Abstract
Whole-cell bacterial bioreporters represent a convenient testing method for quantifying the bioavailability of contaminants in environmental samples. Despite the fact that several bioreporters have been constructed for measuring heavy metals, their application to environmental samples has remained minimal. The major drawbacks of the available bioreporters include a lack of sensitivity and specificity. Here, we report an improvement in the limit of detection of bacterial bioreporters by interfering with the natural metal homeostasis system of the host bacterium. The limit of detection of a Pseudomonas putida KT2440-based Zn/Cd/Pb-biosensor was improved by a factor of up to 45 by disrupting four main efflux transporters for Zn/Cd/Pb and thereby causing the metals to accumulate in the cell. The specificity of the bioreporter could be modified by changing the sensor element. A Zn-specific bioreporter was achieved by using the promoter of the cadA1 gene from P. putida as a sensor element. The constructed transporter-deficient P. putida reporter strain detected Zn2+ concentrations about 50 times lower than that possible with other available Zn-bioreporters. The achieved detection limits were significantly below the permitted limit values for Zn and Pb in water and in soil, allowing for reliable detection of heavy metals in the environment.
Key words: Zn/Cd/Pb-bioreporter, sensor element, specificity, limit of detection
Introduction
While chemical and physical methodologies for monitoring environmental pollution can be powerful, accurate and sensitive, they are also costly and require specialized laboratories. In addition, they fail to provide data on the bioavailability of a pollutant and its effects on living systems. Therefore, whole-cell-based bioassays, which are easily applicable and inexpensive, and which report not only on the presence of a chemical, but also on its bioavailability and its biological effects, are being developed. Whole-cell bioreporters are living microorganisms that produce a specific quantifiable output in response to target chemicals. On the basis of detection specificity, bioreporters can be broadly divided into three groups: nonspecific, semi-specific and specific. Nonspecific bioreporters are useful for measuring the toxicity of samples, but do not provide any information about the identity of the contaminants.1,2 Recombinant semi-specific bioreporters allow for the detection of compounds that cause certain cellular responses, such as stress3–5 or DNA damage6,7 without further characterizing the compound. Specific bioreporters, first described by King et al.8 respond only to a certain compound or class of compounds and therefore permit a quantitative analysis of the contaminant concentration.
In general, specific bacterial bioreporters combine a sensing element, which is responsible for detecting the analyte, and a reporter element, which allows for quantification of the signal.9,10 In this manner, bacterial bioreporters have been developed for heavy metals11 and several organic compounds.9,12
In heavy metal bioreporters, the sensor element usually consists of a specific promoter fused to a reporter gene, and a gene encoding for a transcription factor that in response to certain metal ions activates the promoter. Binding of a metal ion to the transcription factor induces the expression of the reporter gene, thus converting metal sensing into a detectable output signal. The most important characteristics of bacterial bioreporters—specificity (which metals are detected) and sensitivity (what concentrations are detected; characterized by the limit of detection)—depend on the transcription factor used in the sensing element as well as on the microbial metal homeostasis and resistance systems.13 In bacteria, homeostasis of essential metals and resistance to toxic heavy metals are maintained with the help of influx and efflux transporters. Importantly, it is the activity of these transporters that determines the intracellular metal concentration that is ultimately available for detection by the bioreporters.
Despite the importance of whole-cell bioreporters in determining the biologically relevant fraction of metals in the environment, they are rarely used for actual environmental analyses. The wider application of bioreporters has been partly hindered by low sensitivity and a lack of specificity. The amount of bioavailable metal in complex matrices can be less than 1%, even in highly contaminated samples.14,15 Reliable detection of such low concentrations requires improved detection limits. In addition, the ability to specifically characterize the contaminants in a sample requires bioreporters that respond to only one metal.
We hypothesized that, on the removal of specific efflux pumps, bacteria start to accumulate more metal in the cell, and thus, lower extracellular concentrations can be detected. Efflux mechanisms compete with the detection of metal ions and thus lead to a less sensitive response. If all of the metal ions entering the cell were immediately removed, no binding to transcription factors would occur and hence no signal would be generated. However, in the absence of efflux transporters, metal ions accumulate in the cell and the threshold concentration required for the expression of the reporter gene is reached at lower extracellular concentrations. Our approach was to improve the limit of detection (LOD) of Pseudomonas putida-based Cd/Zn/Pb-bioreporters by altering the efflux of respective metals from the cytoplasm, thereby causing metal ions to accumulate in the bacterial cell. In P. putida KT2440, genes for the divalent heavy metal efflux proteins CadA1, CadA2, CzcA1 and CzcA2 were deleted, and the mutant P. putida strain was applied as a Cd/Zn/Pb-bioreporter and tested for improved limit of detection.
