Abstract
Humans and mice with loss-of-function mutations of the genes encoding kisspeptins (Kiss1) or kisspeptin receptors (Kiss1r) are infertile due to hypogonadotropic hypogonadism. Within the hypothalamus, Kiss1 mRNA is expressed in the anteroventral periventricular nucleus (AVPV) and the arcuate nucleus (Arc). In order to better study the different populations of kisspeptin cells we generated Kiss1-Cre transgenic mice. We obtained one line with Cre activity specifically within Kiss1 neurons (line J2-4), as assessed by generating mice with Cre-dependent expression of green fluorescent protein or β-galactosidase. Also, we demonstrated Kiss1 expression in the cerebral cortex and confirmed previous data showing Kiss1 mRNA in the medial nucleus of amygdala and anterodorsal preoptic nucleus. Kiss1 neurons were more concentrated towards the caudal levels of the Arc and higher leptin-responsivity was observed in the most caudal population of Arc Kiss1 neurons. No evidence for direct action of leptin in AVPV Kiss1 neurons was observed. Melanocortin fibers innervated subsets of Kiss1 neurons of the preoptic area and Arc, and both populations expressed MC4R. Specifically in the preoptic area, 18–28% of Kiss1 neurons expressed MC4R. In the Arc, 90% of Kiss1 neurons were glutamatergic, 50% of which also were GABAergic. In the AVPV, 20% of Kiss1 neurons were glutamatergic whereas 75% were GABAergic. The differences observed between the Kiss1 neurons in the preoptic area and the Arc likely represent neuronal evidence for their differential roles in metabolism and reproduction.
Keywords: leptin, melanocortin, Cre-recombinase, reporter mouse, GAD-67, vGluT2
1. Introduction
Humans and mice with loss-of-function mutations of the gene encoding kisspeptins (Kiss1) or kisspeptin receptors (Kiss1r) are infertile due to hypogonadotropic hypogonadism. They show abnormal sexual maturation and decreased circulating levels of sex steroids and gonadotropins (de Roux et al., 2003, Seminara et al., 2003, d’Anglemont de Tassigny et al., 2007, Lapatto et al., 2007). In many species, administration of kisspeptins induce a rapid increase in luteinizing hormone (LH) secretion – an effect mediated via direct action on gonadotropin releasing hormone (GnRH) neurons (Gottsch et al., 2004, Navarro et al., 2004a, Dhillo et al., 2005, Plant et al., 2006).
Kiss1 is expressed in different organs and tissues including pancreas, gonads, placenta and brain (Ohtaki et al., 2001, Colledge, 2008). Within the central nervous system, Kiss1 is expressed in neurons of several hypothalamic nuclei, including the anteroventral periventricular nucleus (AVPV), the anterior periventricular nucleus (PeN) and the arcuate nucleus (Arc) (Gottsch et al., 2004). In these sites, virtually all Kiss1 neurons colocalize sex steroids receptors, particularly estrogen receptor α (ERα) and androgen receptors and are differentially modulated by changing levels of gonadal steroids. For example, high estrogen levels stimulate Kiss1 gene expression in the AVPV and decrease Kiss1 expression in the Arc (Smith et al., 2005a, Smith et al., 2007, Gottsch et al., 2009). In cycling females, increases in estrogen levels induce a GnRH surge, which is observed as an increase in the frequency of pulses and sustained high levels of GnRH secretion; this response in turn triggers an LH surge and ovulation (Levine et al., 1982, Levine and Ramirez, 1982, Moenter et al., 1992, Caraty et al., 1995, Herbison, 2008). Thus, Kiss1 in the AVPV is thought to mediate the estrogen positive feedback action on LH secretion. In contrast, Kiss1 expression in the Arc is high in conditions of low estrogen levels, suggesting that these Kiss1 Arc neurons relay the estrogen negative feedback action on LH secretion (Smith et al., 2005a, Dungan et al., 2006, Popa et al., 2008, Gottsch et al., 2009). As defined in estrogen receptors knockout mice, the estrogen positive and negative feedback action on GnRH secretion are mediated by ERα (Couse et al., 2003, Wintermantel et al., 2006). Therefore binding of estrogen to ERα is thought to drive the presumably distinct kisspeptin action on LH secretion (Smith et al., 2005a, Dungan et al., 2006, Popa et al., 2008, Gottsch et al., 2009). However, the mechanism by which kisspeptin released from different populations of neurons mediate both the negative and positive feedback action of estrogen on LH secretion has not been determined.
A role for kisspeptin in pubertal development has also been described. Hypothalamic levels of Kiss1r and Kiss1 increase during sexual maturation, and administration of kisspeptin to juvenile rodents precipitates puberty (Navarro et al., 2004a, Navarro et al., 2004b, Han et al., 2005). In addition, electrophysiological responses of GnRH neurons to kisspeptin increase across puberty (Han et al., 2005). However, the signals that impinge on Kiss1 neurons to drive their development during puberty initiation are not known. Several studies have suggested that one possible candidate is the adipocyte-derived hormone leptin. Leptin receptors are expressed in a subpopulation of Kiss1 neurons and leptin signaling-deficient mice and humans are hypogonadotropic hypogonadal, remaining in a prepubertal state (Coleman, 1978, Zhang et al., 1994, Tartaglia et al., 1995, Farooqi et al., 1999, Smith et al., 2006). However, leptin’s effects on the various Kiss1 neuronal populations have been inconsistently reported. In one study, obese leptin-deficient (ob/ob) male mice showed decreased expression of Kiss1 in the Arc, which was partially restored by leptin treatment (Smith et al., 2006). In another study, total hypothalamic expression of Kiss1 (which includes the AVPV, the PeN and the Arc) was not changed in ob/ob mice following leptin administration, except when matched with food restricted ob/ob control mice (Luque et al., 2007). In a third study, following fasting (a state of low leptin levels), female rats showed decreases in Kiss1 expression in the AVPV but not in the Arc (Kalamatianos et al., 2008).
Thus, in order to better study and determine the characteristics of specific populations of Kiss1 neurons, we have generated new transgenic mouse lines in which Cre recombinase is expressed within Kiss1 cells. We created reporter mice in which Cre-mediated expression of green fluorescent protein (GFP) or β-galactosidase (βGal) marks the location of Kiss1 neurons. Since the neurotransmitters GABA and glutamate have been identified as fundamental players in GnRH neuron activity and in steroidal feedback control of GnRH secretion (Clarkson and Herbison, 2006, Moenter et al., 2009), we postulated that differences in GABA and glutamate synthesis and neurotransmission reflect the potential roles played by the distinct populations of Kiss1 neurons in GnRH secretion and puberty initiation. In addition, we aimed at defining the subpopulations of Kiss1 neurons directly responsive to leptin and those comprising likely downstream targets of leptin-responsive neurons.
2. Experimental Procedures
2.1 Subjects
Adult male and female Kiss1-Cre, LepR-IRES-Cre/LacZ, MC4R-GFP and C57BL/6 mice were housed in the University of Texas Southwestern Medical Center Animal Resource Center, in a light- (12h on/12h off) and temperature- (21–23°C) controlled environment. They were fed standard chow diet (Harlan Teklad Global Diet), unless otherwise mentioned and had free access to water. All experiments were carried out in accordance with the guidelines established by the National Institute of Health Guide for the Care and Use of Laboratory Animals, as well as with those established by the University of Texas Institutional Animal Care and Use Committee.
2.2 Generation of Kiss1-Cre BAC transgenic mice
We generated several lines of transgenic mice that express Cre recombinase eutopically within kisspeptin-expressing cells, and which we called J2-3, J2-4 and J2-6. These animals were made through the use of various ET-cloning “recombineering” technologies (Lee et al., 2001, Muyrers et al., 2001). We constructed two different Kiss1-Cre transgene-containing bacterial artificial chromosomes (BACs). The original Kiss1 BACs were purchased from BACPAC Resources Center at Children’s Hospital Oakland Research Institute. Both spanned the entire coding region of Kiss1 gene. The first BAC (RP24-186J14) contained approximately 88.03 kb sequence upstream of the Kiss1 start codon and approximately 86.26 kb sequence downstream of the Kiss1 stop codon. The second BAC (RP24-299B2) contained approximately 109.49 kb sequence upstream of the Kiss1start codon and approximately 69.01 kb sequence downstream of the Kiss1 stop codon. These BACs were transformed into EL250 cells by electroporation. EL250 cells were provided by N. Copeland; they contain heat-inducible recE and recT recombinases for homologous recombination and arabinose-inducible Flp-recombinase for site-specific recombination at frt sites (Lee et al., 2001). Next, a DNA fragment containing the coding sequence of Cre recombinase followed by an SV40 polyadenylation (polyA) signal and a kanamycin resistance gene flanked by frt sites (FKF) was inserted into the Kiss1 BACs, at the translational start site of Kiss1, by ET-cloning. The construction of the Cre-polyA-FKF cassette was described previously (Dhillon et al., 2006). This insertion also resulted in the replacement of the entire coding region of Kiss1 and an additional 48-bp downstream of the Kiss1 stop codon. Finally, the kanamycin resistance gene was removed by arabinose induction of Flp-recombinase, and the Cre recombinase coding region was sequenced to ensure that no mutations had been introduced. The Cre-modified Kiss1 BACs were submitted to the UTSW Medical Center Transgenic Core Facility for microinjection into pronuclei of fertilized one-cell stage embryos of C57BL/6 mice. Oligonucleotide primers used to confirm the genotype of mice harboring the Kiss1-Cre transgenes were as follows: M358: 5′-GCTCTGGTGAAGTACGAACTCTGA-3′ and M247: 5′-TGCGAACCTCATCACTCGTTGCAT-3′. The mice used in this study were on a pure C57BL/6 genetic background (Fig. 1).
Figure 1.
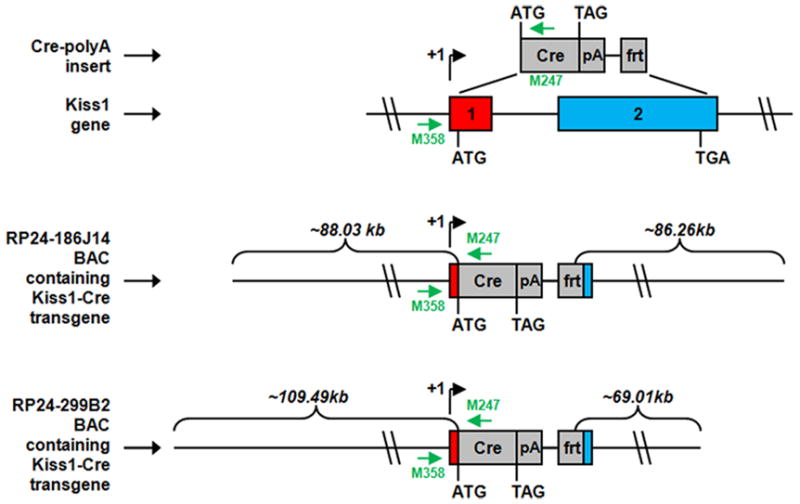
Schematic diagram of the derivation of Kiss1-Cre transgenic mice. In order to generate the transgenes, the entire coding region of Kiss1 and an additional 48-bp downstream of the Kiss1 stop codon were replaced by the coding sequence of Cre recombinase followed by an SV40 polyadenylation signal (pA) and an frt site. The J2-3, J2-4 and J2-6 lines were derived from the RP24-299B2 Kiss1 BAC. Kiss1’s and the transgenes’ transcriptional start sites (+1), Kiss1’s exons (denoted by numbered rectangles), and the start codons (ATG) and stop codons (TGA or TAG) for Kiss1 and Cre, respectively, are all indicated. The amounts of mouse chromosomal sequence present in the BACs and either upstream of Kiss1’s start codon or downstream of Kiss1’s stop codon also are indicated. Small green arrows denote the locations of oligonucleotide primers used for genotyping.
2.3 Validation of Kiss1-Cre mouse models
In order to validate our lines, we crossed the Kiss1-Cre mice with reporter mice that express either GFP [B6.Cg-Tg(ACTB-Bgeo/GFP)21Lbe/J; Jackson Labs] or βGal [B6.129S4-Gt(ROSA)26--Sortm1Sor/J; Jackson Labs] in a Cre-dependent manner (Srinivas et al., 2001, Scott et al., 2009). The lines demonstrating Cre activity (as assessed by Cre-mediated expression of GFP and/or βGal) in the hypothalamic arcuate nucleus (Arc) and anteroventral periventricular nucleus (AVPV, line J2-4) were used in additional histochemical experiments for assessment of colocalization of Cre activity and Kiss1 mRNA expression. Adult (60-day old) males and females from each line (n = 8 females and n = 4 males per line) were perfused in the afternoon (2:00 to 4:00 pm). Females were further divided into two groups: those ovariectomized 15 days prior perfusion (n = 4) and those normally cycling (n = 4). Brains were dissected, cryoprotected overnight and cut in the frontal plane into 25-μm sections on a freezing microtome. Five series were collected into antifreeze solution and stored at −20°C.
2.4 Quantitative RT-PCR (qPCR)
In order to further assess the expression of Kiss1 mRNA, we collected tissue from specific brain sites from wild-type C57BL/6 mice (n=3 males and n=3 females). These sites included olfactory bulb, cerebral cortex, amygdala, hypothalamus, cerebellum and brainstem. Mice were anesthetized (chloral hydrate 7% ip.) and decapitated, brains were removed and blocks containing the specific sites were collected using neuroanatomical landmarks, as reference. Olfactory bulb was collected by an incision made immediately before the anterior fissure at the anterior edge of the frontal cortex. Cerebral cortex was collected by cutting the dorsal cortical mantle, which comprises the motor cortex and medial subfield of the somatosensory cortex. Ventral limits were defined by the corpus callosum. Amygdala was collected following an incision lateral to the optic tract and ventral to the rhinal fissure. Part of the piriform cortex was also included in this block. Hypothalamic blocks were limited by an incision 1 mm anterior to the optic chiasm and another immediately posterior to the mammillary bodies. Lateral limits were defined by the optic tract, and superior limits were defined by the dorsal tip of the third ventricle. We attempted to avoid thalamic tissue, but some contamination may have occurred. The cerebellum was obtained by harvesting the cerebellar cortex, having the forth ventricle as the ventral limit. We expect to have included the cerebellar nuclei as well. The brainstem was collected after incision made immediately caudal to the mammillary bodies and isolation of the cerebellar tissue.
