shares a significant sequence identity with bacterial lipoproteins. A four-amino acid sequence termed the “lipobox” [LVI][ASTVI][GAS]C is well conserved at the N-terminus of bacterial lipoproteins.34 However, no lipobox is observed in p7. Thus, p7 was purified from strain A1 cells through anti-p7 antibody-bound column chromatography, and subjected to N-terminal sequence analysis and mass spectrometry. As a result, p7 was found to contain no lipid moiety. Similar to the case of p7, a large number of function-unknown proteins are classified as lipoproteins based on sequence similarity. Accordingly, lipoproteins should be assigned carefully on the basis of not only primary structure comparison but also lipoprotein-specific program analysis such as LipoP35 and DOLOP.34
p8 (20 kDa) is a predominant protein in the outer membrane of strain A1 cells grown on alginate.24 The protein shares a significant sequence identity with the polyhydroxyalkanoate granule-associated protein of Ralstonia eutropha.36 The p8 gene disruptant exhibits growth retardation in minimal medium containing alginate as the sole carbon source. p8 binds to alginate most efficiently at pH 4.0 with a Kd of 1.3 × 10−7 M. In addition to soluble alginate, p8 shows an affinity for insoluble granules consisting of alginate and metal ions. The alginate-binding ability of p7 and p8 is not much higher than that of cell surface alginate receptors, i.e., flagellin homologues. This suggests that both p7 and p8 can associate with and release the polymer.
The above results indicate that p7 and p8 function as cell surface alginate-binding proteins and that their combined action functions to concentrate alginate in the pits on the strain A1 cell surface.
Outer membrane-bound transporters.
Strain A1 cells grown on alginate inducibly produce four proteins, p1 (78 kDa), p2 (71 kDa), p3 (74 kDa) and p4 (72 kDa), in the outer membrane.25 Homology analysis has revealed that all four proteins are similar to the TonB-dependent outer membrane transporter. Gene disruptants of p1, p2, p3 and p4 exhibit significant growth retardation in alginate-containing medium, suggesting that the outer membrane transporter homologues are involved in alginate assimilation. The TonB-dependent outer membrane transporters characterized thus far incorporate iron-bound siderophores, such as ferrichrome, enterobactin, enterochelin and citrate into the periplasm.37–40 The energy for import by the transporters is provided by a proton motive force generated from an inner membrane complex (TonB-ExbB-ExbD).41 Since alginate is well characterized as a potential iron chelator,42 p1, p2, p3 and/or p4 may function as an outer membrane-bound transporter for import of an iron/alginate complex into the periplasm. In this case, alginate is utilized as a siderophore-like substance. Using homology modeling, p3 has been shown to constitute a tunnel-like β-barrel structure spanning the outer membrane (Fig. 3). After being concentrated by p8 in the pit on the cell surface, the iron/alginate complex is probably transported across the outer membrane to the periplasm by the TonB-dependent transporters, p1, p2, p3 and/or p4.
Periplasmic alginate-binding proteins.
Two proteins, AlgQ1 (59 kDa) and AlgQ2 (59 kDa), are inducibly expressed in the periplasm of strain A1 cells grown on alginate.43 AlgQ1 and AlgQ2 bind to alginate with a Kd of 2.3 × 10−7 M and 1.5 × 10−7 M, respectively, and their binding ability is specific for alginate.43 On the basis of the affinity levels, both proteins, similar to p7 and p8, are assumed to be capable of binding and releasing alginate. This indicates that these periplasmic alginate-binding proteins function as mediators of alginate from the outer membrane (TonB-dependent transporter) to the inner membrane (ABC transporter) (Fig. 3). Genes coding for AlgQ1 and AlgQ2 constitute an alginate import/degradation system clustering in the genome of strain A1 (Fig. 3, bottom).
Both AlgQ1 and AlgQ2 were found to consist of N- and C-domains with an α/β structure by X-ray crystallography (Fig. 3, bottom).43,44 Three linker loops connect the two domains, and alginate is bound to the cleft located between the domains. A drastic conformational change occurs in AlgQ1 and AlgQ2 through alginate binding and release.43,45 The ligand-free proteins exhibit an opened form, whereas the alginate-bound proteins are converted to a closed form by rotation (30°) and translation (0.5 Å) of both domains. This conversion is achieved by the movement of Glu396 followed by exclusion of a water molecule from the binding site.
A hexasaccharide and a longer substrate are predicted to be bound to the active clefts of AlgQ1 and AlgQ2 by structural simulation. Indeed, the clefts in AlgQ1 and AlgQ2 are larger than those in other substrate-binding proteins, such as maltose- binding proteins,43,46 indicating that they enable AlgQ1 and AlgQ2 to accommodate macromolecules. A large number of both positively-charged and aromatic residues are arranged in the clefts of AlgQ1 and AlgQ2 (Fig. 6A).43,45 Positively-charged residues are considered to be crucial for binding to acidic alginate through an electric charge interaction. Stacking interaction is also observed between aromatic and pyranose rings in the proteins and alginate, respectively (Fig. 6B). The nonreducing terminal saccharide of alginate is tightly bound to AlgQ1 and AlgQ2, indicating that the alginate-binding proteins specifically recognize and bind to the substrate nonreducing terminal saccharide, and thereafter deliver the polymer to the inner membrane-bound ABC transporter.
Figure 6.
Structure of the cleft in the alginate-binding protein AlgQ2. (A) Molecular surface in the binding cleft (yellow, aromatic residues; cyan, positively-charged residues). (B) Alginate-binding site. Red sticks indicate unsaturated alginate tetrasaccharide.
Inner membrane-bound ABC transporter.
A typical ABC transporter in gram-negative bacteria consists of four subunits, i.e., two transmembrane subunits and two ATP-binding proteins, and this accompanied by a periplasmic substrate-binding protein.47 Genes coding for ATP-binding proteins, trans membrane domains, and the associated periplasmic binding protein are organized in an operon structure in the bacterial genome. In the case of alginate import by strain A1 cells, five genes for the ATP-binding proteins (AlgS/AlgS), transmembrane domains (AlgM1 and AlgM2), and two alginate-binding proteins (AlgQ1 and AlgQ2) are assembled in the genome to constitute the operon (Fig. 3 and bottom). Evidence for this organization comes from DNA microarray analysis, which shows that the expression level of each gene in alginate-grown strain A1 cells is mutually comparable.