Results
Constructed bioreporter strains.
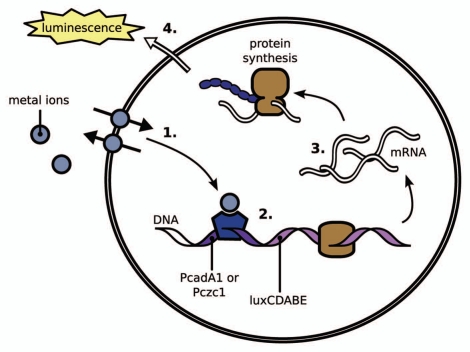
P. putida strains KT2440,16 and KT2440.2431,17 were used as host bacteria. P. putida KT2440 is a ubiquitous saprophytic bacterium endowed with a remarkable adaptability to diverse environments and is thus suitable for the analysis of environmental samples. P. putida KT2440.2431 is a metal transporter-deficient mutant of the strain KT2440. In P. putida KT2440.2431, genes for four Zn/Cd/Pb efflux transporters (P-type ATPases CadA1 and CadA2 and CBA transporters CzcCBA1 and CzcCBA2) have been disrupted, rendering the cell susceptible to heavy metals. As sensor-reporter elements, we used fusions of heavy-metal-regulated cadA1 and czcCBA1 promoters from P. putida KT2440 to bacterial bioluminescence operon luxCDABE in a low copy number plasmid. Altogether, four different bioreporter strains, KT2440(pDNPczc1lux), KT2440.2431(pDNPczc1lux), KT2440(pDNPcadA1lux) and KT2440.2431(pDNPcadA1lux) (Fig. 1), were tested for the response to heavy metals.
Figure 1.
Principle of Pseudomonas putida KT2440 and KT2440.2431 metal bioreporters. (1) Uptake and efflux mechanisms regulate the intracellular metal level; in the mutant strain KT2440.2431, the efflux mechanisms have been removed. (2) Upon binding of the metal ion to the transcription factor, transcription is initiated from the PcadA1 or Pczc1 promoter that is fused to the luxCDABE operon. (3) mRNA for the reporter proteins is produced and subsequently translated into proteins. (4) The bacterial lux operon encodes for luciferase and the proteins necessary for substrate synthesis; therefore, luminescence can be measured directly as no additional substrate is required. Over a certain concentration range, the signal output correlates to the intracellular metal concentration, allowing for quantification of the metal.
Optimization of the assay.
To achieve minimal LOD of the bioreporters within a reasonable testing time, different assay conditions were tested. First, the inducibility of the bioreporters at different growth stages was tested as metal-induced expression from PcadA1 and Pczc1 promoters is known to be dependent on the growth phase of the bacteria.17 The response of the reporter strains to metals was largely dependent on the growth phase (data not shown). The lowest metal concentrations were detected with bacteria grown to the early exponential phase. Therefore, bacteria grown to the exponential phase (OD600 ∼ 0.015…0.017 measured on 96-well plate in 200 µl) were used in further experiments.
In addition, an effect of incubation time (i.e., contact time of the reporter bacteria with the metal) on sensor inducibility was investigated. Clear induction was already visible after 1 hour of incubation. For the wild-type reporter strains, further incubation had no effect on the LOD. However, for the mutant reporter strains, up to 10-fold improvement could be achieved by extending the incubation to 3 hours. For example, the LOD of P. putida KT2440.2431(pDNPczc1lux) for Cd2+ shifted from 0.1 µM to 0.02 µM by extending the incubation from 1 to 3 hours. Although even longer incubation times slightly improved the sensitivity (as low as 0.008 µM LOD for Cd2+ could be achieved), a 3-hour incubation time was used in further experiments to balance the time used for the whole assay. The decrease with time in the LOD of the efflux transporter-deficient strain could be explained by the continuous accumulation of metal ions in the cell and the consequent detection of lower extracellular metal concentrations.
Sensitivity and specificity of the reporter strains.
The constructed bacterial bioreporters were assayed with different divalent heavy metals to determine their sensitivity (LOD) and specificity (metals detected) using the optimized assay conditions. The Zn/Cd/Pb efflux transporter-deficient strain KT2440.2431 was used in parallel with the wild-type strain KT2440.