Total Kiss1 mRNA levels were determined by quantitative RT- PCR (qPCR). RNA was extracted using TRIzol reagent (Invitrogen) and 2 μg of total RNA was incubated in DNase I (Roche Diagnostics) for 30 min at 37°C. Subsequently, cDNA was generated using random hexamers (Roche Diagnostics) and SuperScript II Reverse Transcriptase (Invitrogen). We used SYBR Green PCR master mix (Applied Biosystems) for qPCR analysis, and the assays were performed using an Applied Biosystems Prism 7900 HT sequence detection system. The mRNA contents were normalized to cyclophilin content (forward primer: 5′-TGGAGAGCACCAAGACAGACA-3′; reverse primer: 5′-TGCCGGAGTCGACAATGAT - 3′), and the resulting values were expressed as fold change above control levels. The primers for Kiss1 mRNA quantification were 5′-GGCAAAAGTGAAGCCTGGAT-3′ (forward) and 5′-GATTCCTTTTCCCAGGCATT-3′ (reverse). Data are expressed as mean ± SEM.
2.5 Leptin administration
To identify those hypothalamic Kiss1 neurons directly targeted by circulating leptin, we initially assessed the distribution of leptin-induced phosphorylation of STAT3 (pSTAT3) in C57BL/6 female mice. These mice were treated with recombinant murine leptin (provided by Dr. A.F. Parlow, Harbor-UCLA Medical Center, Torrance, CA-USA; through the National Hormone and Peptide Program) or pyrogen free saline (PFS, Sigma). Estrous cycle was monitored and, on diestrus, mice were fasted for 24h before leptin or PFS treatment. We compared the distribution of pSTAT3 following 2 different pharmacological doses of leptin (2.5 μg/g or 5.0 μg/g) on brains from mice perfused at 3 different time points following leptin administration, as follows: 45 min after injection (2.5 μg/g n=4; 5.0 μg/g n=8; and PFS n=4), 60 min after injection (5.0 μg/g n=4; and PFS n=3) or 120 min after injection (5.0 μg/g n=4; and PFS n=3). Brains were dissected and cryoprotected overnight. Five series of sections (25- μm thickness, frontal plane) containing the entire brain were collected and processed for histology. A set of Kiss1-Cre reporter (GFP or LacZ, line J2-4) male and female mice were also fasted for 24h and treated with leptin or PFS ip. 45 min before perfusion (5.0 μg/g n=4; and PFS n=3) and 120 min before perfusion (5.0 μg/g n=4/line; and PFS n=3). Kiss1-Cre reporter female mice were also monitored for estrous cyclicity and were fasted on diestrus, 24h prior perfusion.
2.6 Single and dual label immunohistochemistry
GFP- or βGal-immunoreactivity (GFP-ir or βGal-ir) was assessed in series of brain sections from both male and female Kiss1-Cre reporter mice (line J2-4). Briefly, sections were incubated in primary anti-GFP (1:5,000, Aves Labs, cat# GFP-1020) or anti-βGal (1:5,000, Abcam, cat# ab9361) antisera (both made in chicken), diluted in PBS + 0.25% Triton X-100 (Sigma) and 2% normal goat serum (Vector Labs.), overnight at room temperature. The next day, sections were rinsed in PBS and incubated in AlexaFluor 488-conjugated goat anti-chicken secondary antisera (1:250, Invitrogen) for 1h. Sections were mounted onto gelatin-coated slides, air dried and coversliped with Fluoromount medium. Series of sections from Kiss1-Cre/GFP reporter mice were also submitted to standard immunoperoxidase reaction (Rondini et al., 2004). Following incubation in anti-GFP antisera (1:20,000), sections were incubated in biotin-conjugated donkey anti-chicken secondary antisera (1:1,000, Jackson Labs.) and avidin-biotin complex. A peroxidase reaction was performed using 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma) as chromogen and 0.03% hydrogen peroxide dissolved in 0.1 M PBS, pH 7.4, for 2–3 min. Sections were mounted onto gelatin-coated slides, dehydrated, delipidated and coverslipped with DPX mounting medium (Sigma-Aldrich).
Series of brain sections from C57BL/6 female mice treated with leptin or saline were submitted to a standard immunoperoxidase reaction to detect pSTAT3 immunoreactivity (pSTAT3-ir). Sections were pretreated with a solution containing 1% hydrogen peroxide and 1% sodium hydroxide, for 20 min. After rinses in PBS, sections were incubated in 0.3% glycine followed by 0.03% lauryl sulfate for 10 min each, and blocked in 3% normal donkey serum diluted in 0.1 M PBS + 0.25% Triton X-100. The sections were then incubated in antisera against pSTAT3 (1:2,000, Cell Signaling, cat# 9131) at 4° C for 48–72h. This was followed by incubation for 1 h in biotin-conjugated donkey anti-rabbit secondary antisera (1:1,000, Jackson Laboratories) and for 1 h in avidin-biotin complex (1:500, Vector Labs). The tissue was then submitted to immunoperoxidase reaction as described except that in this case we used DAB and 0.5% nickel-ammonium sulfate (Fisher Sci.) as chromogens. Sections were mounted onto gelatin-coated slides, dehydrated in increasing concentration of ethanol, delipidated in xylenes and coverslipped with DPX mounting medium (Sigma-Aldrich).
Series of sections from Kiss1-Cre reporter mice (n=3 females from each line) were incubated in anti-ERα antisera (1:10,000, Upstate, antisera made in rabbit, cat#C1355) or antiβ–Endorphin antisera (βEnd 1:2,000, Phoenix Pharmaceutical, antisera made in rabbit, cat# H-022-33), overnight at room temperature, followed by AlexaFluor-594 conjugated donkey anti-rabbit secondary antisera (1:500, Invitrogen). Subsequently, sections were incubated in anti-GFP or anti-βGal antisera (1:5,000) overnight followed by incubation in AlexaFluor-488 conjugated secondary antisera as previously described. Sections were mounted onto gelatin-coated slides, air dried and coversliped with Fluoromount.
To assess whether Kiss1 neurons coexpress melanocortin receptors type 4 (MC4R), we used brain sections from MC4R-GFP female mice, kindly provided by Dr. Joel K. Elmquist (University of Texas Southwestern Medical Center, Dallas, TX). Series of sections were incubated in anti-GFP antisera (1:5,000) overnight at room temperature followed by AlexaFluor-488 conjugated goat anti-chicken secondary antisera (1:250). Subsequently, sections were incubated in anti-kisspeptin antisera (1:2,000, Millipore antisera made in rabbit, cat#AB9754) and AlexaFluor-594 conjugated secondary antisera. Sections were mounted onto gelatin-coated slides, air dried and coverslipped with Fluoromount.
To assess whether Kiss1 neurons are directly responsive to leptin, brain sections from Kiss1-Cre reporter mice (line J2-4) treated with leptin were submitted to dual label immunohistochemistry to detect GFP-ir or βGAl-ir and pSTAT3-ir. In addition, series of brain sections from female LepR-IRES-Cre reporter (LacZ) mice (n = 4) were submitted to dual label immunofluorescence to detect βGal-ir and kisspeptin-ir. The LepR-IRES-Cre mice were kindly provided by Dr. Jeffrey Friedman (Laboratory of Molecular Genetics and Howard Hughes Medical Institute, Rockefeller University, New York, NY).
The antisera used in the present study are all commercially available, have been validated before and used by different laboratories (Scott et al., 2009); (Zhao et al., 2008); (Lee et al., 2009); (Clarkson et al., 2009, Caron et al., 2010).
2.7 In situ hybridization/dual label immunohistochemistry-in situ hybridization
We performed a set of single-label in situ hybridization histochemistry in one series of sections from Kiss1-Cre reporter lines in order to evaluate if Kiss1 gene expression was intact in our Kiss1-Cre mouse lines (n = 3 females on diestrus/line). In addition, single-label in situ hybridization histochemistry was performed in brain sections from C57BL/6 mice to assess whether melanocortin 4 receptors (MC4R) are expressed in the AVPV of female mice. Prior to hybridization, brain sections were mounted onto SuperFrost plus slides (Fisher Scientific) and pretreated in 0.1 M citric acid pH 6.0 under microwave for 10 min, as described (Zigman et al., 2006, Scott et al., 2009). The riboprobes (Kiss1 and MC4R) were generated by in vitro transcription with 35S-UTP. The 35S-labeled probes were diluted in hybridization solution and brains sections were hybridized overnight at 56°C (Zigman et al., 2006). The next day, slides were incubated in 0.002% RNAse A followed by stringency washes. Sections were dehydrated in increasing concentrations of ethanol and delipidated for 15 minutes in xylenes. After washes in 100% and 95% ethanol, tissue was air-dried, and slides were placed in X-ray film cassettes with BMR-2 film (Kodak, Rochester, NY) for 2–3 days. Slides were then dipped in NTB photographic emulsion (Kodak, VWR), and stored in foil-wrapped slide boxes at 4°C for 3–4 weeks. Slides were developed with Dektel developer (Kodak, VWR), counterstained with thionin, dehydrated in increasing concentration of ethanol, cleared in xylenes and coverslipped with Permaslip.
We performed dual label in situ hybridization/immunohistochemistry to determine coexpression of GFP-ir or βGal-ir with Kiss1, vGluT2, GAD-67 or NPY mRNAs (line J2-4). The dual label in situ hybridization procedure was a modification of that previously reported (Liu et al., 2003, Yamamoto et al., 2003, Scott et al., 2009). Series of sections of Kiss1-Cre reporter mice were rinsed with DEPC-treated PBS for one hour before being pretreated with 1% sodium borohydride (Sigma) for 15 minutes. After washes in DEPC-PBS, tissue was briefly rinsed in 0.1 M TEA, pH 8.0 and incubated for 10 minutes in 0.25% acetic anhydride in 0.1 M TEA. The tissue was then rinsed in DEPC-treated 2× SSC before hybridization. The riboprobes (Kiss1, vGluT2, GAD-67 and NPY) were generated by in vitro transcription with 35S-UTP. The 35S-labeled riboprobes were diluted to 106 cpm/ml in hybridization buffer and applied to the tissue. Sections were incubated overnight at 50°C. The next day, sections were incubated in 0.002% RNase A (Roche Applied Bioscience) for 30 minutes at 37°C. Sections were then submitted to stringency washes followed by immunohistochemistry for detection of GFP-ir or βGal-ir, as previously described, using DAB as chromogen. Sections were mounted onto SuperFrost Plus slides, dehydrated in increasing concentrations of ethanol and delipidated for 15 minutes in xylenes. After washes in 100% and 95% ethanol, the tissue was air-dried, and slides were placed in X-ray film cassettes with BMR-2 film (Kodak, Rochester, NY) for 2–3 days. Hybridization signal was detected following standard autoradiographic protocol as previously described. All probes used in this study have been tested and published before (Elias et al., 1999, Elias et al., 2001, Liu et al., 2003, Tong et al., 2007, Donato et al., 2009, Scott et al., 2009).
2.8 Fluorescence Activated Cell sorting (FACS)
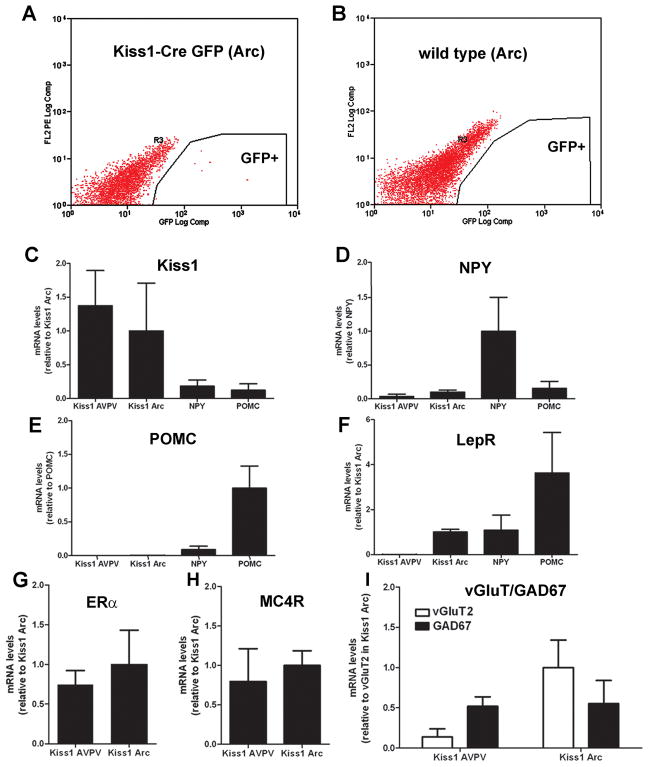
We took advantage of the fluorescence signal transmitted by GFP positive neurons to assess the expression of genes of interest in subpopulations of Kiss1 neurons. Female Kiss1-Cre/GFP mice on diestrus (n = 6) were sacrificed via an overdose of isofluorane. They were quickly decapitated and the brains excised in a manner preserving skull-base hypothalamic structures. The brains were placed in ice cold DEPC-treated PBS on an ice-cold pedestal positioned under a dissecting microscope. Regions comprising the AVPV (incision immediately caudal to the optic chiasm) and the arcuate nucleus (caudal to the optic chiasm to the mammillary body) were acutely dissected away from the rest of the brain tissue using an 11-blade scalpel. Females NPY-hrGFP (n = 3) and POMC-hrGFP (n = 4) kindly provided by Dr. Joel K. Elmquist (University of Texas Southwestern Medical Center, Dallas, TX) were used as control. Wild-type mice were included in each assay as a negative control. Excised regions were placed in cold Hank’s Balanced Salt Solution (HBSS, without calcium or magnesium, Gibco) in a 1.7 ml microcentrifuge tube and kept on ice. The tube was briefly spun down at 13,000 rpm to pellet tissue. HBSS was removed and replaced with a 500 μl of 5% dispase solution in HBSS (v/v, Dispase: BD 354235; HBSS: 14170-112), and the tissue was manually broken up using a pipette and allowed to digest at 37°C for 10 min, with occasional mixing by pipetting. After digestion the tissue was further broken up by manual pipetting and 500 μl of FACS buffer (2.2 ml 45% tissue-culture grade D+glucose; 10 μL 0.5 M EDTA; 30 mg fatty-acid free BSA; in 10 ml HBSS) then was added to neutralize the dispase. The slurry was pelleted at 13,000 rpm for 2 minutes. Supernatant was removed and the pellet was dissolved in 500 μl FACS buffer. Once fully resuspended, the slurry was passed through a cell-strainer topped polypropylene tube (cell strainer tops: BD falcon 5 mL; pp polypropylene tubes: VWR 12×75 mm round bottom).