The strain A1 ABC transporter for alginate is composed of a homodimer of AlgS (40 kDa) as an ATPase and a heterodimer of AlgM1 (37 kDa) and AlgM2 (33 kDa) as a permease (Fig. 3).20 AlgS includes P-loop/Walker and ABC-signature motifs responsible for ATPase activity. AlgS has been shown to contain an α/β structure by X-ray crystallographic analysis.48 The three-dimensional structure of the alginate ABC transporter has been constructed through homology modeling and docking simulation (Fig. 3). Both AlgM1 and AlgM2 include six transmembrane α-helices. A pore formed at the interface between AlgM1 and AlgM2 is considered to function as a passage for alginate.
The recombinant AlgS expressed in E. coli cells adopts a homodimer form and exhibits an ATPase activity; its activity is unchanged in the presence or absence of alginate-bound AlgQ1 and AlgQ2. The ATPase activity in the inner membrane isolated from alginate-grown strain A1 cells is substantially increased in the presence of AlgQ1 or AlgQ2 in complex with alginate,43 suggesting that the structural change induced in the periplasmic binding proteins through alginate binding contributes to the activation of AlgS via mediation by transmembrane domains AlgM1 and AlgM2. As demonstrated for the E. coli maltose import system,49 alginate-bound AlgQ1 or AlgQ2 functions as a trigger for the transport cycle. Once AlgS binds to ATP, the N- and C-domains in the alginate-bound AlgQ1/AlgQ2 open widely followed by release of alginate, and the periplasmic entrance is inducibly activated by the transmembrane domains (AlgM1 and AlgM2). Subsequently, alginate is incorporated into the cytoplasm using the energy generated through ATP hydrolysis by AlgS.
The superchannel, the elegant system evolved in strain A1 cells for alginate import, consists of essentially three sections, i.e., a pit on the cell surface, periplasmic alginate-binding proteins, and the inner membrane-bound ABC transporter (Fig. 3). The cell surface proteins involved in pit formation and function are as follows. Flagellin homologues function as cell surface alginate receptors essential for expressing the superchannel-associated pit. External alginate molecules are concentrated in the pit by cell surface alginate-binding proteins and are subsequently transported to the periplasm by TonB-dependent transporters localized in the outer membrane.
Cytoplasmic System for Alginate Metabolism
Endotype alginate lyases.
Since strain A1 cells can grow on alginate as a sole carbon source, the heteropolysaccharide incorporated in the cytoplasm must be degraded to its constituent monosaccharides by endo- and exotype alginate lyases with different substrate specificities. Three endotype alginate lyases, A1-I (65 kDa), A1-II (25 kDa) and A1-III (40 kDa), are produced in the cytoplasm of strain A1 (Fig. 3).50 A single gene (aly) located in the strain A1 gene cluster for alginate import and degradation encodes these three lyases (Fig. 3, bottom).51 The maturation route for alginate lyases A1-I, A1-II and A1-III is as follows. Po (71 kDa) is first synthesized as a precursor of the gene product, although this precursor exhibits no alginate lyase activity. After cleaving an N-terminal peptide (6 kDa), Po is converted to alginate lyase A1-I. A1-I, which includes N-terminal A1-III and C-terminal A1-II in its molecule, is autocatalytically processed to generate A1-II and A1-III specific for polyG and polyM, respectively. A1-I, a combined form of A1-II and A1-III, exhibits an affinity for both of polyG and polyM. A1-I, A1-II and A1-III all release unsaturated oligosaccharides from alginate, indicating that these lyases exhibit endotype activity.
Polysaccharide lyases are categorized into 21 families (PL-1-21) in the carbohydrate-active enzyme (CAZy) database based on their primary structure.52 Alginate lyases are isolated from various organisms, such as viruses, bacteria, fungi, and marine mollusks,19 and are categorized into the following six families: PL-5, -7, -14, -15, -17 and -18. Among these, most of the bacterial alginate lyases characterized thus far are grouped into two families, PL-5 and -7. Family PL-5 and -7 alginate lyases are known to be specific for polyM and polyG, respectively. In the case of strain A1 endotype alginate lyases, A1-III and A1-II are classified into families PL-5 and -7, respectively, and thus A1-I constitutes a new family, PL-5 + 7. Genes coding for both family PL-5 and -7 alginate lyases are included in the genome of some bacteria such as P. aeruginosa,22 Pseudomonas syringae pv. tomato,53 and Azobacter vinelandii;54 however, these genes are located separately in the respective bacterial genomes. The presence of the peculiar A1-I gene, i.e., a fused gene for family PL 5 + 7 enzymes, suggests that A1-I is an ancestor of alginate lyases, and that subsequently the A1-I gene has divided to family PL-5 and -7 genes through duplication, modification, and translocation. The genome sequence of strain A1 provides clues to the molecular diversity and evolution of alginate lyases as follows.55 Other than the genes coding for A1-I, A1-II and A1-III, a novel alginate lyase (A-II', 31 kDa) gene is included in the genome of strain A1, although its location in the strain A1 genome is far from the gene cluster for alginate import and degradation. A1-II' shares a significant sequence identity with A1-II, indicating that A1-II' is a member of family PL-7 based on its primary structure. Since no A1-II' is expressed even in alginate-grown strain A1 cells, the recombinant A1-II' expressed in E. coli cells was characterized. Despite the sequence identity, there is a significant difference in substrate specificity between A1-II and A1-II'. A1-II' exhibits a broad substrate specificity (polyM, polyG and polyMG), whereas A1-II is specific for polyG. A1-I is located in an intermediate position between families PL-5 and -7 in the phylogenetic tree, suggesting that the A1-II' gene was derived from the A1-II gene through separation from the A1-I gene, and subsequent duplication, modification, and translocation.
