Abstract
In Swingle et al.1 we demonstrate that it is possible to use recombineering to direct a variety of changes in wild-type bacterial cells without the addition of phage-encoded proteins. This discovery is potentially applicable to biological engineering in a wide variety of bacterial species. Here we describe key features of oligo recombination as it is currently understood, and propose strategies for expanding the utility of oligo recombination for bioengineering.
Key words: oligonucleotide recombination, homologous recombination, bacteria, allelic exchange, gene conversion, recombineering, lambda red, RecET
The ability to change bacterial genomic sequences is one of the pillars supporting molecular genetics. The modifications employed depend on the intended outcome, but uniting all is a series of labor-intensive steps that in many cases involve compromises that result in approximations of our true goals. Additionally, the compromises often introduce uncertainties that must be dealt with by additional experiments to adequately address the question. The most common method for modifying genomic sequences, allelic exchange,2 requires cloning steps to generate a mutant allele in a plasmid prior to integration at the cognate site in the genome, followed by resolution to remove extraneous plasmid sequences (requiring additional selection and screening). Because of the difficulty of modifying bacterial genomic sequences there has been an incentive to make this process more efficient and precise.
The range of tools that have been developed to support molecular genetic investigation of bacteria is impressive,3 yet genomic modification in bacteria has lagged behind the highly efficient strategies assisted by homologous recombination using linear DNA in yeast.4–6 The development of similar strategies to enable bacterial genetic manipulation has generally been considered impossible because of the presence of potent nucleases that degrade linear DNA substrates before recombination can take place. These suppositions are supported by studies of mutant strains of Escherichia coli, lacking combinations of nucleases, that are capable of elevated levels of recombination with linear DNA.7,8 These studies are informative and useful in some cases, but not universally applicable because of the requirement that experiments be performed in mutant strains.
The scope of what is possible was broadened considerably in 1989 with the publication of two papers describing the use of the lambda Red and recET genes (sometimes referred to as Red/ET) to facilitate genetic engineering.9,10 The Red/ET genes are derived from bacteriophage and encode recombinases that induce a temporary hyper-recombinogenic state that makes cells capable of homologous recombination with linear DNA substrates. This discovery enabled the very practical technology known as recombineering (genetic engineering by recombination), the principle of which is that the substrate DNA can contain changes that direct alteration of genome sequences.11,12 The substrate DNA can be a double stranded13 (such as a PCR product or restriction fragment) or single stranded (such as an oligo) synthetic sequence.12 There is no question that the Red/ET systems represent a revolution on the genetic engineering front because these systems are capable of facilitating any change that can be incorporated into a linear segment of DNA and tolerated by the recipient genome. However, the distinct limitation of this and similar systems is that they function reliably in a limited range of hosts, preventing their widespread applicability.14 To circumvent this limitation, phage recombinases are now being identified that function in other model bacteria.15–18
In Swingle et al.1 we demonstrate that it is possible to introduce a variety of changes in wild-type bacteria cells without the addition of phage-encoded proteins simply by introducing ssDNA oligos. The discovery occurred while attempting to establish lambda Red assisted recombineering in P. syringae. Recombinase independent oligo recombination was identified using a straightforward assay to detect evidence of recombination, in which, cells were transformed with oligos that are homologous to a specific genomic locus and contain nucleotide changes that confer a selectable phenotype. A recombination frequency can be determined as the portion of resistance cells relative to 108 viable cells, the approximate number of viable cells in a standard electroporation. Using this assay, we did not detect any evidence that lambda Red functioned in P. syringae, but we did observe a low level of recombination in control transformations with cells that did not contain any of the Red genes. This was subsequently tested in other Gram-negative enteric bacteria, including E. coli, Salmonella and Shigella, and in all cases evidence for Red-independent oligo recombination was apparent.
There are a variety of factors that influence these recombination reactions. Chief among them is the concentration of the oligo supplied in the electroporation mixture. We found that the amount of oligo used has a profound effect on the frequency of recombination and that this is likely due to the ability of the DNA to overwhelm the capacity of endogenous nucleases present in the cells that degrade and therefore prevent the transformed oligos from being able to participate in recombination. Given this result, it is likely that oligos containing 5′ phosphothioate linkages would make recombination less dependent on oligo concentration based on the idea that these moieties impede nuclease degradation.19
We also examined the effect of oligo length on recombination frequency. This experiment revealed interesting dynamics that have both theoretical and practical ramifications for oligo recombination.4 We found that there is a length threshold close to 20 nt that, when exceeded, facilitates maximal levels of recombination. Increasing the length beyond this point up to 120 nt does not further increase levels of recombination. The recombination frequency dropped dramatically for oligos shorter than 20 nt and was undetectable below 12 nt. Additional experiments suggest that the recombination frequency is primarily governed by DNA annealing thermodynamics, and that oligos need to anneal with sufficient stability for recombination to be efficient. It is therefore necessary to consider this when designing oligos so that they exceed the minimum length to facilitate robust annealing. This result is also interesting from a theoretical perspective because it establishes an experimental system in which to study DNA annealing characteristics in vivo, including identifying genes whose products affect the process.