The UTSW Flow Cytometry Core facility was used to sort GFP positive (GFP+) neurons. GFP was excited by a 488 nm laser. GFP negative (GFP-) neurons were used as control to properly gate a MoFlo flow cytometry and fluorescence activated cell sorter for GFP+ neurons. Live neurons (as gated by forward and side scatter) that were GFP+ were collected directly into 1.7 ml microfuge tubes containing 500 ul of RNAlater solution (Ambion AM7024). Neurons were spun down at 13,000 rpm for 30 minutes, excess RNAlater was removed, and the neurons were frozen at −80°c until RNA extraction.
RNA extraction was done using the PicoPure RNA Isolation kit (Arcturus). All steps described in the MacroCap LCM instructions were followed, except that 100 ul of extraction buffer and 100ul of 70% ethanol were used instead of the volumes listed. Neurons from multiple animals were pooled on-column in order to achieve at least 1000 GFP+ neurons per pool (pools contained a maximum of two animals, n = 3 pools). RNA was extracted using 12 μl of kit elution buffer. All 12 μl of sample, as well as a standard curve (0.5 ug to 16 ng) of universal RNA was subjected to reverse transcription.
Prior to qPCR, cDNA was pre-amplified for genes of interest (Kiss1, NPY, POMC, ERα, LepR, MC4R, vGluT2, GAD67) using 2xTaqMan PreAmp Master Mix (Applied Biosciences, 4384266), according to the manufacturer’s directions, for 18 cycles. The following primers were used: Kiss1 (primer sequences described above); NPY forward: 5′-CGCTCTGCGACACTACATCA – 3′ reverse: 5′-TCTCAGGGCTGGATCTCTTG – 3′; POMC forward: 5′-TGCTTCAGACCTCCATAGATGTGT – 3′ reverse: 5′-GCGAGAGGTCGAGTTTGCA – 3′; LepR forward: 5′-CCCAGCACAATCCAATCACTAG – 3′ reverse: 5′-CAGACGTAGGATGAATAGATGGACTATC – 3′; MC4R forward: 5′-TGAGCCGAACCCAGAAGAG reverse: 5′-AGGAGCAGGGTCAGAAGCA – 3′; ERα forward: 5′-GCAGATAGGGAGCTGGTTCA - 3′ reverse: 5′-TGGAGATTCAAGTCCCCAAA – 3′; vGluT2 forward: 5′-TCACCCAGATTCCAGGAGGAT – 3′ reverse: 5′-CCCAAAGACCCGGTTAGCA 3′; GAD67 forward: 5′ - GGGCTATGTTCCCCTTTATGTC - 3′ reverse: 5′-TTGGATCGAATGCTCCGTAA - 3′.
Pre-amplified cDNA was diluted 20-fold in molecular grade water in 96-well deep-well liquid handler plates, and 384-well qPCR plates were assembled using a robotic liquid handler (Perkin-Elmer, Multiprobe II Nucleic Acid Workstation). Plates were assayed using SYBR Greener reagent (Invitrogen) and an ABI PRISM 7900 HT instrument. Results were calculated using the efficiency-corrected-ΔCt method as described (Bookout et al., 2006). Briefly, individual PCR primer efficiencies were accounted for to allow gene-to-gene comparisons. PCR efficiencies were calculated from the slope of the standard curve. Results were calculated by correcting all Ct values (concentration of amplified product multiplied by time) for primer efficiency. Relative RNA level of a target gene in each sample was computed by normalizing the efficiency-corrected test gene’s Ct values to efficiency-corrected 18S values. Coefficient of variance across efficiency-corrected Ct values of a sample was used to calculate sum of the squared deviations. Data are expressed as mean ± SEM.
2.9 Data analysis and production of photomicrographs
Brain sections were analyzed using a Zeiss Axioplan microscope or a Zeiss Axioscop2 microscope equipped with the ApoTome system. Dual labeled neurons coexpressing GFP-ir or βGal-ir and pSTAT3-ir, βGal-ir and vGluT2 mRNA, βGal-ir and GAD67, GFP-ir and kisspeptin-ir were quantified. Quantification of dual labeled neurons and percentage of colocalization were determined in the AVPV, in the periventricular nucleus (PeN) and in 3 rostro-to-caudal levels of the Arc (fixed distance among them). Cells were counted in one side (right side) of a determined level of each nucleus (Paxinos and Franklin, 2001). For the experiments with in situ hybridization, we considered cells dual-labeled if the density of silver grains (35S-labeled riboprobe) overlying a brown cytoplasm (βGal-ir) was at least 3 × that observed in the background. For background determination, we used the density of silver grains overlying the superior cerebellar peduncle, where no cell bodies were detected. For the experiments performed to detect pSTAT3-ir in Kiss1-Cre/GFP neurons, we considered cells dual-labeled those with brown cytoplasm surrounded by a fluorescent cytoplasm. For the experiments to detect kisspeptin-ir in MC4-GFP neurons, we considered cells dual labeled those with cytoplasm containing both fluorophores detected under epifluorescence microscope. Data are expressed as mean ± SEM. Comparison between the two groups was carried out using the unpaired two-tailed Student’s t test. Statistical analysis was performed using GraphPad Prism software, and an α value of 0.05 was considered in all analyses.
Leptin-induced pSTAT3 immunoreactive neurons were counted in one side of a determined level of brain nuclei in which we detected subjectively an increase in pSTAT3-ir (Paxinos and Franklin, 2001). Data are expressed as mean ± SEM. One-way ANOVA followed by the pairwise Tukey test were used to compare three groups simultaneously. Statistical analysis was performed using GraphPad Prism software, and an α value of 0.05 was considered in all analyses. Cell counting was corrected for double counting by applying Abercrombie’s formula, N =n(T/T+D), where N = the corrected cell count, n = the observed number of cells, T = section thickness (25 μm), and D = the diameter of the nucleus (Guillery, 2002).
To evaluate apparent innervation or close apposition between putative melanocortin terminals (βEnd-ir) and Kiss1 cell bodies (GFP-ir), we used a Zeiss Axioscop2 microscope equipped with the ApoTome system to collect high-resolution images in the Z-axis (Z-stack of 10 optical sections, 0.4 μm intervals, 63 × magnification) and used the Axiovision 3.1 software (Zeiss) to produce 3D rendering of our images.
Drawings were produced using a camera lucida and the Adobe Illustrator CS3 software was used to incorporate drawings into plates. For the topographic distribution of Kiss1 neurons compared to NPY and POMC neurons, adjacent series of sections (n=4 females, line J2-4) were submitted to two distinct dual labeling procedures: a) dual label immunofluorescence (to reveal immnoreactivity for Kiss1 reporter gene/βGal and βEnd) and dual label immunohistochemistry/in situ hybridization (to reveal immunoreactivity for Kiss1 reporter gene/βGal and NPY mRNA expression). Distribution of individual cells was plotted using a camera lucida. Neuroanatomical landmarks were used to determine the equivalent sections in both procedures. Photomicrographs were produced by capturing images with a digital camera (Axiocam, Zeiss) mounted directly on the microscope. Adobe Photoshop CS3 image-editing software was used to integrate photomicrographs into plates. Only sharpness, contrast and brightness were adjusted.
3. Results
3.1 Production and validation of Kiss1-Cre mouse model
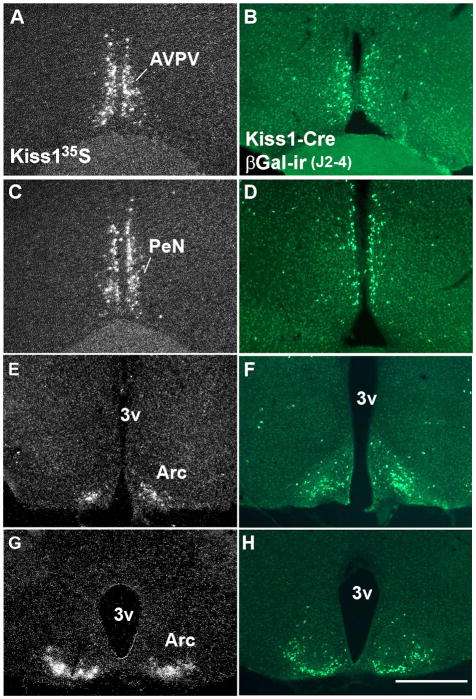
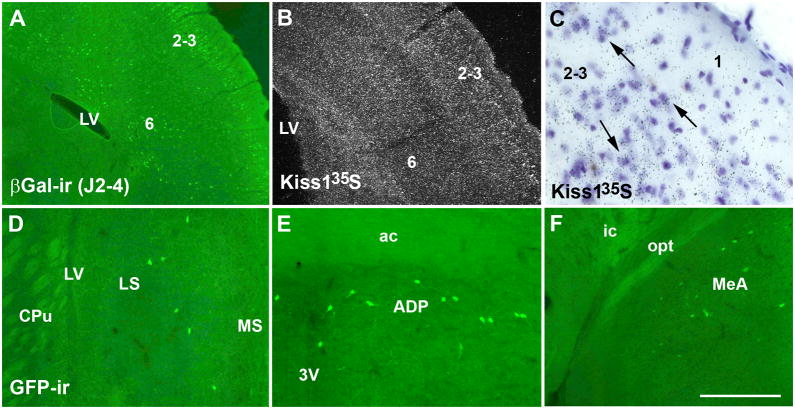
To generate transgenic mice that express Cre-recombinase driven by Kiss1 regulatory elements (Kiss1-Cre mouse model), we constructed two different Kiss1-Cre transgene-containing bacterial artificial chromosomes (BACs) (Fig. 1). We were successful in generating 14 different potential Kiss1-Cre founder mice. After crossing with reporter mice, we identified 3 lines with Cre activity in nuclei previously shown to express Kiss1 mRNA (Gottsch et al., 2004) (Fig. 2A, C, E, G). All 3 lines were originated from founders generated using the RP24-299B2 BAC, and were named J2-3, J2-4 and J2-6. Each of the lines had different amounts of Cre activity noted. In line J2-4, we found GFP- or βGal-immunoreactivity in the AVPV, in the PeN and in the Arc (Fig. 2B, D, F, H). In addition, we also found GFP- or βGal-immunoreactive neurons in the neocortex, in the piriform cortex, in the insular cortex and in the cortical amygdala. In the neocortex, Cre activity was observed mainly in layers 2–3, where Kiss1 mRNA also was observed, and sparcely in layers 5 and 6 (Fig. 3A–C). Very few cells with Cre activity were observed in the lateral septum, the anterodorsal preoptic nucleus, the medial preoptic nucleus, the ventromedial nucleus of the hypothalamus, the medial nucleus of the amygdala and in the nucleus of the solitary tract (Fig. 3D–F).
Figure 2.
Comparative distribution of Kiss1 mRNA in wild-type female mice (perfused on diestrus) and Cre-activity in Kiss1-Cre reporter female mice (line J2-4). A, C, E, G, darkfield photomicrographs of hypothalamic sections showing the distribution of Kiss1 mRNA in the anteroventral periventricular nucleus (AVPV, A), in the anterior periventricular nucleus (PeN, C) and in two rostro-to-caudal levels of the arcuate nucleus (Arc, E, G). B, D, F, H, fluorescent photomicrographs of hypothalamic sections showing the distribution of βGalactosidase immunoreactivity (βGal-ir) in the AVPV (B), in the PeN (D) and in two levels of the Arc (F, H). 3v, third ventricle. Scale bar: A–H = 400 μm.
Figure 3.
Distribution of Cre activity in additional brain sites. A, fluorescent photomicrograph showing βGalactosidase immunoreactivity (βGal-ir) in the cerebral cortex (layers 2–3 and 5–6) of a female mouse from line J2-4. B, darkfield photomicrograph showing the distribution of Kiss1 mRNA in the cerebral cortex (layers 2–3 and 6) of a C57BL/6 female mouse. C, brightfield photomicrograph showing the hybridization signal (silver grains) of Kiss1 riboprobe at cellular level (arrows), in the layers 2–3 of the neocortex. Observe the absence of hybridization signal in the layer 1. D–F, fluorescent photomicrographs showing Cre activity (Kiss1 reporter mice, green fluorescent protein immunoreactivity/GFP-ir) in the lateral septum (LS, D), in the anterodorsal preoptic nucleus (ADP, E), and in the medial nucleus of the amygdala (MeA, F). Abbreviations: 3v, third ventricle; ac, anterior comissure; CPu, caudate putamen; ic, internal capsule; LV, lateral ventricle; MS, medial septum; opt, optic chiasm. Scale bar: A–B = 400 μm, C = 10 μm, D–F = 200 μm.
In line J2-3, we found Cre activity (GFP- or βGal-immunoreactive neurons ) predominantly in neurons of the Arc, and very little in the AVPV and PeN. In line J2-6, Cre activity was found in the AVPV, in the PeN and in the Arc. However, this line also showed high Cre activity in the ventromedial nucleus of the hypothalamus and in the ventral premammillary nucleus. In light of the ectopic Cre expression, this line was not evaluated further in the present study.