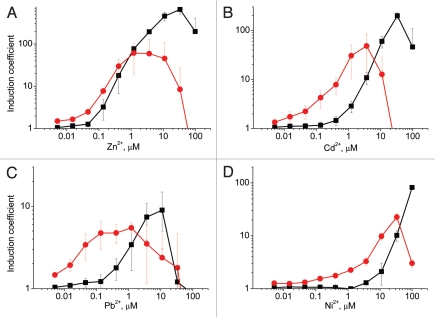
As hypothesized, the removal of efflux transporters affected the LOD of the bioreporters. P. putida KT2440.2431(pDNPczc1lux) could detect Pb2+ at 45-fold, Cd2+ at 12-fold, Ni2+ at 10-fold and Zn2+ at 3-fold lower concentrations than its wild-type counterpart KT2440(pDNPczc1lux) (Fig. 2 and Table 1). For the wild-type strain, Zn2+ was the best inducer, whereas for the mutant strain, Zn2+, Cd2+ and Pb2+ were detected at the same concentrations. For both strains, Ni2+ was a poor inducer as 100-fold higher concentrations were needed for induction. No induction was observed with Hg2+ or Co2+. Toxic concentrations causing a rapid decrease in the luminescence signal were much lower for the mutant strain (Fig. 2), indicating that metals accumulated in this strain.
Figure 2.
Inducibility of P. putida KT2440(pDNPczc1lux) (black squares) and KT2440.2431(pDNPczc1lux) (red circles) with Zn2+ (A), Cd2+ (B), Pb2+ (C) and Ni2+ (D). Data represent the mean ± standard deviation of at least three independent experiments.
Table 1.
Limits of detection of Zn-, Cd- and Pb-sensing bioluminescent bacterial bioreporters
| Bacterial bioreporter | Limit of detection, µM | Reference | ||
| Zn2+ | Cd2+ | Pb2+ | ||
| P. fluorescens OS8::KncadRPcadAluxa | 4 | 0.03 | 0.3 | 28 |
| E. coli MC1061(pSLzntR/pDNPzntAlux)a | 5 | 0.01 | 0.7 | 28 |
| E. coli MC1061(pzntRluc)b | 40 | 0.06 | n.t. | 29 |
| E. coli MG1655(pZNT-lux)b | 1 | 0.01 | 0.03 | 30 |
| S. aureus RN4220(pTO024)c | 1 | 0.01 | 0.03 | 31 |
| B. subtilis BR151(pTO024)c | 1 | 0.03 | n.i. | 31 |
| P. putida KT2440(pDNPczc1lux)d | 0.16 (0.08) | 1.12 (0.49) | 0.90 (0.41) | This study |
| P. putida KT2440.2431(pDNPczc1lux)d | 0.05 (0.02) | 0.09 (0.05) | 0.02 (0.02) | This study |
| P. putida KT2440(pDNPcadA1lux)d | 0.89 (0.63) | n.i. | n.i. | This study |
| P. putida KT2440.2431(pDNPcadA1lux)d | 0.12 (0.08) | n.i. | n.i. | This study |
limit of detection (LOD) determined as a concentration that caused 2 + 6 SDw/Lw-fold induction.
LOD determined as a concentration that caused two-fold induction.
LOD determined as a concentration that caused Lw + 2 SDw-fold induction.
LOD determined as a concentration that caused (1 + 2 CVw)/(1 − 2 CVw)-fold induction; upper estimate of LOD (confidence level 97.7%) and mean LOD (in parenthesis) are presented (see Materials and Methods for details). Lw, luminescence in water; SDw, standard deviation of the signal measured in water; CVw, coefficient of variation of the signal measured in water; n.t., not tested; n.i., not inducible.
Although substantial improvements in LODs were achieved by using the promoter of the czcCBA1 operon as a sensor element, the lack of specificity still remained a problem. Therefore, we tested a promoter of the cadA1 gene, which is known to be induced by Zn2+ and slightly by Cd2+, but not by other metals, 17 as a sensor element. P. putida KT2440(pDNPcadA1lux) was inducible only with Zn2+, and expectedly, the mutant strain KT2440.2431(pDNPcadA1lux) could detect about eight-fold lower Zn2+ concentrations than the wild-type strain (Fig. 3 and Table 1). Although P. putida KT2440.2431(pDNPcadA1lux) could not detect concentrations as low as KT2440.2431 (pDNPczc1lux), the great advantage of the reporter strain harboring the promoter of cadA1 as a sensor element was its specificity—no metals other than Zn2+ induced this reporter strain.
Figure 3.