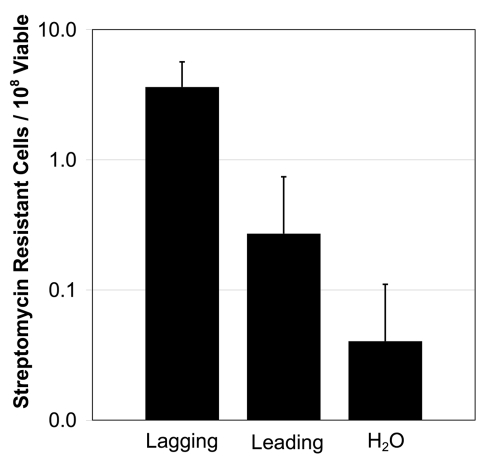
Additional questions regarding the mechanism of oligo recombination remain to be addressed. For example it will be informative to determine whether any part of the process (from transformation to recombination) is in itself mutagenic. To begin to test this, cells were transformed with oligos that were identical to the wild-type sequence except for a silent marker change in a codon's wobble position. The oligos used here were designed to match the rpsL gene, and the rate of streptomycin resistance was monitored (Fig. 1). Streptomycin resistant cells arose more frequently (albeit at a low level) when oligos were added compared to control reactions where oligos were absent. The strand bias that we report in Swingle et al.1 for oligo recombination (the tendency for the oligo matching the lagging strand to recombine more efficiently than its complement) is also apparent in this experiment consistent with the idea that the streptomycin resistance cells were the product of recombination. Sequencing of the rpsL gene of 40 streptomycin resistant isolates arising in reactions with the lagging strand oligo revealed that the majority (32/40) contained the silent marker mutation, indicating that these cells most likely arose due to recombination with the transformed oligos. Our current model is that these streptomycin resistant isolates arose from recombination of oligos that contain misincorpotated bases (i.e., during oligo synthesis). However, at this time we cannot rule out the possibility that the invasion of even a wild-type oligo is mutagenic at a low level (4/108). This further demonstrates the sensitivity of this assay, and we propose that it could be used as a means for analyzing the fidelity of commercial DNA synthesis methods.
Figure 1.
Frequency of streptomycin resistance in P. syringae cells. Cells were electroporated with 5 µg of a 74 nt oligo matching the lagging or leading strand of the rpsL gene or no oligo control (H2O). Then the frequency of streptomycin resistance was determined. Results are the average of three experiments, error bars show standard deviation.
Four genera of Gram-negative bacteria have been used to demonstrate oligo recombination, supporting the possibility that this process is conserved more broadly. A comparison to similar observations of oligo recombination in yeast,20–22 mammalian cells23 and archaea24 suggests the possibility that oligo recombination is universal among all life forms. Establishing that bacteria are capable of oligo recombination holds considerable promise for molecular biologists seeking to engineer sequences in situ. The possible ubiquity encourages development of this system as the basis for new tools to enable genomic modification of previously intractable organisms.
Getting started with oligo recombination requires minimal investment using standard reagents and equipment available in the molecular biology laboratory. It is critical to first identify a gene that can be easily monitored for evidence of recombination. Using current methods, the range of recombination frequencies that are typically observed (∼10−4) require that mutagenesis of the target gene produce a selective phenotype. We have had success using oligo recombination to mutate the rpsL, rpoB and upp genes to confer resistance to streptomycin, rifampacin and 5-flourouracil respectively. Auxotrophic selections are also possible; in Swingle et al.1 we demonstrate situations where oligos direct mutations in the E. coli galK gene allowing cells to utilize galactose as a carbon source. An important consideration when choosing a gene to target is that the mutation conferring the selectable phenotype must fit on an oligo. However, even given this constraint, it is possible to install different types of mutations, including point mutations and deletions. As might be expected oligos encoding nucleotide substitutions recombine more efficiently than deletions, but recombination can be maximized by designing changes that avoid detection by the host encoded methyl directed mismatch repair system.25–27 Considering these prerequisites the oligo can then be designed to include 25 to 30 nt of sequence homology flanking the desired mutation.