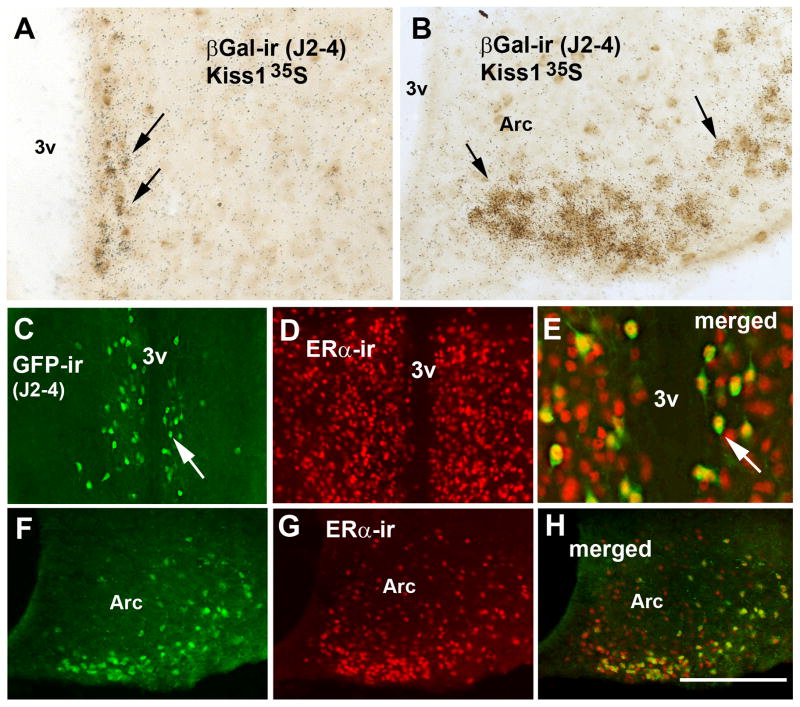
Subsequently, in order to validate the J2-4 line, we performed dual-label histochemistry and assessed the colocalization of Kiss1 mRNA and GFP-ir or βGal-ir in neurons of the AVPV, the PeN and the Arc. Furthermore, to optimize the detection of Kiss1 mRNA, we performed the colocalization studies in both ovariectomized and intact adult female reporter mice (Kiss1-Cre/GFP or Kiss1-Cre/LacZ). No apparent difference between number of GFP-ir and βGal-ir neurons was noticed, in these sites. Virtually all Kiss1 neurons of the AVPV, PeN and Arc expressed Cre-recombinase activity (GFP-ir or βGal-ir, Fig. 4A–B). In addition, we found that few cells of the AVPV, PeN and Arc (7.3 ± 3.7 %) expressed Cre activity alone without binding of the Kiss1 riboprobe. No differences in Cre activity (reporter gene) were observed comparing ovariectomized and non-ovariectomized mice.
Figure 4.
Validation of the Kiss1-Cre mouse model, line J2-4. A–B, brightfield photomicrographs showing the colocalization of βGalactosidase immunoreactivity (βGal-ir, brown cytoplasm) and Kiss1 mRNA (silver grains, arrows) in cells of the anteroventral preoptic nucleus (AVPV, A) and in the arcuate nucleus (Arc, B). C–H, fluorescent photomicrographs showing the colocalization of green fluorescent protein immunoreactivity (GFP-ir, green cytoplasm) and ERα immunoreactivity (ERα -ir, red nuclei) in neurons of the AVPV (female, line J2-4, C–E) and of the Arc (F–H). Note the high number of dual labeled neurons (yellow nuclei) in E and H (arrows in C and E indicate the same cell). Scale bar: A–C = 200 μm. Abbreviations: 3v, third ventricle; ox, optic chiasm; PeN, anterior subdivision of the periventricular nucleus. Scale bar: A–B, E = 100 μm; C–H = 200 μm.
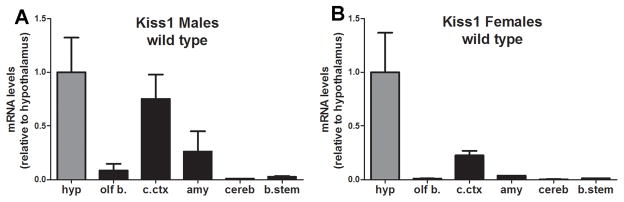
We also assessed Kiss1 mRNA expression in extrahypothalamic sites in which we observed Cre activity. We found low levels of Kiss1 riboprobe hybridization signal in the cerebral cortex (Fig. 3B–C) and amygdala. However, by combining methods (in situ hybridization + immunohistochemistry) we detected inconsistent Kiss1 riboprobe hybridization signal in βGal or GFP immunoreactive neurons in these areas. Thus, in order to assess whether some of these areas represent ectopic expression of Cre-recombinase, we used qPCR to assess Kiss1 mRNA in specific brain sites. We found very little Kiss1 mRNA expression in the brainstem and cerebellum. In contrast, we found moderate to high expression of Kiss1 mRNA in the cerebral cortex and amygdala compared to the hypothalamus (Fig. 5).
Figure 5.
Kiss1 gene expression in male and female wild type mice. A–B, bar graphs showing relative levels of Kiss1 mRNA in various brain sites. Note the moderate levels of Kiss1 mRNA in the cerebral cortex and the low levels in the amygdala. Data shown is mean ± SEM, relative to hypothalamic mRNA levels (Hypothalamus mean Ct values: A = 27.27, B = 25.83; Cerebral Cortex mean Ct values: A = 27.28, B = 27.60). Abbreviations: amy, amygdala; b.stem, brainstem; c.ctx, cerebral cortex; cereb, cerebelo; hyp, hypothalamus; olf b., olfactory bulb.
As an additional step to validate our mouse model, we assessed colocalization between ERα immunoreactivity and Cre activity in our reporter mice. Virtually all neurons in the AVPV, PeN and Arc found to contain Cre activity (GFP-ir or βGal-ir) coexpressed ERα immunoreactivity (Fig. 4C–H).
3.2 Distribution of Kiss1 neurons responsive to leptin
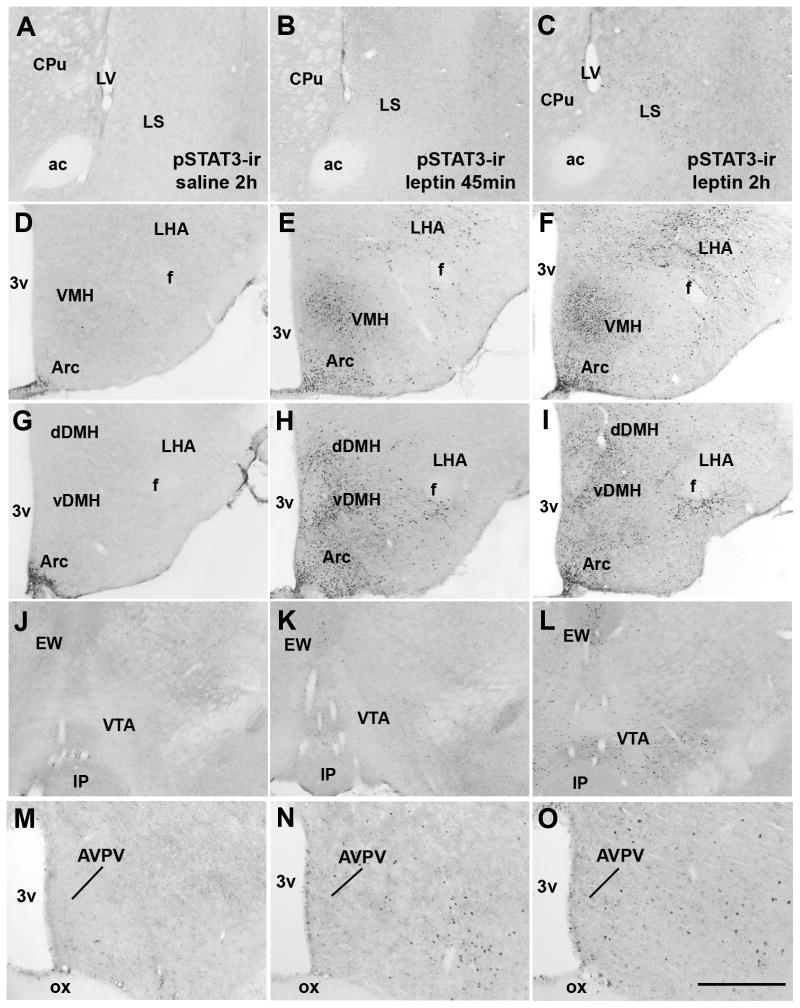
In order to determine whether Kiss1 neurons from the preoptic area and the Arc are directly responsive to leptin we initially performed a systematic analysis of the distribution of leptin receptors and of leptin-induced pSTAT3-ir in female mouse brains. Female mice (on diestrus) treated with 2 different doses of leptin (2.5 μg/g or 5.0 μg/g) and perfused 45 min later showed a similar pSTAT-3-ir distribution to that found in males (Scott et al., 2009, Caron et al., 2010). This includes the medial preoptic area, the Arc, the ventromedial and dorsomedial nuclei of the hypothalamus, and the ventral premammillary nucleus. The same pattern of pSTAT-3 distribution was observed following 60 min of leptin treatment (5.0 μg/g, data not shown). However, mice perfused after 2h of leptin treatment (5.0 μg/g) showed an increase in pSTAT-3-ir in additional (deeper) structures of the brain including the lateral septum, the dorsal subdivision of the dorsomedial nucleus of the hypothalamus, the lateral hypothalamic area and the ventral tegmental area (Fig. 6A–L; Table 1). No apparent difference in the number of leptin-induced pSTAT3-ir was noticed in the Arc, in the ventral subdivision of the dorsomedial nucleus of the hypothalamus and in the ventral premammillary nucleus comparing females perfused after 45 min and those perfused after 2h of leptin treatment. Moreover, we found very little leptin-induced pSTAT3-ir in the AVPV and PeN of female mice following different time points and different doses of ip. leptin (Fig. 6M–O, Fig. 7A).
Figure 6.
Distribution of leptin-induced phosphorylation of STAT3 immunoreactivity (pSTAT3-ir) in the female mouse brain (wild type on diestrus). A–L, brightfield photomicrographs showing the distribution of pSTAT3-ir in various brain sites following intraperitoneal injection of saline or leptin in C57BL/6 mice perfused following 45 min or 2h after the treatment. Note the small number of pSTAT3 immunoreactive neurons in the brains of mice treated with saline (A, D, G, J, M). Also note the increased number of pSTAT3 immunoreactive neurons following 2h of leptin treatment in the lateral septum (LS, B–C), in the lateral hypothalamic area (LHA, E–F, H–I), in the dorsal subdivision of dorsomedial nucleus of the hypothalamus (dDMH, H–I) and in the ventral tegmental area (VTA, K–L). M–O, brightfield photomicrograph showing that very few neurons in the anteroventral periventricular nucleus (AVPV) of female mice display leptin-induced-pSTAT3-ir. Abbreviations: 3v, third ventricle; ac, anterior comissure; CPU, caudate/putamen; EW, Edinger-Westphal nucleus; f, fornix; IP, interpeduncular nucleus; LV, lateral ventricle; ox, optic chiasm; vDMH, ventral subdivision of the dorsomedial nucleus of the hypothalamus; VMH, ventromedial nucleus of the hypothalamus. Scale bar: A–L = 500 μm, M–O = 200 μm.
Table 1.
Number of leptin-induced pSTAT3 immunoreactive neurons in brain nuclei from female mice on diestrus perfused following different time points after leptin administration.
| Regions | atlas levels | Saline 2h | Leptin 45 min | Leptin 2h | significance |
|---|---|---|---|---|---|
| LS | 26 | 0.0 ± 0.0 | 6.320 ± 0.91 | 51.09 ± 4.31* | *P < 0.0001 |
| LHA | 44 | 1.32 ± 0.26 | 32.13 ± 7.82# | 107.2 ± 14.18* | *P < 0.001 |
| DMHd | 46 | 5.26 ± 1.39 | 45.03 ± 3.56# | 102.4 ± 9.95* | *P < 0.0001 |
| VTA | 58 | 0.0 ± 0.0 | 8.43 ± 1.32 # | 54.25 ± 1.84* | *P < 0.0001 |
Values represent mean number of cells ± SEM (n=3 female/treatment). LS: lateral septum; LHA, lateral hypothalamic area; VTA, ventral tegmental area. The atlas level designation corresponds to those described by Paxinos and Franklin, 2001 (Paxinos and Franklin, 2001).
statistically different from saline-treated and leptin-treated 45 min before perfusion;
statistically different from saline-treated group. The atlas level designation corresponds to those described by Paxinos and Franklin, 2001 (Paxinos and Franklin, 2001).
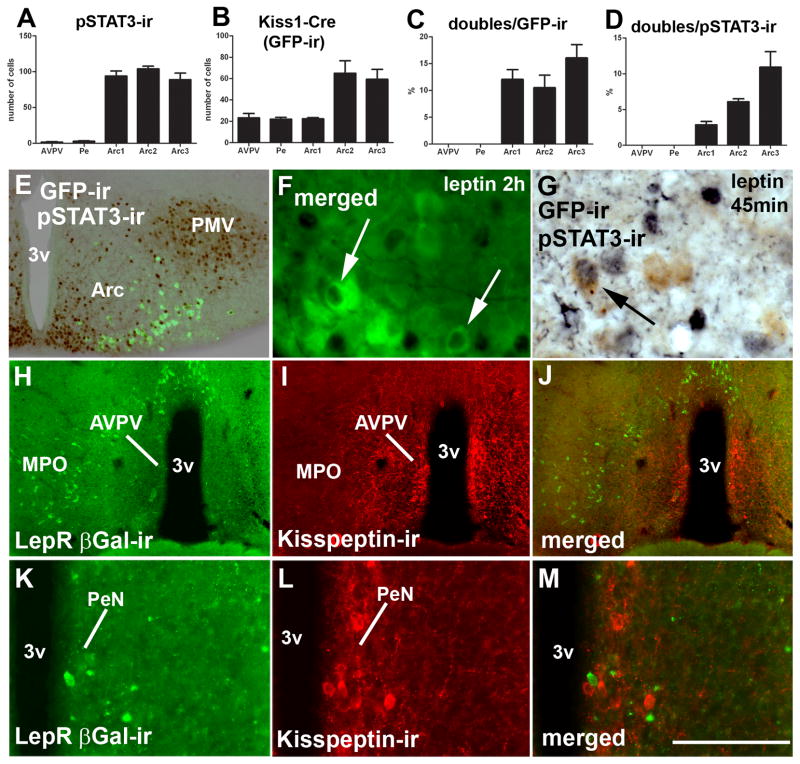
Figure 7.