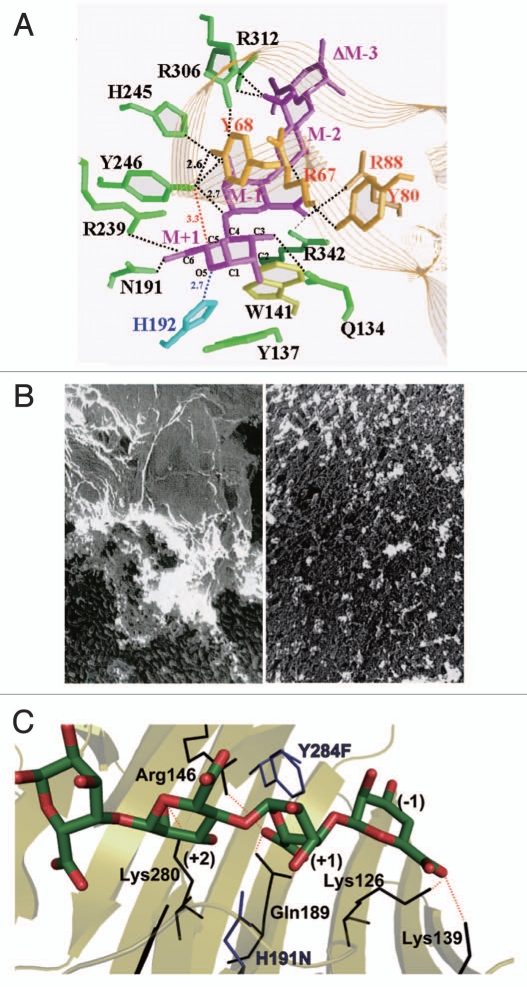
Alginate lyases are considered to be promising biochemicals for the processing of edible seaweed alginate and the removal of bacterial biofilm alginate. However, prior to their application in food and medical areas, the structure and function of these enzymes must be extensively studied in order to ensure safe utilization. Family PL-5 alginate lyase A1-III was found by X-ray crystallography to consist of 12 α-helices constituting an α6/α5-barrel structure as a basic scaffold (Fig. 3).56 A tunnel-like cleft is included in the enzyme molecule and a lid loop consisting of residues 57–90 is located over the cleft. A drastic conformational change occurs in A1-III through substrate binding. That is, there is a significant difference in the lid loop structure between substrate-free and bound A1-III. The loop is situated approximately 16.8 Å over the glycosidic linkage to be cleaved in the ligand-free form, whereas this distance is changed to 8.6 Å through substrate binding.57,58 Accordingly, the ligand-free form has an open conformation, whereas the substrate-bound form has a closed conformation. The lid loop movement induces the activation of the catalytic center Tyr246 through the formation of a Tyr68–Tyr246 pair, binding of substrates, and release of products. Reactions catalyzed by polysaccharide lyases are divided to three steps:59 first, neutralization or removal of the negative charge on the C6 carboxylate anion by residue A; second, the abstraction of the C5 proton at subsite +1 by a general base (residue B); and third, proton donation to the O4 atom at subsite +1 by a general acid (residue C). In the case of A1-III reactions, His192 was found to function as residue A, and Tyr246 as both residues B and C by structural determination of the alginate tetrasaccharide-bound A1-III (Fig. 7A). The catalysis by a single Tyr as an active residue is considered to be a novel β-elimination reaction.58,60
Figure 7.
Structure and function of alginate lyases. (A) Active site in A1-III complexed with unsaturated alginate tetrasaccharide. (B) Removal of alginate biofilm by A1-III (left, P. aeruginosa cells covered with biofilm; right, P. aeruginosa cells exposed after treatment with A1-III). (C) Active site in A1-II' complexed with unsaturated alginate tetrasaccharide.
The pathogenic bacterium P. aeruginosa produces a capsule- like biofilm that contains alginate.61 A firm barrier of the biofilm covers the bacterial cells (Fig. 7B, left), protecting them from attack by antimicrobial agents and phagocytes. Consequently, biofilm-dependent bacterial infections are considered difficult to treat. Removal of the bacterial biofilm by degrading enzymes thus constitutes a novel therapy for biofilm-dependent bacterial infectious diseases. Since A1-III shows a potent activity against the bacterial alginate (Fig. 7B, right), this enzyme is considered to be a promising candidate for the treatment of such diseases.62
The three-dimensional structure of family PL-7 alginate lyase A1-II' was determined at 1.0 Å resolution by X-ray crystallography.63 The enzyme has a β-sandwich structure consisting of two β-sheets (Fig. 3). An active site is situated in the glove-like cleft covered with two flexible loops (residues 133–145 and 193–203).63 The movement of these loops is crucial for substrate accommodation at the active site.64 Structural determinants in A1-II' for a broad substrate specificity were identified through structural comparison between M- and G-trisaccharide-bound A1-II' molecules. The substrate's carboxyl groups are recognized by specific residues, whereas the substrate's hydroxyl groups are appropriately bound to residues in a nonspecific manner. The crystal structure of A1-II' in complex with alginate tetrasaccharide indicated that the catalytic reaction mechanism of the enzyme occurs as follows (Fig. 7C): Gln189 as residue A neutralizes the negative charge of the carboxyl group; His191 as residue B abstracts the C5 proton; and Tyr284 as residue C donates the proton to the glycosidic bond to be cleaved.
Exotype alginate lyases.
Alginate incorporated in the cytoplasm is depolymerized by A1-I, A1-II and A1-III to unsaturated di, tri and tetrasaccahhrides with different M/G ratios as final products.50 The resultant alginate oligosaccharides are degraded to the constituent monosaccharides by alginate lyase A1-IV (86 kDa).65 In contrast to endotype A1-I, A1-II and A1-III, A1-IV exhibits an exotype activity, i.e., it releases only monosaccharide from alginate oligosaccharides. In addition to alginate oligosaccharides, alginate in a polymeric form is also degraded to its constituent monosaccharides by A1-IV. A1-IV is peculiar in that the enzyme acts on alginate poly- and oligosaccharides in the saturated form, as well as on unsaturated oligosaccharides, indicating that A1-IV can degrade both saturated and unsaturated alginate poly- and oligosaccharides with various M/G ratios.66 Due to instability, the pyranose ring of the resultant monosaccharides opens nonenzymatically to generate an α-keto acid (4-deoxy-l-erythro-5-hexoseulose uronic acid). The gene coding for A1-IV is located downstream from the AlgQ2 gene for periplasmic alginate-binding protein, indicating that the A1-IV gene is included in the gene cluster for alginate import and degradation (Fig. 3, and bottom).20,51,65
The increasing number of bacterial genome sequence analyses indicates that putative genes similar to the A1-IV gene occur in the genomes of some bacteria. The strain A1 genome also encodes a A1-IV homologue, A1-IV′ (90 kDa). Although A1-IV' catalyzes the cleavage of glycosidic bonds in alginate molecules through the β-elimination reaction, unsaturated di- and trisaccharides are released as the main products in the enzyme reaction. This implies that A1-IV′ is an endotype alginate lyase.67 On the other hand, another A1-IV homologue, Atu3025 (88 kDa), from Agrobacterium tumefaciens has been identified as an exotype alginate lyase.68 Accumulated sequence information indicates that A1-IV and other A1-IV homologues constitute a novel polysaccharide lyase family, PL-15, and points to the evolutionary route of alginate lyases in the PL-15 family.67 Similar to A1-II′, no A1-IV′ is expressed in strain A1 cells grown on alginate, suggesting that the A1-IV′ gene is derived from the A1-IV gene and that A1-IV is the original member of the PL-15 family.