Conceptually, carrying out the experiment is not very different from introducing a plasmid by genetic transformation. However, optimizing the transformation process is useful since the recombination reactions presumably require a direct interaction between the mutagenic oligo and the homologous location in the recipient genome. It is necessary to introduce a large amount of DNA into the cells so that a sufficient number of molecules are present to undergo recombination and saturate ssDNA nucleases. Therefore, electroportation is the favored method for introducing the oligo into the cell. Chemical methods may work, but it is not clear that this method can introduce enough oligo to have the titrating effect. The data presented in Swingle et al.1 suggest that electroporation mixtures containing between 5 and 20 µg of oligo may be sufficient. As an alternative to adding large amounts of oligo, we predict that the addition of thiol moieties to the 5′ end of the oligo should compensate for high oligo concentrations. We should also note that lengthening the time cells incubate after electroporation can improve the number of recombinants obtained in an experiment. Extending this incubation prior to selection is important for markers like rpsL where the wild-type (streptomycin sensitive) allele is dominant to the mutant (streptomycin resistant) allele. Permitting cells to recover for longer periods of time allows the mutant allele to segregate completely from other wild-type copies present in the cells by resolving the merodiploid state induced by the presence of other partially replicated chromosomes.28 Mutations that are dominant also seem to benefit from longer recovery incubations. This may be due to a requirement for sufficient time to initiate or complete the recombination process and allow for protein expression.
What is next for oligo recombination? As alluded to above, the frequency of oligo recombination limits the use of this technology to mutations that result in a selectable phenotype. As our understanding of oligo recombination grows and methods are improved, we may find strategies that permit integration of mutations without selection. For the time being, we can still take advantage of oligo recombination. One potentially powerful strategy is to use this system to eliminate a counter selectable marker (e.g., rpsL-streptomycin, sacB-sucrose, tetR-fusaric acid) in the genome. The counter selectable marker could be integrated into the genome by the traditional allelic exchange approach and then excised using oligo recombination, simultaneously introducing mutations encoded by the oligo (i.e., point mutants, deletion derivatives, or epitope/affinity tags).
Finally, even though genetic transformation is a prerequisite for oligo recombination we recognize that this system could be used to develop a transformation protocol without the need for validated replicons. This solves the chicken-or-egg problem that may be faced when studying a new microbe, namely that in order to observe transformation a plasmid must stably replicate. In lieu of a plasmid, tranformants can be observed by selecting recombinant genomic alleles. After a transformation protocol is established, then the search for useful replicons can begin.
Acknowledgements
We thank Don Court and Jim Sawitzke for their contributions to this manuscript and for the many invaluable discussions that led to the discovery and characterization of oligo recombination in bacteria.
Note to: Swingle B, Markel E, Costantino N, Bubunenko MG, Cartinhour S, Court DL. Oligonucleotide recombination in Gram-negative bacteria. Mol Microbiol. 2010;75:138–148. doi: 10.1111/j.1365-2958.2009.06976.x.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/12098
References
- 1.Swingle B, Markel E, Costantino N, Bubunenko MG, Cartinhour S, Court DL. Oligonucleotide recombination in Gram-negative bacteria. Mol Microbiol. 2010;75:138–148. doi: 10.1111/j.1365-2958.2009.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ried JL, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 3.Neidhart FC, editor. Escherichia coli and salmonella: cellular and molecular biology. Washington, DC: ASM Press; 1996. [Google Scholar]
- 4.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 5.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 7.Winans SC, Elledge SJ, Krueger JH, Walker GC. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutra BE, Sutera VA, Jr, Lovett ST. RecA-independent recombination is efficient but limited by exonucleases. Proc Natl Acad Sci USA. 2007;104:216–221. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 11.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annual Review of Genetics. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 12.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 15.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 16.van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol Microbiol. 2008;67:1094–1107. doi: 10.1111/j.1365-2958.2008.06109.x. [DOI] [PubMed] [Google Scholar]
- 17.van Kessel JC, Marinelli LJ, Hatfull GF. Recombineering mycobacteria and their phages. Nat Rev Microbiol. 2008;6:851–857. doi: 10.1038/nrmicro2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci USA. 2008;105:1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moerschell RP, Tsunasawa S, Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc Natl Acad Sci USA. 1988;85:524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto T, Moerschell RP, Wakem LP, Ferguson D, Sherman F. Parameters affecting the frequencies of transformation and co-transformation with synthetic oligonucleotides in yeast. Yeast. 1992;8:935–948. doi: 10.1002/yea.320081104. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto T, Moerschell RP, Wakem LP, Komar-Panicucci S, Sherman F. Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics. 1992;131:811–819. doi: 10.1093/genetics/131.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell CR, Keown W, Lowe L, Kirschling D, Kucherlapati R. Homologous recombination involving small single-stranded oligonucleotides in human cells. New Biol. 1989;1:223–227. [PubMed] [Google Scholar]
- 24.Grogan DW, Stengel KR. Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli/lambdaRed and Sulfolobus acidocaldarius recombination systems. Mol Microbiol. 2008;69:1255–1265. doi: 10.1111/j.1365-2958.2008.06356.x. [DOI] [PubMed] [Google Scholar]
- 25.Kramer B, Kramer W, Fritz HJ. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984;38:879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- 26.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker BO, Marinus MG. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]