Distribution of Kiss1 neurons responsive to leptin. A, bar graph showing number of leptin-induced phosphorylation of STAT3 immunoreactivity (pSTAT3-ir) in hypothalamic nuclei which express Kiss1 reporter gene (anteroventral periventricular nucleus/AVPV, anterior periventricular nucleus/PeN, 3 rostro-to-caudal levels of the arcuate nucleus Arc1-3); B, bar graph showing the number of neurons expressing Kiss1 reporter gene (green fluorescent protein immunoreactivity/GFP-ir); C, percentage of Kiss1 neurons expressing pSTAT3-ir; D, percentage of pSTAT3-ir neurons expressing Kiss1 reporter gene. Females on diestrus from line J2-4 were used in this experiment. E, fluorescent and brightfield photomicrographs showing distribution of leptin-induced pSTAT3-ir and Kiss1 neurons (reporter gene), in a caudal level of the Arc. F–G, fluorescent and brightfield photomicrographs showing Kiss1 neurons (reporter gene) expressing leptin-induced pSTAT3-ir (black nucleus, arrows). H–M, fluorescent photomicrographs showing the distribution of leptin receptor reporter gene (LepR, βGalactosidase immunoreactivity/βGal-ir) in the preoptic area. Note the reduced number of neurons expressing LepR reporter gene in the AVPV (H–J) and PeN (K–M), and the absence of colocalization between LepR and kisspeptin immunoreactivity. Abbreviations: 3v, third ventricle; MPO, medial preoptic nucleus; ox, optic chiasm; PMV, ventral premammillary nucleus. Scale bar: E–F = 200 μm; G = 50 μm; H–J, 400 μm; K–M, 100 μm.
To assess whether Kiss1 neurons are directly responsive to leptin we used mice from the J2-4 line, which express the reporter gene in neurons of the preoptic area and the Arc. We found no colocalization of Cre activity and pSTAT3-ir in the AVPV and PeN (Fig. 7B–D). However, we did observe pSTAT3-ir within nearly 15% of Kiss1 neurons of the Arc (Fig. 7D). We also found that 2 to 10% of Arc neurons which express leptin-induced pSTAT3-ir coexpress Kiss1 (Fig. 7B–D, E–G). We noticed a topographic distribution of leptin-responsive Kiss1 neurons, such that higher numbers of dual labeled neurons were found toward the caudal aspect of the Arc (Fig. 7C–D, Fig. 8).
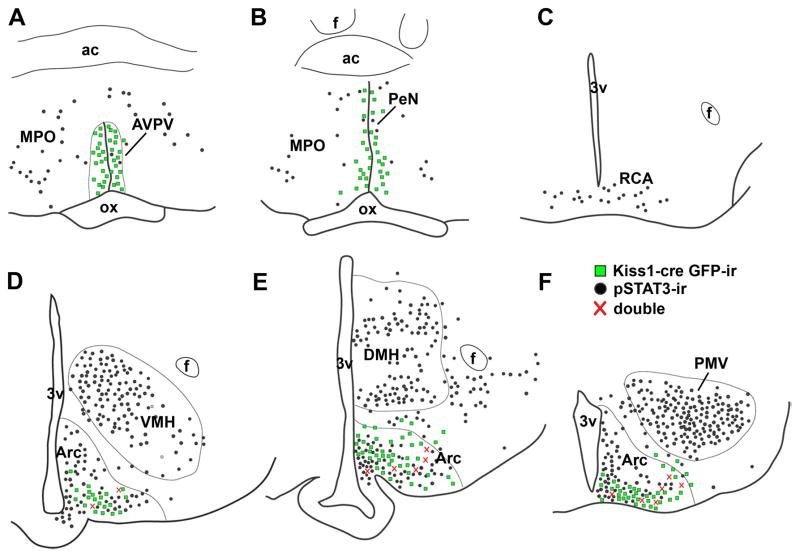
Figure 8.
Distribution of Kiss1 neurons expressing leptin-induced phosphorylation of STAT3 immunoreactivity (pSTAT3-ir). A–F, line drawings of two rostro-to-caudal levels of the preoptic area (A–B), of the retrochiasmatic area (RCA, C) and three rostro-to-caudal levels of the arcuate nucleus (Arc, D–F). We found no colocalization of Kiss1-reporter gene (green fluorescent protein immunoreactivity/GFP-ir, green squares) and leptin-induced pSTAT3-ir (black dots) in the anteroventral periventricular nucleus (AVPV) and in the anterior periventricular nucleus (Pe). Colocalization of Kiss1-reporter gene and leptin-induced pSTAT3-ir was only found in the Arc. We also noticed an increase in number of dual labeled neurons (red crosses) toward the caudal levels of the Arc. Females on diestrus from line J2-4 were used in this experiment. Abbreviations: 3v, third ventricle; ac, anterior comissure; DMH, dorsomedial nucleus of the hypothalamus; f, fornix, MPO, medial preoptic nucleus; ox, optic chiasm; PMV, ventral premammillary nucleus; VMH, ventomedial nucleus of the hypothalamus.
Additionally, we used leptin receptor (LepR) reporter mice (Scott et al., 2009) to identify neurons that express LepRs in the female mouse brain. The pattern of distribution of the LepR reporter gene was very similar to that found in male mice (Scott et al., 2009). Similar to our findings above with leptin-induced pSTAT3-ir, we found very few neurons expressing the LepR reporter gene in the AVPV and PeN of which none coexpressed kisspeptin-ir (Fig. 7H–M).
3.3 Kiss1 neurons are distinct from NPY and POMC neurons
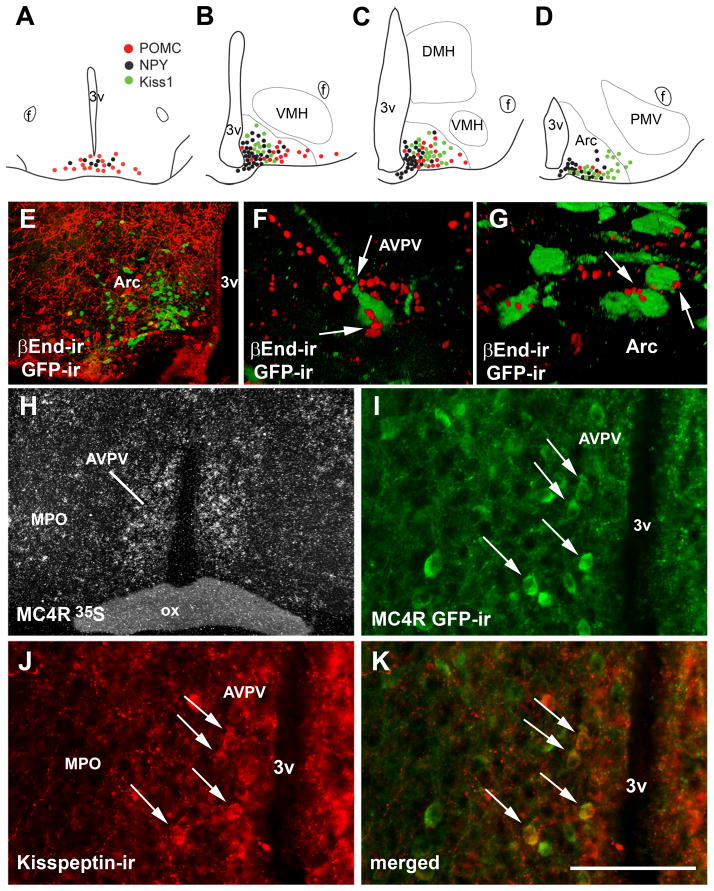
We found that Kiss1, NPY and POMC comprise segregated populations of cells within the Arc (Figure 9A–E). We also noticed that Kiss1, NPY and POMC neurons display a distinct topographic organization throughout the rostro-caudal axis of the mouse Arc. POMC neurons were concentrated in the rostral and tuberal levels, NPY neurons were distributed through the entire extension of the Arc being more concentrated in the intermediate levels, and Kiss1 neurons were found in the intermediate and caudal levels of the Arc (Fig. 9A–D). In addition, we observed βEndorphin (a POMC product)-immunoreactive fibers in close apposition with subsets of Kiss1 neurons of the AVPV and the Arc (Fig. 9F–G).
Figure 9.
Topographic distribution of Kiss1 neurons and their innervation by melanocortins. A–D, schematic drawings showing the topographic distribution of Kiss1 neurons (green dots, line J2-4), proopiomelanocortin neurons (red dots) and neuropeptide Y neurons (black dots) in the retrochiasmatic area (RCA) and 3 rostro-to-caudal levels of the arcuate nucleus (Arc). E, fluorescent photomicrograph showing the segregated distribution of neurons expressing Kiss1 reporter gene (green fluorescent protein immunoreactivity/GFP-ir, green cytoplasm) and βEndorphin immunoreactivity (βEnd-ir, a POMC product, red cytoplasm). F–G, fluorescent photomicrographs showing close appositions between βEnd immunoreactive fibers and GFP immunoreactive neurons, in the anteroventral periventricular nucleus (AVPV, F) and in the Arc (G). H, darkfield photomicrograph showing expression of melanocortin 4 receptor (MC4R) mRNA (hybridization signal, silver grains) in the AVPV of a female mouse. I–K, fluorescent photomicrographs showing the colocalization of MC4R reporter gene (GFP-ir) and kisspeptin immunoreactivity (kisspeptin-ir) in the AVPV (arrows). Abbreviations: 3v, third ventricle; MPO medial preoptic nucleus; ox, optical chiasm. Scale bar: A–D = 600 μm; E, H = 400 μm; I–K = 100 μm, F–G, 50 μm.
3.4 Kiss1 neurons in the AVPV coexpress MC4R
Moderate to dense MC4R expression (mRNA and reporter gene) was noticed in the AVPV of female mice (Figure 9H–I) whereas low MC4R expression was observed in the Arc (data not shown). Using the MC4R-GFP mouse model, we found that 20% of MC4R neurons in the AVPV and PeN coexpresses kisspeptin-ir (21.5 ± 1.5% in the AVPV and 19.9 ± 2.6% in the PeN) whereas 17.3 ± 1.5% of kisspeptin neurons in the AVPV and 28.4 ± 4.9% of kisspeptin neurons in the PeN coexpresses MC4R reporter gene (GFP-ir, Figure 9I–K).
3.5 Kiss1 neurons differentially express vGluT2 and GAD-67 mRNAs
We used the expression of the glutamic acid decarboxylase (a GABA synthesizing enzyme) and vesicular glutamate transporter type 2 (vGluT2) mRNAs as markers for GABA and glutamate expression, respectively. The preoptic area and the Arc expressed moderate to high density of GAD67 and vGluT2 mRNAs (Fig. 10A–D) (Esclapez et al., 1993, Ziegler et al., 2002). In the preoptic area (AVPV and PeN), approximately 20% of Kiss1 neurons (Kiss1-Cre LacZ, line J2-4) coexpressed vGluT2 mRNA and 75% coexpressed GAD-67 mRNA (Fig. 10E). In the Arc, 90% of Kiss1 neurons coexpressed vGluT2 mRNA (Fig. 10F) and 50% coexpressed GAD-67 mRNA (Table 2).
Figure 10.
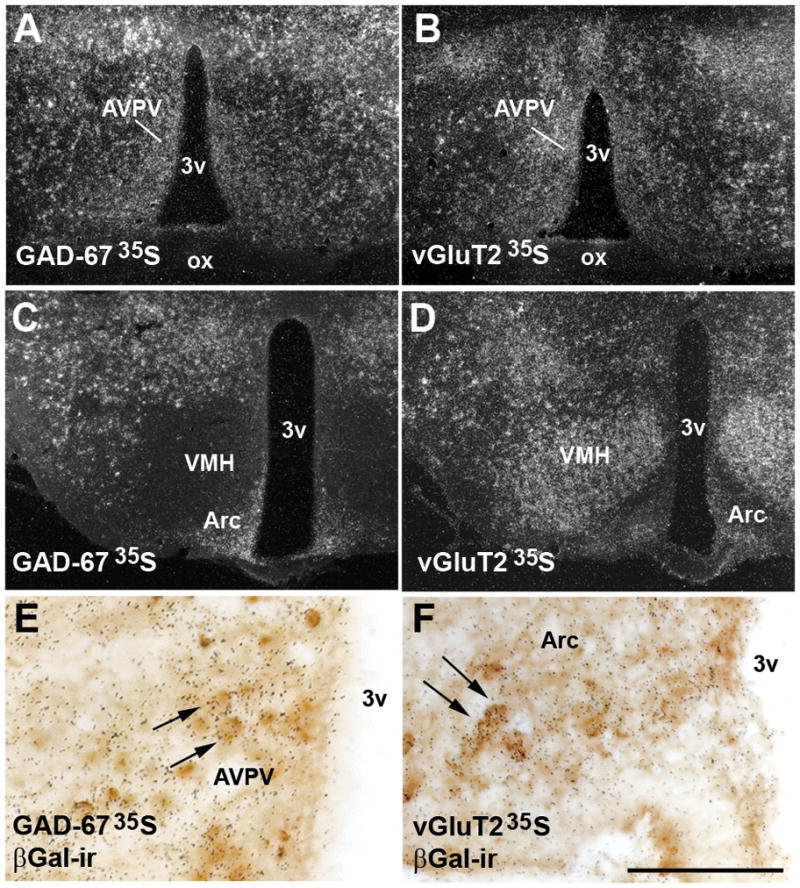
Kiss1 neurons differentially express glutamate and GABA. A, C, darkfield photomicrographs showing the distribution of glutamic acid decarboxylase (GAD-67) mRNA (hybridization signal, silver grains) in the anteroventral preoptic nucleus (AVPV, A) and in the arcuate nucleus (Arc, C). C, brightfield photomicrograph showing colocalization of GAD-67 mRNA (silver grains) and Kiss1 reporter gene (βGalactosidase immunoreactivity/βGal-ir, brown cytoplasm, arrows) in the AVPV (line J2-4). B, D, darkfield photomicrographs showing the distribution of vesicular glutamate transporter type 2 (vGluT2) mRNA (hybridization signal, silver grains) in the AVPV (B) and in the Arc (D). F, brightfield photomicrograph showing colocalization of vGluT2 mRNA (silver grains) and Kiss1 reporter gene (βGal-ir, brown cytoplasm, arrows) in the Arc (line J2-4). Abbreviations: 3v, third ventricle; DMH, dorsomedial nucleus of the hypothalamus; f, fornix; MPO medial preoptic nucleus; ox, optic chiasm; PMV, ventral premammillary nucleus; VMH, ventromedial nucleus of the hypothalamus.. Scale bar: A–B, D–E = 400 μm; C, F = 100 μm.
