Perhaps best known as the genomics pioneer who defied all the odds to decode the book of life and publish its contents years ahead of the government funded Human Genome Project's projected completion date; J. Craig Venter has done it again—this time in the nascent field of synthetic biology.
In the May issue of Sciencexpress, Venter (an associate editor on BioBugs) and colleagues (including John Glass—also a member of the BioBugs editorial board) reported the design, synthesis and assembly of Mycoplasma mycoides JCVI-syn1.01—the first bacterial cell to contain a completely synthetic genome, and as such the first citizen of synthetic biology.
The story which led to the development of JCVI-syn1.0 has its origins as far back as 1995, when Venter and his team published the sequence of Mycoplasma genitalium.2 A pathogen isolated from the epithelia of primate genital and respiratory tracts; M. genitalium possesses one of the smallest genomes of any free-living organism and, as such, represented the perfect target genome to recreate. Of the 485 genes encoded by the ∼600,000 bp M. genitalium genome, approximately 100 were found to be non-essential—suggesting a minimal genome size of ∼400 genes. To confirm that this did, in fact, constitute the minimum requirement for life, Venter and co-workers first had to synthesise the minimal genome, and then transplant it into a recipient cell to ‘boot-up—two steps which had never before been achieved in the laboratory.
The first breakthrough came in 2007, when the team finally cracked the chromosomal transplantation step.3 By replacing the genome of the bacterium Mycoplasma capricolum with the native chromosome of a different species of Mycoplasma (M. mycoides), Venter and colleagues had, in essence, changed one species of bacteria into another. Proof that the transplantation was a success was achieved when the recipient cell (M. capricolum) began to adopt the physical characteristics of its chromosomal donor (M. mycoides).
The final hurdle, the chromosomal synthesis step, was overcome the following year with the publication of a paper entitled “complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome”.4 All the elements were now in place for Venter and his team to create the first synthetic life-form. However, the slow growth rate of the target organism M. genitalium (which has a doubling time of 16 hours) meant-that a single experiment could take several weeks to complete and so, despite its tiny genome, M. genitalium was replaced with the larger (in DNA terms) but faster growing M. mycoides.
The genome assembly process consisted of a painstaking assembly of 1,078 sequence cassettes, each of 1,080 bp in length (the 80 bp overlaps the adjacent cassette, facilitating correctly orientated sequence assembly). Yeast cells were employed to assemble the DNA in stages; the first stage involved taking 10 cassettes at a time to build 110, 10,000 bp segments. In the second stage, these 10,000 bp segments were taken 10 at a time to produce 11, 100,000 bp segments. In the final step, all 11, 100 kb segments were assembled into a complete synthetic genome, and propagated as a single yeast artificial chromosome. The complete synthetic M. mycoides genome was then isolated from the yeast cell and transplanted into restriction deficient M. capricolum recipient cells (which accept the foreign DNA without breaking it down). Following uptake, the synthetic genome begins to encode all the proteins required for life, including restriction enzymes which degrade the native M. capricolum genome. Following transplantation and replication on a nutrient agar plate to form a colony (representing >30 divisions or >109 fold dilution), the resulting ‘synthetic’ cells do not contain any protein molecules that were present in the recipient cell. In essence the DNA ‘software’ builds its own ‘hardware’.
To distinguish the synthetic genome from native DNA, the researchers incorporated four ‘watermark’ sequences. Replacing one or more cassettes in regions experimentally demonstrated or predicted not to interfere with cell viability, these watermarks contain strings of bases that, in code, spell out a web address to send emails to if you can successfully crack the new code, the names of 46 authors and other key contributors as well as three famous quotations. One of which by James Joyce, perfectly encapsulates the ups and downs of a the 15 year project—“To live, to err, to fall, to triumph, to recreate life out of life.”
At a cost of ∼$40 million, and countless man hours, Venter and his team have achieved the unimaginable—they have created (or perhaps more correctly ‘recreated’) life from scratch; converting a digitized DNA sequence, stored in a computer file, into a living entity capable of growth and self replication.
The next step will undoubtedly be to return to the minimum genome of M. genitalium—the ideal platform for analyzing the function of every essential gene in a cell, and from there; who knows? Perhaps the creation of completely artificial life forms—an upgrade to JCVI-syn2.0—entirely new species designed for specific roles from biodegradation to biomedicine… the possibilities are endless.
However, the ethical, legal and social implications of this new science cannot be ignored, and in a passage reminiscent of that famous understatement from Watson and Crick's double helix paper of 19535 “it has not escaped our notice…”; Venter and colleagues conclude that “this work will continue to raise philosophical issues that have broad societal and ethical implications.” That it will…
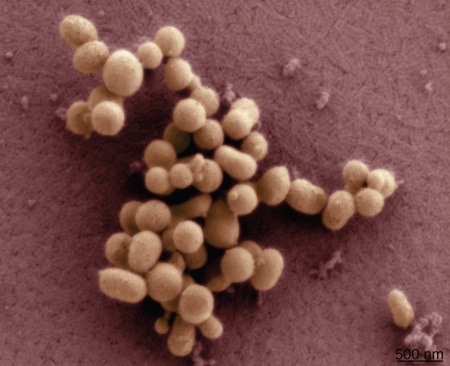
Figure 1.
Scanning electron micrographs of M. mycoides JCVI-syn1. Image courtesy of Tom Deerinck and Mark Ellisman of the National Center for Microscopy and Imaging Research at the University of California at San Diego.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/12465
References
- 1.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 2.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, et al. The minimal gene complement of mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 3.Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA, 3rd, et al. Genome transplantation in bacteria: changing one species to another. Science. 2007;317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 4.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, et al. Complete chemical synthesis, assembly, and cloning of a mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 5.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]