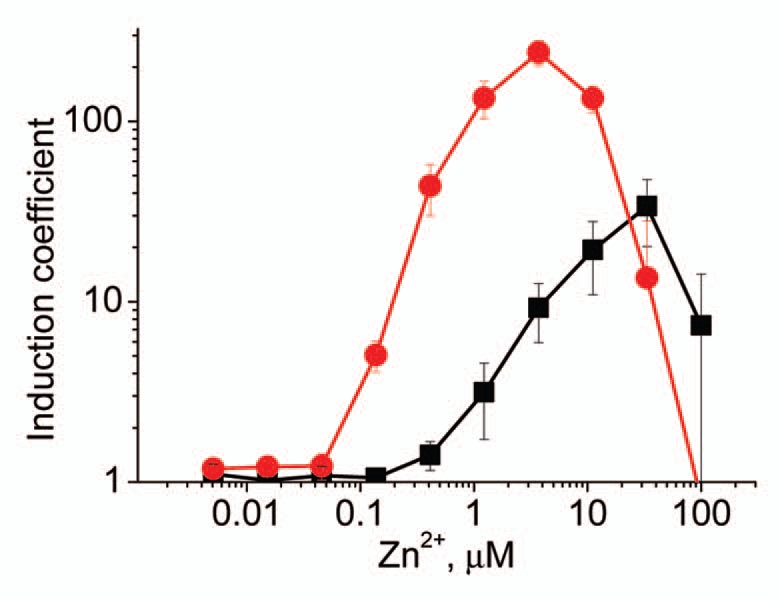
Inducibility of P. putida KT2440(pDNPcadA1lux) (black squares) and KT2440.2431(pDNPcadA1lux) (red circles) with Zn2+. Data represent the mean ± standard deviation of three independent experiments.
Performance of the bioreporters in Zn-contaminated soil samples.
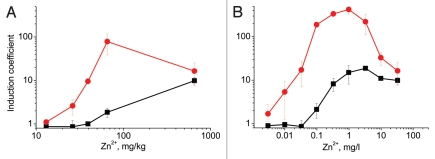
The performance of the constructed bioreporter strains was tested on soil samples intentionally contaminated with a known amount of Zn2+. In the measurements of undiluted soil samples wild-type P. putida KT2440(pDNPcad1lux) could reliably detect zinc from the soil that contained 65 mg/kg of Zn2+; whereas, the more sensitive transporter-deficient strain KT2440.2431(pDNPcad1lux) could detect as low concentrations as 20 mg/kg of Zn2+ (Fig. 4A).
Figure 4.
Performance of P. putida KT2440(pDNPcadA1lux) (black squares) and KT2440.2431(pDNPcadA1lux) (red circles) in Zn-contaminated soil samples. (A) Induction of the bioreporters as a function of Zn2+ concentration in the soil samples. The x-axis shows the spiked Zn concentration in the soil. Each measurement point corresponds to a soil sample spiked with different concentration of Zn. (B) Induction of the bioreporters in the dilution series of the soil sample spiked with 654 mg/kg of Zn2+. The x-axis shows the actual Zn concentration in the diluted samples. Data represent the mean ± standard deviation of three independent experiments.
In highly contaminated samples metal concentrations become toxic to the cell and measurements of undiluted samples do not give correct estimation of the bioavailable amount of the metal. In such cases additional diluting of the sample is required in order to reach concentrations which are close to the LOD of the bioreporter. The transporter-deficient reporter strain KT2440.2431(pDNPcadA1lux) could detect about 10-fold lower Zn concentrations than the wild-type strain KT2440(pDNPcadA1lux) in the dilution series of soil spiked with 654 mg/kg of Zn2+ (Fig. 4B). Thus, as was the case with standard metal solutions, transporter-deficient bioreporter performed better also in actual environmental samples.
Discussion
Dozens of bioreporters have been constructed, and their suitability for testing environmental samples has been demonstrated.13 Despite this research, bioreporters are not yet widely used in environmental applications, partly due to their high LODs and lack of specificity. We approached the sensitivity and specificity issues by disrupting the metal homeostasis of the host bacteria and testing different sensor elements.