Table 2.
Percentage of βGal immunoreactive neurons coexpressing GAD-67 or vGluT2 mRNA.
| Regions | atlas levels | % GAD-67/total βGal | % vGluT2/total βGal |
|---|---|---|---|
| AVPV | 28 | 77.4 ± 10.7 | 26.9 ± 13.2 |
| Pe | 30 | 75.1 ± 4.1 | 14.6 ± 2.1 |
| Arc1 | 44 | 57.0 ± 12.8 | 90.7 ± 1.4 |
| Arc2 | 48 | 48.4 ± 4.8 | 85.3 ± 1.7 |
| Arc3 | 52 | 43.6 ± 2.5 | 90.9 ± 0.75 |
Values represent mean percentage of dual labeled cells/total βGal immunoreactive neurons ± SEM (n=3 female, line J2-4). AVPV: anteroventral periventricular nucleus, Pe: anterior periventricular nucleus, Arc1 to Arc3: 3 rostral to caudal levels of the arcuate nucleus. The atlas level designation corresponds to those described by Paxinos and Franklin, 2001 (Paxinos and Franklin, 2001).
3.6 Comparative analysis of gene expression in subpopulations of Kiss1 neurons
We took advantage of the fluorescence signal emitted by Kiss1-Cre neurons from Kiss1-Cre GFP reporter mice to further evaluate our findings. POMC-hrGFP, NPY-hrGFP and wild-type mice were used as controls. Cells with intense fluorescence were collected as the GFP-enriched pools (Fig. 11A). No fluorescence was detected in control wild-types (Fig. 11B). As an initial control, Kiss1, NPY and POMC mRNAs were evaluated in each of the genotypes. As expected, Kiss1 mRNA was very high in GFP-sorted cells from the AVPV and Arc and very low in NPY and POMC cells. The same pattern was observed for NPY and POMC GFP-sorted cells. NPY mRNA was very high in NPY cells and barely detectable in GFP-sorted cells from the Arc of NPY-hrGFP and POMC-hrGFP mice. Similarly, high levels of NPY mRNA or POMC-hrGFP mRNA were amplified only from the GFP-enriched pools isolated from the Arc of NPY-hrGFP mice or POMC-hrGFP mice, respectively (Figure 11C–E). In agreement with our histological data, we found LepR expression in Kiss1 neurons in the Arc but not in those from the AVPV. LepR mRNA was also enriched in NPY and POMC neurons, as expected (Figure 11F). Kiss1 neurons from AVPV and Arc expressed ERα and MC4R mRNAs (Figure 11G–H). These data confirm our colocalization results for the AVPV and indicate that MC4R is also expressed in Kiss1 neurons of the Arc. GAD67 and vGluT2 mRNAs were also detected in both populations of Kiss1 neurons. GAD67 mRNA was 3-fold higher than vGluT2 mRNA in the AVPV, whereas in the Arc VGluT2 mRNA was 2-fold higher than GAD67 mRNA (Figure 11I).
Figure 11.
Expression of genes of interest in GFP neurons collected by fluorescence activated cells sorting (FACS). A–B, graphical representation of FACS of cells from the arcuate nucleus indicating cells collected as part of the GFP-enriched pools (Kiss1-Cre/GFP line J2-4). Observe in B that no fluorescent cell was detected in wild-types. C–F, bar graphs showing relative expression levels of Kiss1 mRNA (A), NPY mRNA (B), POMC mRNA (C) and LepR mRNA (D) in Kiss1-GFP neurons from the AVPV/PeN and Arc, from NPY-GFP neurons and from POMC-GFP neurons. G–I, bar graphs showing relative expression levels of ERα mRNA (E), MC4R mRNA (F), vGluT2 and GAD67 mRNA (F) in Kiss1-GFP neurons from the AVPV/PeN and Arc. Data shown is mean ± SEM, relative mRNA levels compared to Kiss1 mRNA in Kiss1 positive neurons of the Arc (A, mean Ct value = 21.67), to NPY mRNA in NPY positive neurons (B, mean Ct value = 20.54), to POMC mRNA in POMC positive neurons (C, mean Ct value = 17.95), to Kiss1 mRNA in Kiss1 positive neurons of the Arc (D, mean Ct value = 25.31), to ERα mRNA in Kiss1 positive neurons of the Arc (E, mean Ct value = 22.21), to MC4R mRNA in Kiss1 positive neurons of the Arc (F, mean Ct value = 23.27), to vGluT2 mRNA in Kiss1 positive neurons of the Arc.
4. Discussion
In the present study, we describe the generation and validation of a Kiss1-Cre transgenic mouse line (J2-4) which displayed Cre activity in areas known to express Kiss1 mRNA. We then generated Kiss1-reporter mice which allowed us to identify kisspeptin neurons in the mouse brain. We validated our mouse model through colocalization of the reporter gene and Kiss1 mRNA in the preoptic area (AVPV and Pe) and Arc, and confirmed the expression of Kiss1 in other sites, such as the medial nucleus of amygdala and anterodorsal preoptic nucleus (Gottsch et al., 2004). In addition, we observed Cre activity (reporter gene) in the neocortex. Because previous studies have not reported Kiss1 gene expression in this site, we performed additional experiments using two different methodologies (in situ hybridization and qPCR). In both cases, we found moderate expression of Kiss1 in the cerebral cortex. However, our findings must be interpreted with caution as immunohistochemistry to detect the biological active peptide Kp-10 (Kisspeptin 10) have not shown consistent neuronal labeling in this site. Subsequently, we used our reporter mice to identify neurochemical differences between subpopulations of Kiss1 neurons in the preoptic area and Arc. Clear differences were noticed. Whereas Kiss1 neurons in both AVPV and Arc may be directly modulated by estrogen and melanocortins, as indicated by the coexpression of ERα and MC4R within those neurons, leptin acted directly upon Kiss1 neurons of the Arc only, and not upon Kiss1 neurons of the AVPV. Of note, subsets of Kiss1 neurons in the preoptic area expressed MC4R and subsets of Kiss1 neurons in the Arc were direct targets of leptin. Furthermore, a high proportion of Kiss1 neurons in the AVPV were GABAergic (75%), whereas in the Arc they were predominantly glutamatergic (90%). In the Arc, a high percentage of Kiss1 neurons also expressed GAD-67 (50%).
4.1 Sites of Kiss1 expression in the mouse brain
In the current study, our Kiss1 reporter mice were useful in confirming known regions of vigorous (AVPV, PeN and Arc) and less intense (medial nucleus of amygdala and anterodorsal preoptic nucleus) Kiss1 expression (Gottsch et al., 2004). Our Kiss1 reporter mice also showed an intriguing high expression of Cre-recombinase in the neocortex. Our findings indicate that, while cortical Kiss1 expression may not be high from in situ hybridization histochemistry for Kiss1 mRNA alone, Kiss1 expression levels within the cerebral cortex as determined by qPCR and as determined by examination of Kiss1 gene-driven Cre activity do seem confirmatory. However, Cre expression may also reflect transgene ectopic expression and, therefore, further studies will be necessary in order to attest this finding and to assess whether developmental changes, gender and/or physiological conditions contribute to the cortical expression of Kiss1 mRNA and to the apparent elevated expression of Cre activity in the cerebral cortex of these mouse models.
The validation of our Kiss1-Cre mouse line (J2-4) was performed in males and females. Due to the sexually dimorphic nature of the AVPV and of Kiss1 neurons in the AVPV (Bleier et al., 1982, Kauffman et al., 2007) we assessed Cre expression in histological sections from normally cycling females and from those that had been ovariectomized. The line J2-4 was used due to the apparent specific expression of the Kiss1 reporter gene within Kiss1 neurons of the Arc and AVPV/PeN. Lines J2-3 and J2-6 were not examined further in light of the absence of consistent Cre activity in the AVPV/PeN in line J2-3 and some ectopic expression of Kiss1-reporter gene in line J2-6. The manipulation of sex hormone levels was designed to take advantage of previous work demonstrating regulation of Kiss1 expression by sex steroids in both the AVPV and Arc (Smith et al., 2005a, Smith et al., 2005b). In particular, low levels of sex steroids induce an increase in Kiss1 expression in Arc neurons and a decrease in Kiss1 expression in the AVPV neurons. Thus, Cre expression in AVPV neurons (line J2-4) was validated in normally cycling females, whereas Cre expression in Arc neurons was validated in ovariectomized females. Of note, changes in circulating levels of gonadal steroids did not alter the expression of the Kiss1 reporter gene and therefore, we were and will be able to identify Kiss1 neurons in our Kiss1-Cre reporter mice without need to manipulate the levels of gonadal steroids. This is another advantage of using the Kiss1-Cre mouse model instead of standard histological techniques to study the physiological and neurochemical properties of Kiss1 neurons. Moreover, these mouse lines will be of great value to assess the physiological differences between Kiss1 neurons in males versus females.
4.2 Leptin action in kisspeptin neurons
Several reports support the concept that kisspeptin neurons are fundamental players in the initiation of puberty. However, the developmental cues that stimulate the Kiss1 system during pubertal development are still unclear. Studies in rodents and humans have indicated that a female’s fat reserve must exceed a critical threshold to allow the onset of puberty, for normal sexual maturation and for the maintenance of cyclicity (Kennedy, 1969, Frisch and McArthur, 1974, Frisch, 1997). In this respect, the adipocyte-derived hormone leptin has been extensively investigated as a potential signal. Mice lacking leptin (ob/ob) or LepR (db/db) exhibit low LH levels, incomplete development of reproductive organs and do not undergo puberty (Coleman, 1978, Zhang et al., 1994, Tartaglia et al., 1995). Leptin administration to ob/ob mice induces puberty, maturation of reproductive organs and restores fertility (Barash et al., 1996, Chehab et al., 1996, Chehab et al., 1997, Cunningham et al., 1999, Chehab, 2000). Together, these findings have suggested that leptin action in kisspeptin neurons may be the link between individual energy reserves and the initiation of puberty. LepRs are expressed in Kiss1 neurons in the Arc (Smith et al., 2006) but have not been previously described in the AVPV of rats or mice (Mercer et al., 1996, Fei et al., 1997, Elmquist et al., 1998, Scott et al., 1999, Caron et al., 2010). However, a recent study did show pSTAT3-ir in the AVPV of female rats after intracerebroventricular injection of leptin (Quennell et al., 2009), and LepRs were observed in preoptic area Kiss1 neurons of ewes (Backholer et al., 2010).
Here, we have used our reporter mice to better clarify leptin’s action on Kiss1 neurons. Our observation of leptin-induced pSTAT3-ir in Arc Kiss1 neurons, although in diminished proportion, confirms previous findings (Smith et al., 2006). The reason for the difference in the percentage of Kiss1 neurons expressing leptin-induced pSTAT3-ir (the present study) and the percentage of Kiss1 neurons coexpressing LepR is probably due to the use of different techniques (double in situ hybridization versus reporter gene and pSTAT3-ir). Of note, we found no evidence for direct leptin action on Kiss1 neurons of the preoptic area (AVPV or PeN). In this case, we used a combination of 3 different methodologies (reporter genes, immunohistochemistry and qPCR of GFP-sorted cells). Thus, our findings indicate that if leptin has any effects on Kiss1 neurons of the preoptic area of mice, they must be achieved via an indirect route.
One possibility is the NPY and POMC neurons of the Arc, as their role in mediating leptin’s effect on energy homeostasis is well established (Niswender et al., 2004, Elmquist et al., 2005). Thus, we initially determined if the chemical profiles of NPY and/or POMC Arc neurons might include kisspeptin. Using our Kiss1 reporter mice to assess coexpression of Kiss1 in NPY and/or POMC neurons, we showed that these Arc Kiss1 neurons were segregated from NPY and POMC Arc neurons and were located within the most caudal levels of the Arc. This observation will be critical for further studies on the physiological properties and developmental features of kisspeptin neurons.
4.3 Kisspeptin neurons are potential targets of melanocortin system
Melanocortin 4 receptors (MC4R) are important regulators of energy homeostasis (Huszar et al., 1997, Vaisse et al., 1998, Yeo et al., 1998). LepRs are found in melanocortin neurons, and leptin modulates melanocortin expression and activity of MC4R-containing downstream neurons (Cheung et al., 1997, Thornton et al., 1997, Mizuno and Mobbs, 1999, Cone, 2005). Although it is well established that part of leptin action on energy homeostasis is mediated by the melanocortin system, whether melanocortins mediate leptin’s effects on reproduction is controversial. In ob/ob mice, administration of melanocortin agonist (MTII) produced no amelioration of their reproductive malfunction. Likewise, administration of melanocortin antagonist (SHU9119) did not block leptin’s effect to restore fertility (Hohmann et al., 2000). However, in normally fed or leptin-treated fasted rats, SHU9119 decreased the steroid-induced LH surge (Watanobe et al., 1999). POMC or MC4R loss-of-function cause no evident reproductive deficits (Huszar et al., 1997, Vaisse et al., 1998, Yeo et al., 1998), whereas constitutive blockade of melanocortin receptors, as occurs in obese Ay mice, is associated with decreased fertility (Granholm et al., 1986). Together, these observations suggest that the melanocortin system is not required for sexual maturation or fertility, but disruption of the system may cause reproductive deficits.
We then assessed whether kisspeptin neurons might be a target of melanocortins. Terminals from POMC cells were found in close apposition with Kiss1 neurons of the preoptic area and the Arc. This is in line with previous studies showing that small to moderate expression of MC4R is found in the AVPV and in the Arc of male mice (Liu et al., 2003). Here we show that MC4R is also expressed in the AVPV and Arc of female mice and confirmed these POMC innervations in MC4R-GFP reporter mice, in which we identify coexpression of GFP-ir and kisspeptin-ir in a subset of AVPV and PeN neurons. Quantitative PCR on GFP-sorted Arc neurons from Kiss1 reporter mice further confirmed MC4R expression within Arc Kiss1 neurons. Additional studies will be necessary to precisely determine the distribution and percentage of Kiss1 neurons responsive to melanocortins in the Arc. Of note, a recent study in ewes showed that intracerebroventricular delivery of melanocortin agonist (MTII) induced an increase in LH secretion and Kiss1 expression only in the preoptic area, not in the Arc (Backholer et al., 2009). Whether this was a direct effect of melanocortins on Kiss1 neurons is not clear, but our finding indicates that, in mice, the melanocortin system may directly regulate Kiss1 neurons of the preoptic area.