As described above, in addition to two silent genes for A1-II′ and A1-IV′, four genes encoding three endotype alginate lyases and one exotype alginate lyase are included in the genome of strain A1. These alginate lyases are categorized into families PL-5, -7, -5 + 7 and -15, which contain almost all the bacterial alginate lyases. On the basis of this gene profile, it can be postulated that strain A1 is the original producer of alginate lyases.
α-keto acid-metabolizing enzymes.
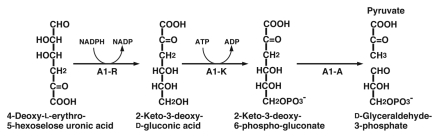
Monosaccharides generated from alginate through the reactions of endo- and exotype alginate lyases are nonenzymatically converted to an α-keto acid (4-deoxy-l-erythro-5-hexoseulose uronic acid). Although the metabolic pathway for α-keto acid derived from alginate has been postulated in P. aeruginosa,69 the genes coding for the enzymes involved in the pathway remain to be clarified. In strain A1 cells grown on alginate, NADPH-dependent reductase (A1-R, 26 kDa) was found to act on α-keto acid derived from the polysaccharide.70 The enzyme is specific for α-keto acid derived from alginate and converts 4-deoxy-l-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-d-gluconic acid (KDG) (Fig. 8). Since a large number of bacteria and yeasts show no growth in nutrient-rich media containing α-keto acid, this compound is considered to be highly toxic to both of prokaryotes and eukaryotes. For example, no growth of E. coli cells is observed in LB medium in the presence of α-keto acid, whereas the cells show a significant growth in medium containing KDG produced from α-keto acid by the action of A1-R. This clearly indicates that A1-R is crucial in strain A1 cells for metabolizing alginate and detoxifying α-keto acid.
Figure 8.
The proposed pathway for α-keto acid metabolism in strain A1 cells.
Molecular cloning of the A1-R gene has indicated that the enzyme is a member of the short-chain dehydrogenase/reductase (SDR) family.71 Although the enzyme gene is located far from the gene cluster for alginate import and degradation, DNA micro array analysis demonstrates that, similar to alginate-related genes in the cluster, the enzyme gene is inducibly expressed in the presence of alginate. The crystal structure of the recombinant A1-R expressed in E. coli cells has been determined at 1.6 Å resolution by X-ray crystallography.72 The enzyme has a three-layer (α/β/α) structure as a basic scaffold, as is typically observed in the SDR family enzymes. A Rossmann fold for coenzyme binding was also identified in A1-R. Indeed, NADP was found to be bound to the Rossmann fold through structural determination of A1-R in complex with NADP.72
On the basis of a comparison with the pseudomonad pathway, the KDG molecule is suggested to be converted to 2-keto-3-deoxy-6-phospho-gluconate by 2-keto-3-deoxygluconokinase (A1-K, 35 kDa), and subsequently to d-glyceraldehdye-3-phosphate and pyruvate by 2-keto-3-deoxy-6-phospho-gluconate aldolase (A1-A, 24 kDa) (Fig. 8).73
Environmental and Bioenergy Applications
Bioremediation through molecular transplantation of the superchannel.
In countries around the world, environmental pollution has given rise to a range of serious social problems.11 Soil, air, rivers, seas, waste streams and various sites have been contaminated by the widespread use of chemicals, such as solvents, herbicides, insecticides and many industrial chemicals. A large number of organisms, including humans, suffer from these environmental pollutants as a result of their entry into associated food chains. For example, dioxin is regarded as a typical hazardous compound generated in combustion processes and in certain chemical syntheses of haloaromatics.74 Several physicochemical processes such as high-temperature incineration have been used to eliminate not only dioxin but also other hazardous compounds, although these methods are not always economically viable for the treatment of pollutants when these are present at low concentrations and in soluble/insoluble forms. Bioremediation, a more efficient and convenient biological method using microorganisms and plants, has the potential to augment the removal of hazardous compounds from contaminated sites.
Sphingomonas wittichii RW1 (strain RW1), the genome of which has been completely sequenced, can assimilate dibenzofuran, an analogue of dioxin.75 The bacterium has been improved to show an elevated activity for dioxin degradation by metabolic engineering. Thus, the enhancement of import activity of this toxic compound was attempted in strain RW1 cells. The strain A1 superchannel may provide pollutant-degrading sphingomonads with the elevated pollutant import activity. The methods for the transplantation of the superchannel to other bacteria were sought to breed transformants with an elevated transport activity. The lack of an ABC transporter for alginate import through disruption of the AlgS or AlgM1 gene causes failure of pit formation on the strain A1 cell surface and, consequently, an inability to assimilate alginate. This accordingly suggests that the strain A1 gene cluster for alginate import and degradation affects pit formation on the cell surface.20
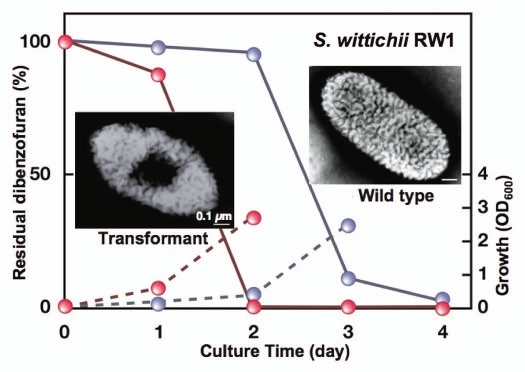
The ability to express the strain A1 superchannel was transferred to strain RW1 cells through introduction of the gene cluster for alginate import and degradation.76 The wild-type cells of strain RW1 formed no pit, whereas a pit was clearly observed on the cell surface of the strain RW1 transformant containing the strain A1 gene cluster (Fig. 9). The transformant cells exhibited a rapid growth on dibenzofuran and only 2 days were required for complete degradation of the compound. The rapid growth was due to the enhanced import of dibenzofuran into the transformant cells. In contrast, the wild-type cells of strain RW1 exhibited a significantly retarded growth on dibenzofuran and took twice as long (4 days) to degrade this toxin. The transformant cells can also lower the concentration of dibenzofuran contaminant in the soil. Other than strain RW1, transplantation of the strain A1 pit has enhanced the efficacy of polypropylene glycol-degrading Sphingomonas subarctica and polydextrose-degrading Sphingomonas sanguis. These transformants rapidly degraded the corresponding macromolecules through constitutive formation of the pit.76
Figure 9.