In this study, we constructed new transporter-deficient bacterial bioreporter strains P. putida KT2440.2431(pDNPczc1lux) and KT2440.2431(pDNPcadA1lux), which, depending on the metal, could detect 3–45-fold lower metal concentrations as compared to their wild-type counterparts. The improvement in LODs was achieved due to the disruption of metal efflux transporters and consequent accumulation of metals in the cell. P. putida KT2440.2431(pDNPczc1lux), whose upper estimate of LOD for zinc was 0.05 µM (3.3 µg/l), is the most sensitive bacterial sensor for zinc constructed thus far (Table 1). Although the LOD for lead and cadmium remained on the same level as those for already existing bioreporters (Table 1), the success of metal homeostasis interruption indicates that improved LOD for these metals could also be achieved by modifying already existing bioreporter strains. A similar strategy has been shown to work for Cu/Ag-bioreporters, but not for As-bioreporters. The removal of CopA, a Cu/Ag-exporting ATPase, from E. coli rendered a bioreporter employing the copA promoter as a sensor system 15-fold more sensitive to Cu and eight-fold more sensitive to Ag.18 On the other hand, Tauriainen et al.19 compared the performance of two E. coli As-bioreporters—one with an intact As detoxification system and another containing a deletion in the chromosomal arsenite-resistance operon. They found no differences in performance of these bioreporters, showing that the lack of the ars operon did not cause a remarkable accumulation of As in the cell.
Almost all bioreporters constructed to date detect only intracellular metal. In addition to causing the accumulation of metals in the cell by removing efflux transporters, permeabilization of the membrane or additional metal uptake mechanisms could also improve the LOD of bacterial bioreporters. The influence of uptake transporters has been tested on a Hg-bioreporter.20 In addition to detoxification proteins, the Hg-resistance operon encodes for a protein that mediates the uptake of Hg2+ ions into the cell. Employing this protein in the bioreporter construct improved the sensitivity of the Hg-bioreporter two-fold.20 Although specific uptake systems only exist for a limited number of metals, the proteins responsible for the uptake of most metals are known. Overexpression of these systems in combination with the removal of efflux transporters would thus be one approach to improving the LOD of bacterial bioreporters.
In addition to improving the LOD of Zn/Cd/Pb-bioreporters, we aimed at more specific reporters by changing the sensor element. P. putida KT2440.2431(pDNPcadA1lux), which carried the cadA1 promoter as a sensor element, proved to be Zn2+-specific. Although P. putida KT2440.2431(pDNPcadA1lux) was somewhat less sensitive than P. putida KT2440.2431(pDNPczc1lux) (Table 1), its specificity is a great advantage over other bioreporters. Restricted specificity allows for more accurate quantification of a certain contaminant regardless of other contaminants in the sample. The existence of a Zn-specific sensor element is peculiar, as all of the transcription factors and resistance mechanisms known for Zn2+ thus far cross-react with Cd2+ and Pb2+ (see Table 1).17,21–24 However, as only three sensor elements (the promoters of zntA from E. coli, cadA from P. putida and cadA from S. aureus) have been previously used for the construction of Zn/Cd/Pb-bioreporters, it is still possible that an open-minded search for new heavy-metal-regulated promoters would yield bioreporters with different specificities.
Although the LOD is an important characteristic of bacterial bioreporters, no common method for calculating the LOD has been established in the literature (see Table 1). Several of the methods applied for calculating the LOD lack any statistical basis. However, as bacterial bioreporters are living systems, some variability in the signal within one experiment and between independent experiments is natural. Therefore, a statistically sound method for determining LOD should be used. IUPAC suggests that LOD for analytical methods is determined as the concentration which has a response equal to the mean response plus two or three standard deviations of the blank samples depending on the confidence level desired.26 However, this method eliminates only the false positives and does not take into account the variability in the signal measured for the analyte. We used here the method recommended for immunoassays.27 This method considers the variability in the signal for both the blank and also the analyte. The concentration of the analyte where the confidence interval does not overlap with the confidence interval of the blank is considered as the LOD.27 Thus, the method eliminates both the false positives and the false negatives. In addition to considering the variation within the experiment we took into account the variability between the independent experiments and reported the upper estimate of LOD in addition to reporting the mean LOD. If only the mean LOD is used for characterizing the bioreporter, 50% of the experiments actually have a higher LOD than initially reported. However, the upper estimate of LOD guarantees that with a probability of 97.7% the LOD in a single experiment is below the reported level. Therefore, it is important to consider the variability of the method when reporting the characteristics of the bioreporter.
The new bioreporters constructed in this work, P. putida KT2440.2431(pDNPczc1lux) and KT2440.2431-(pDNPcadA1lux), are suitable for measuring zinc, cadmium or lead from environmental samples, as their upper estimate of LOD remained well below the limit values of water and soil contaminants set by the European Union (Table 2). Our results show that the limit of detection of bacterial heavy metal bioreporters could be improved by removing heavy metal efflux pumps, thereby artificially increasing the intracellular concentration of the metal. This study indicates that the disruption of metal homeostasis in bacteria is a feasible approach for the development of bacterial bioreporters with increased sensitivity. The use of hyper-sensitive bioreporters in environmental risk assessment opens the possibility for the detection of low-level pollution.
Table 2.