4.4 Differential expression of GAD-67 and vGluT2 in subsets of Kiss1 neurons
We also sought to further elucidate the potential roles of GABA and gluatamate in kisspeptin neurons neurotransmission. It has been determined that the balance and interplay between GABA and glutamate neurotransmission regulates GnRH neuron activity and secretion (Clarkson and Herbison, 2006, Moenter et al., 2009). Neurons containing GABA and glutamate in the AVPV express ERα and project to GnRH neurons (Eyigor et al., 2004, Ottem et al., 2004). Receptors for GABA and glutamate are also enriched in GnRH neurons and their pharmacological manipulation causes significant changes in GnRH and LH secretion (Brann and Mahesh, 1994, Gore, 2001, Mahesh and Brann, 2005, Todman et al., 2005). Although GABA and glutamate are classical inhibitory and excitatory neurotransmitters, respectively, previous work has suggested that GABAergic action upon GnRH neurons is excitatory across sexual development and, although controversial, that GABA also exerts excitatory effects on GnRH neurons in adult life (DeFazio et al., 2002, Han et al., 2002, Han et al., 2004, Moenter and DeFazio, 2005). In the present study we show that a high proportion of Kiss1 neurons in the preoptic area expresses GAD67 and therefore are potentially GABAergic. In the Arc, Kiss1 neurons are nearly 90% glutamatergic but, notably, 50% of Kiss1/vGluT2 neurons of the Arc also coexpress GAD67. A similar pattern was observed in the AVPV of female rats in which a high percentage of neurons coexpress GAD67 and vGluT2 (Ottem et al., 2004). Interestingly, estrogen modifies the number of GABAergic and glutamatergic vesicles in terminals contacting GnRH neurons and modulates GnRH responses to GABA (Ottem et al., 2004, Moenter et al., 2009, Pielecka-Fortuna and Moenter). Future studies will evaluate whether GABA and/or glutamate neurotransmission are important players in Kiss1 function.
In the present study we describe the generation of a novel Kiss1-Cre mouse model. This mouse model enabled us to generate reporters for Kiss1 neurons and to determine the chemical profiles of different populations of Kiss1 neurons. Future studies will take advantage of the Kiss1-specific expression of Cre recombinase to manipulate gene expression in Kiss1 neurons. This will allow us to further elucidate the mechanisms by which these neurons are influenced by changing metabolic states and by which they control reproduction and sexual maturation.
Acknowledgments
We acknowledge the assistance of Robert Hammer and the UTSW Medical Center Transgenic Core Facility in generating the Kiss1-Cre transgenic animals, as well as of Angela Mobley and the UTSW Flow Cytometry Core Facility for the FACS procedure. We also thank Dr. Joyce Repa (UTSW) for Kiss1, Cyclophilin and GAD67 primers. We thank Dr. Jeffrey Friedman (Rockefeller University, New York) for kindly providing the LepR-IRES-Cre mice and Dr. Joel K. Elmquist (UTSW Medical Center, Dallas-TX) for kindly providing the MC4R-GFP, NPY-hrGFP and POMC-hrGFP mice.
Grants information: This work was supported by the NIH grants 1R01HD061539 (to C.F.E.), K08-DK-068069-01A2 and 1R01DA024680 (to J.M.Z.), GM007062 (to A.L.B), by the Coordination of the Advancement of Higher Education/CAPES-Brazil (to L.O.M.), by the National Council for Scientific and Technological Development (CNPq-Brazil) 201804/2008-5 (to R.F.), by the UTSW Medical Center Regent’s Scholar Research Award and President’s Council Research Award (to C.F.E.) and by the UTSW Medical Center Disease-Oriented Clinical Scholars Award (to J.M.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Backholer K, Smith J, Clarke IJ. Melanocortins May Stimulate Reproduction by Activating Orexin Neurons in the Dorsomedial Hypothalamus and Kisspeptin Neurons in the Preoptic Area of the Ewe. Endocrinology. 2009;150:5488–5497. doi: 10.1210/en.2009-0604. [DOI] [PubMed] [Google Scholar]
- Barash IA, Cheung CC, Weigle DS, Ren H, Kabigting EB, Kuijper JL, Clifton DK, Steiner RA. Leptin is a metabolic signal to the reproductive system. Endocrinology. 1996;137:3144–3147. doi: 10.1210/endo.137.7.8770941. [DOI] [PubMed] [Google Scholar]
- Bleier R, Byne W, Siggelkow I. Cytoarchitectonic sexual dimorphisms of the medial preoptic and anterior hypothalamic areas in guinea pig, rat, hamster, and mouse. J Comp Neurol. 1982;212:118–130. doi: 10.1002/cne.902120203. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical Profiling of Nuclear Receptor Expression Reveals a Hierarchical Transcriptional Network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann DW, Mahesh VB. Excitatory amino acids: function and significance in reproduction and neuroendocrine regulation. Front Neuroendocrinol. 1994;15:3–49. doi: 10.1006/frne.1994.1002. [DOI] [PubMed] [Google Scholar]
- Caraty A, Evans NP, Fabre-Nys CJ, Karsch EJ. The preovulatory gonadotrophin-releasing hormone surge: a neuroendocrine signal for ovulation. J Reprod Fertil Suppl. 1995;49:245–255. [PubMed] [Google Scholar]
- Caron E, Sachot C, Prevot V, Bouret SG. Distribution of leptin-sensitive cells in the postnatal and adult mouse brain. The Journal of Comparative Neurology. 2010;518:459–476. doi: 10.1002/cne.22219. [DOI] [PubMed] [Google Scholar]
- Chehab FF. Leptin as a regulator of adipose mass and reproduction. Trends Pharmacol Sci. 2000;21:309–314. doi: 10.1016/s0165-6147(00)01514-5. [DOI] [PubMed] [Google Scholar]
- Chehab FF, Lim ME, Lu R. Correction of the sterility defect in homozygous obese female mice by treatment with the human recombinant leptin. Nat Genet. 1996;12:318–320. doi: 10.1038/ng0396-318. [DOI] [PubMed] [Google Scholar]
- Chehab FF, Mounzih K, Lu R, Lim ME. Early onset of reproductive function in normal female mice treated with leptin. Science. 1997;275:88–90. doi: 10.1126/science.275.5296.88. [DOI] [PubMed] [Google Scholar]
- Cheung CC, Clifton DK, Steiner RA. Proopiomelanocortin neurons are direct targets for leptin in the hypothalamus. Endocrinology. 1997;138:4489–4492. doi: 10.1210/endo.138.10.5570. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Herbison AE. Development of GABA and glutamate signaling at the GnRH neuron in relation to puberty. Mol Cell Endocrinol. 2006;254–255:32–38. doi: 10.1016/j.mce.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Tassigny XdAd, Colledge WH, Caraty A, Herbison AE. Distribution of Kisspeptin Neurones in the Adult Female Mouse Brain. Journal of Neuroendocrinology. 2009;21:673–682. doi: 10.1111/j.1365-2826.2009.01892.x. [DOI] [PubMed] [Google Scholar]
- Coleman DL. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia. 1978;14:141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- Colledge WH. GPR54 and kisspeptins. Results Probl Cell Differ. 2008;46:117–143. doi: 10.1007/400_2007_050. [DOI] [PubMed] [Google Scholar]
- Cone RD. Anatomy and regulation of the central melanocortin system. Nat Neurosci. 2005;8:571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- Couse JF, Yates MM, Walker VR, Korach KS. Characterization of the hypothalamic-pituitary-gonadal axis in estrogen receptor (ER) Null mice reveals hypergonadism and endocrine sex reversal in females lacking ERalpha but not ERbeta. Mol Endocrinol. 2003;17:1039–1053. doi: 10.1210/me.2002-0398. [DOI] [PubMed] [Google Scholar]
- Cunningham MJ, Clifton DK, Steiner RA. Leptin’s actions on the reproductive axis: perspectives and mechanisms. Biol Reprod. 1999;60:216–222. doi: 10.1095/biolreprod60.2.216. [DOI] [PubMed] [Google Scholar]
- d’Anglemont de Tassigny X, Fagg LA, Dixon JPC, Day K, Leitch HG, Hendrick AG, Zahn D, Franceschini I, Caraty A, Carlton MBL, Aparicio SAJR, Colledge WH. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proceedings of the National Academy of Sciences. 2007;104:10714–10719. doi: 10.1073/pnas.0704114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio RA, Heger S, Ojeda SR, Moenter SM. Activation of A-type gamma-aminobutyric acid receptors excites gonadotropin-releasing hormone neurons. Mol Endocrinol. 2002;16:2872–2891. doi: 10.1210/me.2002-0163. [DOI] [PubMed] [Google Scholar]
- Dhillo WS, Chaudhri OB, Patterson M, Thompson EL, Murphy KG, Badman MK, McGowan BM, Amber V, Patel S, Ghatei MA, Bloom SR. Kisspeptin-54 stimulates the hypothalamic-pituitary gonadal axis in human males. J Clin Endocrinol Metab. 2005;90:6609–6615. doi: 10.1210/jc.2005-1468. [DOI] [PubMed] [Google Scholar]
- Dhillon H, Zigman JM, Ye C, Lee CE, McGovern RA, Tang V, Kenny CD, Christiansen LM, White RD, Edelstein EA, Coppari R, Balthasar N, Cowley MA, Chua S, Jr, Elmquist JK, Lowell BB. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron. 2006;49:191–203. doi: 10.1016/j.neuron.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Donato J, Jr, Silva RJ, Sita LV, Lee S, Lee C, Lacchini S, Bittencourt JC, Franci CR, Canteras NS, Elias CF. The Ventral Premammillary Nucleus Links Fasting-Induced Changes in Leptin Levels and Coordinated Luteinizing Hormone Secretion. J Neurosci. 2009;29:5240–5250. doi: 10.1523/JNEUROSCI.0405-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungan HM, Clifton DK, Steiner RA. Minireview: kisspeptin neurons as central processors in the regulation of gonadotropin-releasing hormone secretion. Endocrinology. 2006;147:1154–1158. doi: 10.1210/en.2005-1282. [DOI] [PubMed] [Google Scholar]
- Elias CF, Aschkenasi C, Lee C, Kelly J, Ahima RS, Bjorbaek C, Flier JS, Saper CB, Elmquist JK. Leptin differentially regulates NPY and POMC neurons projecting to the lateral hypothalamic area. Neuron. 1999;23:775–786. doi: 10.1016/s0896-6273(01)80035-0. [DOI] [PubMed] [Google Scholar]
- Elias CF, Lee CE, Kelly JF, Ahima RS, Kuhar M, Saper CB, Elmquist JK. Characterization of CART neurons in the rat and human hypothalamus. J Comp Neurol. 2001;432:1–19. doi: 10.1002/cne.1085. [DOI] [PubMed] [Google Scholar]
- Elmquist JK, Bjorbaek C, Ahima RS, Flier JS, Saper CB. Distributions of leptin receptor mRNA isoforms in the rat brain. J Comp Neurol. 1998;395:535–547. [PubMed] [Google Scholar]
- Elmquist JK, Coppari R, Balthasar N, Ichinose M, Lowell BB. Identifying hypothalamic pathways controlling food intake, body weight, and glucose homeostasis. The Journal of Comparative Neurology. 2005;493:63–71. doi: 10.1002/cne.20786. [DOI] [PubMed] [Google Scholar]
- Esclapez M, Tillakaratne NJ, Tobin AJ, Houser CR. Comparative localization of mRNAs encoding two forms of glutamic acid decarboxylase with nonradioactive in situ hybridization methods. J Comp Neurol. 1993;331:339–362. doi: 10.1002/cne.903310305. [DOI] [PubMed] [Google Scholar]
- Eyigor O, Lin W, Jennes L. Identification of Neurones in the Female Rat Hypothalamus That Express Oestrogen Receptor-Alpha and Vesicular Glutamate Transporter-2. Journal of Neuroendocrinology. 2004;16:26–31. doi: 10.1111/j.1365-2826.2004.01109.x. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, Jebb SA, Langmack G, Lawrence E, Cheetham CH, Prentice AM, Hughes IA, McCamish MA, O’Rahilly S. Effects of Recombinant Leptin Therapy in a Child with Congenital Leptin Deficiency. N Engl J Med. 1999;341:879–884. doi: 10.1056/NEJM199909163411204. [DOI] [PubMed] [Google Scholar]
- Fei H, Okano HJ, Li C, Lee GH, Zhao C, Darnell R, Friedman JM. Anatomic localization of alternatively spliced leptin receptors (Ob-R) in mouse brain and other tissues. Proc Natl Acad Sci U S A. 1997;94:7001–7005. doi: 10.1073/pnas.94.13.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch RE. Critical fatness hypothesis. Am J Physiol. 1997;273:E231–232. doi: 10.1152/ajpendo.1997.273.1.E231. [DOI] [PubMed] [Google Scholar]
- Frisch RE, McArthur JW. Menstrual cycles: fatness as a determinant of minimum weight for height necessary for their maintenance or onset. Science. 1974;185:949–951. doi: 10.1126/science.185.4155.949. [DOI] [PubMed] [Google Scholar]
- Gore AC. Gonadotropin-releasing hormone neurons, NMDA receptors, and their regulation by steroid hormones across the reproductive life cycle. Brain Research Reviews. 2001;37:235–248. doi: 10.1016/s0165-0173(01)00121-7. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Navarro VM, Zhao Z, Glidewell-Kenney C, Weiss J, Jameson JL, Clifton DK, Levine JE, Steiner RA. Regulation of Kiss1 and Dynorphin Gene Expression in the Murine Brain by Classical and Nonclassical Estrogen Receptor Pathways. J Neurosci. 2009;29:9390–9395. doi: 10.1523/JNEUROSCI.0763-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granholm NH, Jeppesen KW, Japs RA. Progressive infertility in female lethal yellow mice (Ay/a; strain C57BL/6J) J Reprod Fertil. 1986;76:279–287. doi: 10.1530/jrf.0.0760279. [DOI] [PubMed] [Google Scholar]
- Guillery RW. On counting and counting errors. J Comp Neurol. 2002;447:1–7. doi: 10.1002/cne.10221. [DOI] [PubMed] [Google Scholar]