Bioremediation through molecular transplantation of the superchannel. Strain RW1 cells with (transformant, red) and without (wild-type, gray) the strain A1 gene cluster were grown in minimal medium containing dibenzofuran as the sole carbon source. Solid and broken lines indicate residual dibenzofuran and cell growth, respectively.
Bioenergy generation from alginate by sphingomonads.
Starch and cellulose become important biomasses for a new energy source since fossil fuels are considered to be exhausted near future. Starch is the major carbohydrate in corn grain. This polysaccharide consists of glucose units linked via α-1,4 and/or α-1,6 glycosidic bonds, and it is readily degraded to glucose by glycoside hydrolases such as amylases. The resultant glucose is the best monosaccharide for microbial fermentation to produce ethanol. The utilization of corn grain for bioethanol production does, however, have a disadvantage in that it results in a sharp rise in crop prices due to the competition with foodstuffs. As alternatives to starch, cellulose or lignocellulose abundant in land plants are thought to be potential sources of biomass for biofuel production.77 Cellulose is also a glucose polymer in the crystalline form, although its glucose units are linked by β-1,4 glycosidic bonds. In order to utilize cellulose or lignocellulose as a resource for biofuel production, technical problems regarding how to remove lignin and hydrolyze cellulose still must be overcome. Thermochemical pretreatment, e.g., sulfuric acid treatment at high temperature, is conducted to improve the enzymatic degradation of the polysaccharide through the removal of lignin and hydrolysis of the β-glycosidic linkages. However, this treatment is occasionally expensive and also has a harmful effect on the environment.
Marine biomass is, therefore, one of new promising bioenergy sources. Alginate is a major component of brown seaweeds, suggesting that strain A1 cells can be a candidate for bioconversion of alginate to a new energy. In fact, the culture broth of strain A1 cells in the alginate medium includes a trace of ethanol. Thus, the bacterium was improved to enhance the ethanol-producing activity. The α-keto acid produced nonenzymatically from alginate monosaccharides is further degraded to pyruvate and D-glyceraldehyde-3-phosphate through the successive reactions of cytoplasmic reductase A1-R, kinase A1-K, and aldolase A1-A.73 The tricarbon compounds, pyruvate and d-glyceraldehyde-3-phosphate, are converted to ethanol by the cells of Saccharomyces cerevisiae or Zymomonas mobilis. The E. coli cells transformed with Z. mobilis genes for pyruvate decarboxylase and alcohol dehydrogenase II produce ethanol from glucose.78 Strain A1 cells were also transformed with the two Z. mobilis genes, and the ethanol production ability was examined. The transformant produced ethanol at a concentration of approximately 1% when grown statically on alginate.73 The culture conditions and organization of the introduced gene are currently being optimized for the increased production of biofuels.
Conclusion and Perspective
During a long research in microbiology, strain A1 is the first bacterium with an ability to form a mouth-like pit on the cell surface and incorporate macromolecules through “superchannel.”79 This direct import system of macromolecules in strain A1 cells is superior to the ordinary extracellular degrading enzyme-dependent system for macromolecule assimilation in that, in the latter case, all the degraded products cannot be incorporated within the cells and macromolecule-degrading enzyme must be excreted extracellularly.
One of the unexpected findings of the research on strain A1 is that flagellin homologues are localized on the cell surface, and that these homologues exhibit a potent alginate-binding ability. These flagellins are not, however, involved in the formation of flagella. In order to investigate the molecular diversity and evolution of bacterial flagellins, E. coli flagellin (FliC) was purified and characterized. Similar to the strain A1 flagellin homologues, FliC can also associate with alginate, suggesting that alginate binding is common to the bacterial flagellins.25 Other than flagellins that function in cell motility, a plant-pathogenic bacterium, P. syringae, produces flagellum-associated flagellins that induce a hypersensitive reaction in plants.80 The flagellins of human pathogens such as Salmonella and Listeria cause a host innate immune response through interaction with the host's Toll-like receptors.81 The intrinsic function of these flagellins is, however, the formation of flagella. Strain A1 cells produce flagellin homologues not for flagellum formation but for alginate binding. Strain A1 flagellin homologues are exclusively localized on the cell surface. This provides valuable clues as to the origin, evolution, and function of the flagella. One possibility is that a flagellin may have arisen initially as a cell surface protein capable of recognizing external nutrients, and that this protein gradually evolved into flagellar proteins, or vice versa.
Research in bioremediation has focused to a large extent on the isolation of microorganisms capable of degrading environmental pollutants and on determining the degradation pathways through gene/enzyme analysis. As described in the introduction, among the pollutant-degrading microbes isolated and characterized to date, sphingomonads are some of the most promising bacteria with respect bioremediation.11 Although sphingomonads can be enhanced for bioremediation by overexpressing pollutant-degrading enzymes through introduction of the corresponding genes, the import of pollutants into the bacterial cells has thus far not been elevated. The strain A1 superchannel is considered to have the potential to enhance the permeability of various compounds across cell membranes. In fact, strain RW1 cells transformed with strain A1 gene cluster for alginate import and degradation show an elevated activity for dibenzofuran import.76 From a practical viewpoint, such results clearly indicate that molecular transplantation of the strain A1 superchannel to pollutant-degrading sphingomonads can be considered as a promising biotechnological approach to tackling environmental pollution.