Limit values of zinc, cadmium and lead in the European Union, compared to the upper estimates of the limit of detection of the bacterial bioreporter P. putida KT2440.2431(pDNPczc1lux)
| Metal | Limit value | Upper estimate of the limit of detection, mg/l | |
| Surface water, mg/la | Soil, mg/kgb | ||
| Zn | 3 | 150…300 | 0.003 |
| Cd | 0.005 | 1…3 | 0.010 |
| Pb | 0.05 | 50…300 | 0.004 |
Council Directive 75/440/EEC
Council Directive 86/278/EEC.
Materials and Methods
Bacterial strains and growth conditions.
As bioreporters, P. putida strains KT2440,16 and KT2440.2431,17 were used. Both strains were transformed with the sensor plasmids pDNPcadA1lux and pDNPczc1lux.17 These plasmids contain a luxCD-ABE operon from Photorhabdus luminescens under the control of the cadA1 and czcCBA1 promoters from P. putida, respectively.
Bacteria were grown in a heavy metal MOPS (HMM) medium25 supplemented with 0.05% caseine hydrolysate, 0.4% glucose and 12 µg/ml tetracycline. For maintaining the strains, HMM plates containing 1.5% agar were used.
Induction assay.
Inducibility of the constructed strains was tested with different heavy metals. Solutions of CdCl2 (stock solution 1 M), ZnCl2 (0.1 M), HgCl2 (0.1 M), NiSO4 (1 M), CoCl2 (1 M) and Pb (CH3COO)2 (0.05 M) at different concentrations were prepared in MilliQ water (Millipore, France). First, metal dilutions were prepared with a Biomek NXP (Beckman Coulter, Fullerton) robot using a dilution factor of 1:3. Then, 20 µl of the metal dilution was dispensed to a white 384-well plate (Thermo Labsystems, Finland), and an equal amount of bacterial suspension was added using a Biomek NXP pipetting robot. The plates were incubated for 3 h at 30°C, and the luminescence was measured with a Victor Wallac 1420 multilabel counter (PerkinElmer, Finland). The samples in every assay were analyzed in four parallels, i.e., each metal concentration was plated in four wells. At least three independent assays were performed for every strain. Induction of the bioreporters was reported as an induction coefficient (IC), calculated as follows: IC = LM/LW, where LM is the luminescence in the metal solution and LW is the background luminescence measured in MilliQ water.
Testing conditions.
The growth and test conditions were optimized to achieve a satisfactory limit of detection within a minimal time. The cells were grown overnight in HMM medium, diluted 100-fold in fresh medium and then grown at 30°C with shaking. Aliquots of bacteria were removed at different time points and used in the induction assays. Growth curves were determined by measuring the OD600 values at 1-h intervals, and growth phases were assigned according to different stages of the growth curve. OD600 was measured with Victor Wallac 1420 on 96-well plate in 200 µl. The optimal incubation time was determined by measuring induction plates at 1-h intervals.
Determination of the limit of detection.
The limit of detection for bacterial bioreporters was determined as the metal concentration which induced the reporter to above the threshold level LODIC, calculated according to the equation LODIC = (1 + 2 CVw)/(1 − 2 CVw). Here CVw is coefficient of variation of the signal determined in water (background or blank). The signal in water was always measured in at least 12 parallels, but as it was not reasonable to measure samples in as many parallels it was assumed that CV for samples is the same as for water. This equation assures that, within the confidence interval of 97.7% the lowest signal from the sample, LM − 2 SDM, is higher than the highest value for the blank within the same confidence interval, LW + 2 SDW, thus eliminating both false positives and false negatives. The lowest detectable concentration was determined separately for each measurement (according to the LODIC of each independent measurement). An upper estimate of LOD was determined as the mean plus two standard deviations of the mean, calculated from the independent measurements. This corresponds to a 97.7% confidence level, i.e., the probability that the LOD determined from a single experiment is below this upper limit is 97.7%. In order to facilitate the comparison to previously published bioreporters also the mean LOD was presented.
Analyses of the soil samples.
Artificial Zn-contaminated soil samples were made by spiking industrially produced garden soil (Rainbow, Finland) with different Zn2+ concentrations. One gram of soil (dry weight) was mixed with 9 ml of ZnCl2 solution (final concentrations ranging from 6.54 to 654 mg of Zn2+ per 1 kg of soil) and then shaken on vortex for 1 h. Non-spiked soil sample (MilliQ water added instead of ZnCl2 solution) was used as a background control to take into account potential changes in the luminescence not caused by zinc. This includes potential interference of the sample matrix (stimulation or quenching of the luminescence) due to the presence of nutrients, toxicants or turbidity of the sample, and changes in the luminescence due to the changes in the physiological state of the cell. Samples were analyzed immediately and stored for further measurements at −20°C.