- Han S-K, Gottsch ML, Lee KJ, Popa SM, Smith JT, Jakawich SK, Clifton DK, Steiner RA, Herbison AE. Activation of Gonadotropin-Releasing Hormone Neurons by Kisspeptin as a Neuroendocrine Switch for the Onset of Puberty. J Neurosci. 2005;25:11349–11356. doi: 10.1523/JNEUROSCI.3328-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Abraham IM, Herbison AE. Effect of GABA on GnRH neurons switches from depolarization to hyperpolarization at puberty in the female mouse. Endocrinology. 2002;143:1459–1466. doi: 10.1210/endo.143.4.8724. [DOI] [PubMed] [Google Scholar]
- Han SK, Todman MG, Herbison AE. Endogenous GABA release inhibits the firing of adult gonadotropin-releasing hormone neurons. Endocrinology. 2004;145:495–499. doi: 10.1210/en.2003-1333. [DOI] [PubMed] [Google Scholar]
- Herbison AE. Estrogen positive feedback to gonadotropin-releasing hormone (GnRH) neurons in the rodent: The case for the rostral periventricular area of the third ventricle (RP3V) Brain Research Reviews. 2008;57:277–287. doi: 10.1016/j.brainresrev.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann JG, Teal TH, Clifton DK, Davis J, Hruby VJ, Han G, Steiner RA. Differential role of melanocortins in mediating leptin’s central effects on feeding and reproduction. Am J Physiol Regul Integr Comp Physiol. 2000;278:R50–59. doi: 10.1152/ajpregu.2000.278.1.R50. [DOI] [PubMed] [Google Scholar]
- Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, Lee F. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- Kalamatianos T, Grimshaw SE, Poorun R, Hahn JD, Coen CW. Fasting reduces KiSS-1 expression in the anteroventral periventricular nucleus (AVPV): effects of fasting on the expression of KiSS-1 and neuropeptide Y in the AVPV or arcuate nucleus of female rats. J Neuroendocrinol. 2008;20:1089–1097. doi: 10.1111/j.1365-2826.2008.01757.x. [DOI] [PubMed] [Google Scholar]
- Kauffman AS, Gottsch ML, Roa J, Byquist AC, Crown A, Clifton DK, Hoffman GE, Steiner RA, Tena-Sempere M. Sexual differentiation of Kiss1 gene expression in the brain of the rat. Endocrinology. 2007;148:1774–1783. doi: 10.1210/en.2006-1540. [DOI] [PubMed] [Google Scholar]
- Kennedy GC. Interactions between feeding behavior and hormones during growth. Ann N Y Acad Sci. 1969;157:1049–1061. doi: 10.1111/j.1749-6632.1969.tb12936.x. [DOI] [PubMed] [Google Scholar]
- Lapatto R, Pallais JC, Zhang D, Chan Y-M, Mahan A, Cerrato F, Le WW, Hoffman GE, Seminara SB. Kiss1/Mice Exhibit More Variable Hypogonadism than Gpr54/Mice. Endocrinology. 2007;148:4927–4936. doi: 10.1210/en.2007-0078. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee CE, Elias CF, Elmquist JK. Expression of the diabetes-associated gene TCF7L2 in adult mouse brain. The Journal of Comparative Neurology. 2009;517:925–939. doi: 10.1002/cne.22199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JE, Pau KY, Ramirez VD, Jackson GL. Simultaneous measurement of luteinizing hormone-releasing hormone and luteinizing hormone release in unanesthetized, ovariectomized sheep. Endocrinology. 1982;111:1449–1455. doi: 10.1210/endo-111-5-1449. [DOI] [PubMed] [Google Scholar]
- Levine JE, Ramirez VD. Luteinizing hormone-releasing hormone release during the rat estrous cycle and after ovariectomy, as estimated with push-pull cannulae. Endocrinology. 1982;111:1439–1448. doi: 10.1210/endo-111-5-1439. [DOI] [PubMed] [Google Scholar]
- Liu H, Kishi T, Roseberry AG, Cai X, Lee CE, Montez JM, Friedman JM, Elmquist JK. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci. 2003;23:7143–7154. doi: 10.1523/JNEUROSCI.23-18-07143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque RM, Kineman RD, Tena-Sempere M. Regulation of Hypothalamic Expression of KiSS-1 and GPR54 Genes by Metabolic Factors: Analyses Using Mouse Models and a Cell Line. Endocrinology. 2007;148:4601–4611. doi: 10.1210/en.2007-0500. [DOI] [PubMed] [Google Scholar]
- Mahesh VB, Brann DW. Regulatory role of excitatory amino acids in reproduction. Endocrine. 2005;28:271–280. doi: 10.1385/ENDO:28:3:271. [DOI] [PubMed] [Google Scholar]
- Mercer JG, Hoggard N, Williams LM, Lawrence CB, Hannah LT, Trayhurn P. Localization of leptin receptor mRNA and the long form splice variant (Ob-Rb) in mouse hypothalamus and adjacent brain regions by in situ hybridization. FEBS Lett. 1996;387:113–116. doi: 10.1016/0014-5793(96)00473-5. [DOI] [PubMed] [Google Scholar]
- Mizuno TM, Mobbs CV. Hypothalamic Agouti-Related Protein Messenger Ribonucleic Acid Is Inhibited by Leptin and Stimulated by Fasting. Endocrinology. 1999;140:814–817. doi: 10.1210/endo.140.2.6491. [DOI] [PubMed] [Google Scholar]
- Moenter SM, Brand RM, Midgley AR, Karsch FJ. Dynamics of gonadotropin-releasing hormone release during a pulse. Endocrinology. 1992;130:503–510. doi: 10.1210/endo.130.1.1727719. [DOI] [PubMed] [Google Scholar]
- Moenter SM, Chu Z, Christian CA. Neurobiological Mechanisms Underlying Oestradiol Negative and Positive Feedback Regulation of Gonadotrophin-Releasing Hormone Neurones. Journal of Neuroendocrinology. 2009;21:327–333. doi: 10.1111/j.1365-2826.2009.01826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenter SM, DeFazio RA. Endogenous gamma-aminobutyric acid can excite gonadotropin-releasing hormone neurons. Endocrinology. 2005;146:5374–5379. doi: 10.1210/en.2005-0788. [DOI] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Stewart AF. Techniques: Recombinogenic engineering--new options for cloning and manipulating DNA. Trends Biochem Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- Navarro VM, Castellano JM, Fernandez-Fernandez R, Barreiro ML, Roa J, Sanchez-Criado JE, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology. 2004a;145:4565–4574.2. doi: 10.1210/en.2004-0413. [DOI] [PubMed] [Google Scholar]
- Navarro VM, Fernandez-Fernandez R, Castellano JM, Roa J, Mayen A, Barreiro ML, Gaytan F, Aguilar E, Pinilla L, Dieguez C, Tena-Sempere M. Advanced vaginal opening and precocious activation of the reproductive axis by KiSS-1 peptide, the endogenous ligand of GPR54. J Physiol. 2004b;561:379–386. doi: 10.1113/jphysiol.2004.072298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender KD, Baskin DG, Schwartz MW. Insulin and its evolving partnership with leptin in the hypothalamic control of energy homeostasis. Trends in Endocrinology and Metabolism. 2004;15:362–369. doi: 10.1016/j.tem.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, Ishibashi Y, Watanabe T, Asada M, Yamada T, Suenaga M, Kitada C, Usuki S, Kurokawa T, Onda H, Nishimura O, Fujino M. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- Ottem EN, Godwin JG, Krishnan S, Petersen SL. Dual-phenotype GABA/glutamate neurons in adult preoptic area: sexual dimorphism and function. J Neurosci. 2004;24:8097–8105. doi: 10.1523/JNEUROSCI.2267-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin C. The mouse brain in stereotaxic coordinates. Sydney: Academic Press; 2001. [Google Scholar]
- Pielecka-Fortuna J, Moenter SM. Kisspeptin Increases {gamma}-Aminobutyric Acidergic and Glutamatergic Transmission Directly to Gonadotropin-Releasing Hormone Neurons in an Estradiol-Dependent Manner. Endocrinology. 2010;151:291–300. doi: 10.1210/en.2009-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant TM, Ramaswamy S, Dipietro MJ. Repetitive activation of hypothalamic G protein-coupled receptor 54 with intravenous pulses of kisspeptin in the juvenile monkey (Macaca mulatta) elicits a sustained train of gonadotropin-releasing hormone discharges. Endocrinology. 2006;147:1007–1013. doi: 10.1210/en.2005-1261. [DOI] [PubMed] [Google Scholar]
- Popa SM, Clifton DK, Steiner RA. The Role of Kisspeptins and GPR54 in the Neuroendocrine Regulation of Reproduction. Annual Review of Physiology. 2008;70:213. doi: 10.1146/annurev.physiol.70.113006.100540. [DOI] [PubMed] [Google Scholar]
- Quennell JH, Mulligan AC, Tups A, Liu X, Phipps SJ, Kemp CJ, Herbison AE, Grattan DR, Anderson GM. Leptin Indirectly Regulates Gonadotropin-Releasing Hormone Neuronal Function. Endocrinology. 2009;150:2805–2812. doi: 10.1210/en.2008-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondini TA, Baddini SP, Sousa LF, Bittencourt JC, Elias CF. Hypothalamic cocaine- and amphetamine-regulated transcript neurons project to areas expressing gonadotropin releasing hormone immunoreactivity and to the anteroventral periventricular nucleus in male and female rats. Neuroscience. 2004;125:735–748. doi: 10.1016/j.neuroscience.2003.12.045. [DOI] [PubMed] [Google Scholar]
- Scott CJ, Rawson JA, Pereira AM, Clarke IJ. Oestrogen receptors in the brainstem of the female sheep: relationship to noradrenergic cells and cells projecting to the medial preoptic area. J Neuroendocrinol. 1999;11:745–755. doi: 10.1046/j.1365-2826.1999.00370.x. [DOI] [PubMed] [Google Scholar]
- Scott MM, Lachey JL, Sternson SM, Lee CE, Elias CF, Friedman JM, Elmquist JK. Leptin targets in the mouse brain. J Comp Neurol. 2009;514:518–532. doi: 10.1002/cne.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- Smith JT, Acohido BV, Clifton DK, Steiner RA. KiSS-1 neurones are direct targets for leptin in the ob/ob mouse. J Neuroendocrinol. 2006;18:298–303. doi: 10.1111/j.1365-2826.2006.01417.x. [DOI] [PubMed] [Google Scholar]
- Smith JT, Clay CM, Caraty A, Clarke IJ. KiSS-1 messenger ribonucleic acid expression in the hypothalamus of the ewe is regulated by sex steroids and season. Endocrinology. 2007;148:1150–1157. doi: 10.1210/en.2006-1435. [DOI] [PubMed] [Google Scholar]
- Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005a;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- Smith JT, Dungan HM, Stoll EA, Gottsch ML, Braun RE, Eacker SM, Clifton DK, Steiner RA. Differential regulation of KiSS-1 mRNA expression by sex steroids in the brain of the male mouse. Endocrinology. 2005b;146:2976–2984. doi: 10.1210/en.2005-0323. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin C-S, William C, Tanabe Y, Jessell T, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Developmental Biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Cheung CC, Clifton DK, Steiner RA. Regulation of hypothalamic proopiomelanocortin mRNA by leptin in ob/ob mice. Endocrinology. 1997;138:5063–5066. doi: 10.1210/endo.138.11.5651. [DOI] [PubMed] [Google Scholar]
- Todman MG, Han SK, Herbison AE. Profiling neurotransmitter receptor expression in mouse gonadotropin-releasing hormone neurons using green fluorescent protein-promoter transgenics and microarrays. Neuroscience. 2005;132:703–712. doi: 10.1016/j.neuroscience.2005.01.035. [DOI] [PubMed] [Google Scholar]
- Tong Q, Ye C, McCrimmon RJ, Dhillon H, Choi B, Kramer MD, Yu J, Yang Z, Christiansen LM, Lee CE, Choi CS, Zigman JM, Shulman GI, Sherwin RS, Elmquist JK, Lowell BB, Lowell BB. Synaptic glutamate release by ventromedial hypothalamic neurons is part of the neurocircuitry that prevents hypoglycemia. Cell Metab. 2007;5:383–393. doi: 10.1016/j.cmet.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- Watanobe H, Schiöth HB, Wikberg JES, Suda T. The Melanocortin 4 Receptor Mediates Leptin Stimulation of Luteinizing Hormone and Prolactin Surges in Steroid-Primed Ovariectomized Rats. Biochemical and Biophysical Research Communications. 1999;257:860–864. doi: 10.1006/bbrc.1999.0547. [DOI] [PubMed] [Google Scholar]
- Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne H-J, Todman MG, Korach KS, Greiner E, Pérez CA, Schütz G, Herbison AE. Definition of Estrogen Receptor Pathway Critical for Estrogen Positive Feedback to Gonadotropin-Releasing Hormone Neurons and Fertility. Neuron. 2006;52:271–280. doi: 10.1016/j.neuron.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kishi T, Lee CE, Choi BJ, Fang H, Hollenberg AN, Drucker DJ, Elmquist JK. Glucagon-like peptide-1-responsive catecholamine neurons in the area postrema link peripheral glucagon-like peptide-1 with central autonomic control sites. J Neurosci. 2003;23:2939–2946. doi: 10.1523/JNEUROSCI.23-07-02939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Farooqi I, Aminian S, Halsall D, Stanhope R, O’Rahilly S. Nat Genet. 1998. A frameshift mutation in MC4R associated with dominantly inherited human obesity; p. 20. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue [published erratum appears in Nature 1995 Mar 30;374(6521):479] [see comments] Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JLR. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. The Journal of Comparative Neurology. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler DR, Cullinan WE, Herman JP. Distribution of vesicular glutamate transporter mRNA in rat hypothalamus. The Journal of Comparative Neurology. 2002;448:217–229. doi: 10.1002/cne.10257. [DOI] [PubMed] [Google Scholar]
- Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol. 2006;494:528–548. doi: 10.1002/cne.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]