In the future, biofuels such as bioethanol and biodiesel oil, are expected to become new energy sources that replace fossil fuels such as petroleum and coal.82 Currently, bioethanol production processes are being actively investigated, and the production of ethanol from sugar cane and corn grain has already been developed on an industrial scale.83 To the best of our knowledge, ethanol-fermenting microbes such as S. cerevisiae and Z. mobilis convert monosaccharides to ethanol. Saccharification of biomass is therefore one of the most important steps in biofuel production. Polysaccharide-degrading enzymes such as glycoside hydrolases and lyases play an important role in the saccharification of biomass. Aquatic plants such as marine and nonmarine algae have recently been considered as a potential resource for biofuel production.84 For example, gulfweed, a type of seaweed, is abundant in the sea and readily cultivable. Alginate, a major part of this marine biomass, constitutes 30–60% of dried seaweed.85 Strain A1 cells are suitable for biofuel production from alginate because the bacterium can assimilate alginate as a sole carbon source. The exotype alginate lyase A1-IV degrades alginate with various M/G compositions to the constituent monosaccharides, indicating that the enzyme is also suitable for the saccharification of alginate. The recombinant strain A1 cells with a high ethanol-producing activity now become a target of patent for a novel microbe converting marine biomass to bioenergy.73
Acknowledgements
This work was supported by Grants-in-Aid to K.M., B.M. and W.H. and by the Targeted Proteins Research Program (to W.H.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Part of this work was also supported by the Program of Basic Research Activities for Innovative Biosciences (PROBRAIN) of Japan (to K.M.).
Abbreviations
- A1-I, A1-II, A1-III, A1-IV, A1-II' and A1-IV'
strain A1 alginate lyases
- ABC
ATP-binding cassette
- AlgM1 and AlgM2
permease domains of strain A1 alginate ABC transporter
- AlgQ1 and AlgQ2
strain A1 periplasmic alginate-binding proteins
- AlgS
ATP-binding protein of strain A1 alginate ABC transporter
- CAZy
carbohydrate-active enzyme
- FliC
Escherichia coli flagellin
- G
α-l-guluronate
- GSL
glycosphingolipids
- Inc
incompatibility
- KDG
2-keto-3-deoxy-d-gluconic acid
- LPS
lipopolysaccharides
- M
β-d-mannuronate
- p1, p2, p3 and p4
TonB-dependent outer membrane transporters
- p5 and p6
strain A1 flagellin homologues as cell surface alginate receptors
- p7 and p8
strain A1 cell surface alginate-binding proteins
- SDR
short-chain dehydrogenase/reductase
- SPR
surface plasmon resonance
- strain A1
Sphingomonas sp. A1
- strain RW1
Sphingomonas wittichii RW1
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/10322
References
- 1.White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol. 1996;7:301–306. doi: 10.1016/s0958-1669(96)80034-6. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki S, Moriguchi R, Sekiya K, Nakai T, Ono E, Kume K, et al. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J Bacteriol. 1994;176:284–290. doi: 10.1128/jb.176.2.284-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yabuuchi E, Yano I, Oyaizu H, Hashimoto Y, Ezaki T, Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34:99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi M, Hamana K, Hiraishi A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int J Syst Evol Microbiol. 2001;51:1405–1417. doi: 10.1099/00207713-51-4-1405. [DOI] [PubMed] [Google Scholar]
- 5.Maruyama T, Park HD, Ozawa K, Tanaka Y, Sumino T, Hamana K, et al. Sphingosinicella microcystinivorans gen. nov., sp. nov., a microcystin-degrading bacterium. Int J Syst Evol Microbiol. 2006;56:85–89. doi: 10.1099/ijs.0.63789-0. [DOI] [PubMed] [Google Scholar]
- 6.Hisano T, Yonemoto Y, Yamashita T, Fukuda Y, Kimura A, Murata K. Direct uptake of alginate molecules through a pit on the bacterial cell surface: a novel mechanism for the uptake of macromolecules. J Ferment Bioeng. 1995;79:538–544. [Google Scholar]
- 7.Hisano T, Kimura N, Hashimoto W, Murata K. Pit structure on bacterial cell surface. Biochem Biophys Res Commun. 1996;220:979–982. doi: 10.1006/bbrc.1996.0526. [DOI] [PubMed] [Google Scholar]
- 8.Fredrickson JK, Balkwill DL, Drake GR, Romine MF, Ringelberg DB, White DC. Aromatic-degrading Sphingomonas isolates from the deep subsurface. Appl Environ Microbiol. 1995;61:1917–1922. doi: 10.1128/aem.61.5.1917-1922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laskin AI, White DC. Preface to special issue on Sphingomonas. J Ind Microbiol Biotechnol. 1999;23:231. doi: 10.1038/sj.jim.2900748. [DOI] [PubMed] [Google Scholar]
- 10.Field JA, Sierra-Alvarez R. Microbial degradation of chlorinated dioxins. Chemosphere. 2008;71:1005–1018. doi: 10.1016/j.chemosphere.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 11.Stolz A. Molecular characteristics of xenobiotic-degrading sphingomonads. Appl Microbiol Biotechnol. 2009;81:793–811. doi: 10.1007/s00253-008-1752-3. [DOI] [PubMed] [Google Scholar]
- 12.Fialho AM, Moreira LM, Granja AT, Popescu AO, Hoffmann K, Sá-Correia I. Occurrence, production and applications of gellan: current state and perspectives. Appl Microbiol Biotechnol. 2008;79:889–900. doi: 10.1007/s00253-008-1496-0. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto W, Murata K. α-L-Rhamnosidase of Sphingomonas sp. R1 producing an unusual exopolysaccharide of sphingan. Biosci Biotechnol Biochem. 1998;62:1068–1074. doi: 10.1271/bbb.62.1068. [DOI] [PubMed] [Google Scholar]
- 14.Gacesa P. Alginates. Carbohydr Polym. 1988;8:161–182. [Google Scholar]
- 15.Hashimoto W, Maruyama Y, Itoh T, Mikami B, Murata K. Bacterial system for alginate uptake and degradation. In: Rehm B, editor. Alginates: Biology and Applications. Vol. 13. Heidelberg: Springer; 2009. pp. 73–94. Microbiology Monographs. [Google Scholar]
- 16.Jensen A. Present and future needs for algae and algal polysaccharides. Hydrobiologica. 1993;260/261:15–23. [Google Scholar]