For the induction assay 50 µl of the soil suspension was plated into a 96-well microplate (Thermo Labsystems, Finland) and mixed with an equal volume of a bacterial suspension at an early exponential growth phase (OD600 ∼ 0.015…0.020). Plates were then incubated for 3 h at 30°C, and luminescence was measured with a Victor Wallac 1420 multilabel counter. All measurements were obtained using two replicates in three independent experiments. Sample specific IC was calculated by dividing luminescence measured in spiked sample with luminescence measured in non-spiked sample at respective dilution. The soil sample was considered to induce the bioreporter if higher IC than LODIC (see above) was achieved.
Note that the measurement protocol causes 20-fold dilution of the initial sample (10-fold in preparation of suspension and additional two-fold in actual measurement). The soil suspensions which induced the bioreporters were further diluted up to 10,000-fold with MilliQ water in order to achieve IC close to LODIC.
Acknowledgements
This study was financially supported by the EnSTe Graduate School and Academy of Finland. We thank Dr. Teemu Hynninen for fruitful discussions and Leena Pitkänen for a critical reading of the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/10902
References
- 1.Lampinen J, Virta M, Karp M. Comparison of Gram positive and Gram negative bacterial strains cloned with different types of luciferase genes in bioluminescence cytotoxicity tests. Environmental toxicology and water quality. 1995;10:157–166. [Google Scholar]
- 2.Boyd EM, Killham K, Wright J, Rumford S, Hetheridge M, Cumming R, et al. Toxicity assessment of xenobiotic contaminated groundwater using lux modified Pseudomonas fluorescens. Chemosphere. 1997;35:1967–1985. doi: 10.1016/s0045-6535(97)00271-3. [DOI] [PubMed] [Google Scholar]
- 3.Van Dyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, LaRossa RA. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkin S, Smulski DR, Vollmer AC, Van Dyk TK, LaRossa RA. Oxidative stress detection with Escherichia coli harboring a katG'::lux fusion. Appl Environ Microbiol. 1996;62:2252–2256. doi: 10.1128/aem.62.7.2252-2256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechor O, Smulski DR, Van Dyk TK, LaRossa RA, Belkin S. Recombinant microorganisms as environmental biosensors: pollutants detection by Escherichia coli bearing fabA'::lux fusions. J Biotechnol. 2002;94:125–132. doi: 10.1016/s0168-1656(01)00423-0. [DOI] [PubMed] [Google Scholar]
- 6.Ptitsyn LR, Horneck G, Komova O, Kozubek S, Krasavin EA, Bonev M, et al. A biosensor for environmental genotoxin screening based on an SOS lux assay in recombinant Escherichia coli cells. Appl Environ Microbiol. 1997;63:4377–4384. doi: 10.1128/aem.63.11.4377-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vollmer AC, Belkin S, Smulski DR, Van Dyk TK, LaRossa RA. Detection of DNA damage by use of Escherichia coli carrying recA'::lux, uvrA'::lux or alkA'::lux reporter plasmids. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King JMH, Digrazia PM, Applegate B, Burlage R, Sanseverino J, Dunbar P, et al. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1990;249:778. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- 9.Köhler S, Belkin S, Schmid RD. Reporter gene bioassays in environmental analysis. Fresenius J Anal Chem. 2000;366:769–779. doi: 10.1007/s002160051571. [DOI] [PubMed] [Google Scholar]
- 10.Belkin S. Microbial whole-cell sensing systems of environmental pollutants. Curr Opin Microbiol. 2003;6:206–212. doi: 10.1016/s1369-5274(03)00059-6. [DOI] [PubMed] [Google Scholar]
- 11.Magrisso S, Erel Y, Belkin S. Microbial reporters of metal bioavailability. Microbial Biotechnology. 2008;1:320–330. doi: 10.1111/j.1751-7915.2008.00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keane A, Phoenix P, Ghoshal S, Lau PC. Exposing culprit organic pollutants: a review. J Microbiol Methods. 2002;49:103–119. doi: 10.1016/s0167-7012(01)00382-7. [DOI] [PubMed] [Google Scholar]
- 13.Hynninen A, Virta M. Whole-cell bioreporters for the detection of bioavailable metals. Adv Biochem Eng Biotechnol. 2009 doi: 10.1007/10_2009_9. [DOI] [PubMed] [Google Scholar]