- 17.May TB, Chakrabarty AM. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 18.Schweizer HP, Boring JR., III Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect Immun. 1973;3:762–767. doi: 10.1128/iai.3.6.762-767.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong TY, Preston LA, Schiller NL. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles and applications. Annu Rev Microbiol. 2000;54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- 20.Momma K, Okamoto M, Mishima Y, Mori S, Hashimoto W, Murata K. A novel bacterial ATP-binding cassette (ABC) transporter system that allows uptake of macromolecules. J Bacteriol. 2000;182:3998–4004. doi: 10.1128/jb.182.14.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto W, Momma K, Maruyama Y, Yamasaki M, Mikami B, Murata K. Structure and function of bacterial super-biosystem responsible for import and depolymerization of macromolecules. Biosci Biotechnol Biochem. 2005;69:673–692. doi: 10.1271/bbb.69.673. [DOI] [PubMed] [Google Scholar]
- 22.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 23.Harada KM, Aso Y, Hashimoto W, Mikami B, Murata K. Sequence and analysis of the 46.6-kb plasmid pA1 from Sphingomonas sp. A1 that corresponds to the typical IncP-1β plasmid backbone without any accessory gene. Plasmid. 2006;56:11–23. doi: 10.1016/j.plasmid.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 24.He J, Nankai H, Hashimoto W, Murata K. Molecular identification and characterization of an alginate-binding protein on the cell surface of Sphingomonas sp. A1. Biochem Biophys Res Commun. 2004;322:712–717. doi: 10.1016/j.bbrc.2004.07.184. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto W, He J, Wada Y, Nankai H, Mikami B, Murata K. Proteomics-based identification of outer-membrane proteins responsible for import of macromolecules in Sphingomonas sp. A1: alginate-binding flagellin on the cell surface. Biochemistry. 2005;44:13783–13794. doi: 10.1021/bi050873b. [DOI] [PubMed] [Google Scholar]
- 26.Turley EV, Noble PW, Bourguignon LYW. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277:4589–4592. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama Y, Momma M, Mikami B, Hashimoto W, Murata K. Crystal structure of a novel bacterial cell-surface flagellin binding to a polysaccharide. Biochemistry. 2008;47:1393–1402. doi: 10.1021/bi701872x. [DOI] [PubMed] [Google Scholar]
- 28.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 29.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 30.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Hirano T, Minamino T, Macnab RM. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J Mol Biol. 2001;312:359–369. doi: 10.1006/jmbi.2001.4963. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto W, Ochiai A, Momma K, Itoh T, Mikami B, Maruyama Y, et al. Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ. Biochem Biophys Res Commun. 2009;381:16–21. doi: 10.1016/j.bbrc.2009.01.186. [DOI] [PubMed] [Google Scholar]
- 33.He J, Ochiai A, Fukuda Y, Hashimoto W, Murata K. A putative lipoprotein of Sphingomonas sp. A1 binds alginate rather than a lipid moiety. FEMS Microbiol Lett. 2008;288:221–226. doi: 10.1111/j.1574-6968.2008.01354.x. [DOI] [PubMed] [Google Scholar]
- 34.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, et al. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol. 2006;188:2761–2773. doi: 10.1128/JB.188.8.2761-2773.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juncker AS, Willenbrock H, von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanley SZ, Pappin DJ, Rahman D, White AJ, Elborough KM, Slabas AR. Re-evaluation of the primary structure of Ralstonia eutropha phasin and implications for polyhydroxyalkanoic acid granule binding. FEBS Lett. 1999;447:99–105. doi: 10.1016/s0014-5793(99)00235-5. [DOI] [PubMed] [Google Scholar]
- 37.Coulton JW, Mason P, Cameron DR, Carmel G, Jean R, Rode HN. Protein fusions of β-galactosidase to the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol. 1986;165:181–192. doi: 10.1128/jb.165.1.181-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pressler U, Staudenmaier H, Zimmermann L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–2724. doi: 10.1128/jb.170.6.2716-2724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundrigan MD, Kadner RJ. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 40.Koedding J, Howard P, Kaufmann L, Polzer P, Lustig A, Welte W. Dimerization of TonB is not essential for its binding to the outer membrane siderophore receptor FhuA of Escherichia coli. J Biol Chem. 2004;279:9978–9986. doi: 10.1074/jbc.M311720200. [DOI] [PubMed] [Google Scholar]
- 41.Larsen RA, Thomas MG, Postle K. Proton motive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol. 1999;31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 42.Sreeram KJ, Shrivastava HY, Nair BU. Studies on the nature of interaction of iron(III) with alginates. Biochim Biophys Acta. 2004;1670:121–125. doi: 10.1016/j.bbagen.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005;44:5053–5064. doi: 10.1021/bi047781r. [DOI] [PubMed] [Google Scholar]
- 44.Momma K, Mikami B, Mishima Y, Hashimoto W, Murata K. Crystal structure of AlgQ2, a macro molecule (alginate)-binding protein of Sphingomonas sp. A1 at 2.0 Å resolution. J Mol Biol. 2002;316:1061–1069. doi: 10.1006/jmbi.2001.5393. [DOI] [PubMed] [Google Scholar]
- 45.Mishima Y, Momma K, Hashimoto W, Mikami B, Murata K. Crystal structure of AlgQ2, a macro molecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-Å resolution. J Biol Chem. 2003;278:6552–6559. doi: 10.1074/jbc.M209932200. [DOI] [PubMed] [Google Scholar]
- 46.Quiocho FA, Spurlino JC, Rodseth LE. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 47.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 48.Mishima Y, Momma K, Hashimoto W, Mikami B, Murata K. Crystallization and preliminary X-ray analysis of AlgS, a bacterial ATP-binding-cassette (ABC) protein specific to macromolecule import. Acta Crystallogr D Biol Crystallogr. 2001;57:884–885. doi: 10.1107/s090744490100525x. [DOI] [PubMed] [Google Scholar]
- 49.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 50.Yoon H-J, Hashimoto W, Miyake O, Okamoto M, Mikami B, Murata K. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr Purif. 2000;19:84–90. doi: 10.1006/prep.2000.1226. [DOI] [PubMed] [Google Scholar]
- 51.Murata K, Inose T, Hisano T, Abe S, Yonemoto Y, Yamashita T, et al. Bacterial alginate lyase: enzymology, genetics and application. J Ferment Bioeng. 1993;76:427–437. [Google Scholar]
- 52.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Setubal JC, dos Santos P, Goldman BS, Ertesvåg H, Espin G, Rubio LM, et al. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol. 2009;191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyake O, Ochiai A, Hashimoto W, Murata K. Origin and diversity of alginate lyases of families PL-5 and -7 in Sphingomonas sp. strain A1. J Bacteriol. 2004;186:2891–2896. doi: 10.1128/JB.186.9.2891-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon H-J, Mikami B, Hashimoto W, Murata K. Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 Å resolution. J Mol Biol. 1999;290:505–514. doi: 10.1006/jmbi.1999.2883. [DOI] [PubMed] [Google Scholar]
- 57.Yoon H-J, Hashimoto W, Miyake O, Murata K, Mikami B. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution. J Mol Biol. 2001;307:9–16. doi: 10.1006/jmbi.2000.4509. [DOI] [PubMed] [Google Scholar]
- 58.Mikami B, Suzuki S, Yoon H-J, Miyake O, Hashimoto W, Murata K. X-ray structural analysis of alginate lyase A1-III mutants/substrate complexes: activation of a catalytic tyrosine residue by a flexible lid loop. Acta Crystallogr A. 2002;58:271. [Google Scholar]
- 59.Linker A, Meyer K, Hoffman P. The production of unsaturated uronides by bacterial hyaluronidases. J Biol Chem. 1956;219:13–25. [PubMed] [Google Scholar]
- 60.Yoon H-J, Choi Y-J, Miyake O, Hashimoto W, Murata K, Mikami B. Effect of His192 mutation on the activity of alginate lyase A1-III from Sphingomonas species A1. J Microbiol Biotechnol. 2001;11:118–123. [Google Scholar]
- 61.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 62.Sakakibara H, Tamura T, Suzuki T, Hisano T, Abe S, Murata K. Preparation and properties of alginate lyase modified with poly(ethylene glycol) J Pharm Sci. 2002;91:1191–1199. doi: 10.1002/jps.10110. [DOI] [PubMed] [Google Scholar]
- 63.Yamasaki M, Ogura K, Hashimoto W, Mikami B, Murata K. A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J Mol Biol. 2005;352:11–21. doi: 10.1016/j.jmb.2005.06.075. [DOI] [PubMed] [Google Scholar]
- 64.Ogura K, Yamasaki M, Mikami B, Hashimoto W, Murata K. Substrate recognition by family 7 alginate lyase from Sphingomonas sp. A1. J Mol Biol. 2008;380:373–385. doi: 10.1016/j.jmb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Hashimoto W, Miyake O, Momma K, Kawai S, Murata K. Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate. J Bacteriol. 2000;182:4572–4577. doi: 10.1128/jb.182.16.4572-4577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyake O, Hashimoto W, Murata K. An exotype alginate lyase in Sphingomonas sp. A1: overexpression in Escherichia coli, purification and characterization of alginate lyase IV (A1-IV) Protein Expr Purif. 2003;29:33–41. doi: 10.1016/s1046-5928(03)00018-4. [DOI] [PubMed] [Google Scholar]
- 67.Hashimoto W, Miyake O, Ochiai A, Murata K. Molecular identification of Sphingomonas sp. A1 alginate lyase (A1-IV') as a member of novel polysaccharide lyase family 15 and implications in alginate lyase evolution. J Biosci Bioeng. 2005;99:48–54. doi: 10.1263/jbb.99.48. [DOI] [PubMed] [Google Scholar]
- 68.Ochiai A, Hashimoto W, Murata K. A biosystem for alginate metabolism in Agrobacterium tumefaciens strain C58: molecular identification of Atu3025 as an exotype family PL-15 alginate lyase. Res Microbiol. 2006;157:642–649. doi: 10.1016/j.resmic.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Preiss J, Ashwell G. Alginic acid metabolism in bacteria I. Enzymatic formation of unsaturated oligosaccharides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J Biol Chem. 1962;237:309–316. [PubMed] [Google Scholar]
- 70.Ochiai A, Takase R, Hashimoto W, Murata K. Molecular identification of reductase involved in detoxifying α-keto acid derived from alginate monosaccharide. Abstract for the annual meeting of the Society for Biotechnology, Japan. 2008:182. [Google Scholar]
- 71.Kavanagh KL, Jörnvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takese R, Ochiai A, Mikami B, Hashimoto W, Murata K. X-ray crystal structure of α-keto acid reductase involved in alginate metabolism. Abstract for the 458th regular meeting of Kansai Branch of Japan Society for Bioscience, Biotechnology, and Agrochemistry. 2009:18. [Google Scholar]
- 73.Takeda H, Ochiai A, Hashimoto W, Murata K. Molecular breeding of bacteria for bioethanol production from alginate. Abstract for the annual meeting of Japan Society for Bioscience, Biotechnology, and Agrochemistry. 2009:98. [Google Scholar]
- 74.Chang YS. Recent developments in microbial biotransformation and biodegradation of dioxins. J Mol Microbiol Biotechnol. 2008;15:152–171. doi: 10.1159/000121327. [DOI] [PubMed] [Google Scholar]
- 75.Wittich RM, Wilkes H, Sinnwell V, Francke W, Fortnagel P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1992;58:1005–1010. doi: 10.1128/aem.58.3.1005-1010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aso Y, Miyamoto Y, Harada KM, Momma K, Kawai S, Hashimoto W, et al. Engineered membrane super-channel improves bioremediation potential of dioxin-degrading bacteria. Nat Biotechnol. 2006;24:188–189. doi: 10.1038/nbt1181. [DOI] [PubMed] [Google Scholar]
- 77.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 78.Ohta K, Beall DS, Mejia JP, Shanmugam KT, Ingram LO. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991;57:893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murata K, Kawai S, Mikami B, Hashimoto W. Superchannel of bacteria: biological significance and new horizons. Biosci Biotechnol Biochem. 2008;72:265–277. doi: 10.1271/bbb.70635. [DOI] [PubMed] [Google Scholar]
- 80.Shimizu R, Taguchi F, Marutani M, Mukaihara T, Inagaki Y, Toyoda K, et al. The ΔfliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol Genet Genomics. 2003;269:21–30. doi: 10.1007/s00438-003-0817-3. [DOI] [PubMed] [Google Scholar]
- 81.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 82.Gray KA, Zhao L, Emptage M. Bioethanol. Curr Opin Chem Biol. 2006;10:141–146. doi: 10.1016/j.cbpa.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 83.Brehmer B, Bals B, Sanders J, Dale B. Improving the corn-ethanol industry: studying protein separation techniques to obtain higher value-added product options for distillers grains. Biotechnol Bioeng. 2008;101:49–61. doi: 10.1002/bit.21881. [DOI] [PubMed] [Google Scholar]
- 84.Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol. 2008;19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 85.d'Ayala GG, Malinconico M, Laurienzo P. Marine derived polysaccharides for biomedical applications: chemical modification approaches. Molecules. 2008;13:2069–2106. doi: 10.3390/molecules13092069. [DOI] [PMC free article] [PubMed] [Google Scholar]