- 14.Kahru A, Ivask A, Põllumaa L, Kasemets K, Kurvet I, François M, et al. Biotests and biosensors in ecotoxicological risk assessment of field soils polluted with zinc, lead and cadmium. Environ Toxicol Chem. 2005;24:2973–2982. doi: 10.1897/05-002r1.1. [DOI] [PubMed] [Google Scholar]
- 15.Ivask A, Francois M, Kahru A, Dubourguier H-C, Virta M, Douay F. Recombinant luminescent bacterial sensors for the measurement of bioavailability of cadmium and lead in soils polluted by metal smelters. Chemosphere. 2004;55:147–156. doi: 10.1016/j.chemosphere.2003.10.064. [DOI] [PubMed] [Google Scholar]
- 16.Bagdasarian M, Lurz R, Ruckert B, Franklin FC, Bagdasarian MM, Frey J, et al. Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 17.Leedjärv A, Ivask A, Virta M. Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J Bacteriol. 2008;190:2680–2689. doi: 10.1128/JB.01494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoyanov JV, Magnani D, Solioz M. Measurement of cytoplasmic copper, silver and gold with a lux biosensor shows copper and silver, but not gold, efflux by the CopA ATPase of Escherichia coli. FEBS Lett. 2003;546:391–394. doi: 10.1016/s0014-5793(03)00640-9. [DOI] [PubMed] [Google Scholar]
- 19.Tauriainen S, Virta M, Chang W, Karp M. Measurement of firefly luciferase reporter gene activity from cells and lysates using Escherichia coli arsenite and mercury sensors. Anal Biochem. 1999;272:191–198. doi: 10.1006/abio.1999.4193. [DOI] [PubMed] [Google Scholar]
- 20.Selifonova O, Burlage R, Barkay T. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl Environ Microbiol. 1993;59:3083–3090. doi: 10.1128/aem.59.9.3083-3090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon KP, Misra TK, Silver S. Regulation of the cadA cadmium resistance determinant of Staphylococcus aureus plasmid pI258. J Bacteriol. 1991;173:7643–7649. doi: 10.1128/jb.173.23.7643-7649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rensing C, Sun Y, Mitra B, Rosen BP. Pb(II)- translocating P-type ATPases. J Biol Chem. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- 23.Brocklehurst KR, Megit SJ, Morby AP. Characterisation of CadR from Pseudomonas aeruginosa: a Cd(II)-responsive MerR homologue. Biochem Biophys Res Commun. 2003;308:234–239. doi: 10.1016/s0006-291x(03)01366-4. [DOI] [PubMed] [Google Scholar]
- 24.Lee SW, Glickmann E, Cooksey DA. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl Environ Microbiol. 2001;67:1437–1444. doi: 10.1128/AEM.67.4.1437-1444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaRossa RA, Smulski DR, Van Dyk TK. Interaction of lead nitrate and cadmium chloride with Escherichia coli K-12 and Salmonella typhimurium global regulatory mutants. J Ind Microbiol. 1995;14:252–258. doi: 10.1007/BF01569936. [DOI] [PubMed] [Google Scholar]
- 26.IUPAC, author. Compendium of Chemical Terminology. 2nd ed. Oxford: Blackwell Scientific Publications; 1997. Compiled by McNaught AD, Wilkinson A. XML on-line corrected version: http://goldbook.iupac.org (2006) created by Nic M, Jirat J, Kosata B. [Google Scholar]
- 27.Biddlecombe RA, Law B. Validation of an immuno assay. In: Law B, editor. Immunoassay: A Practical Guide. Bristol, PA: Taylor & Francis; 1996. pp. 176–178. [Google Scholar]
- 28.Ivask A, Rõlova T, Kahru A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol. 2009;9:41. doi: 10.1186/1472-6750-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivask A, Virta M, Kahru A. Construction and use of specific luminescent recombinant bacterial sensors for the assessment of bioavailable fraction of cadmium, zinc, mercury and chromium in the soil. Soil Biol Biochem. 2002;34:1439–1447. [Google Scholar]
- 30.Riether K, Dollard M-A, Billard P. Assessment of heavy metal bioavailability using Escherichia coli zntAp::lux and copAp::lux-based biosensors. Appl Microbiol Biotechnol. 2001;57:712–716. doi: 10.1007/s00253-001-0852-0. [DOI] [PubMed] [Google Scholar]
- 31.Tauriainen S, Karp M, Chang W, Virta M. Luminescent bacterial sensor for cadmium and lead. Biosens Bioelectron. 1998;13:931–938. doi: 10.1016/s0956-5663(98)00027-x. [DOI] [PubMed] [Google Scholar]